Abstract
Free full text

The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men
Abstract
Protein lysine deacetylases play a pivotal role in numerous biological processes and are divided into the Rpd3/Hda1 and sirtuin families, each having members in diverse organisms including prokaryotes. In vertebrates, the Rpd3/Hda1 family contains eleven members, traditionally referred to as histone deacetylases (HDACs) 1–11 and grouped into distinct classes. While most class I HDACs are subunits of multiprotein nuclear complexes crucial for transcriptional repression and epigenetic landscaping, class II members regulate cytoplasmic processes or act like signal transducers trafficking between the cytoplasm and the nucleus. This review focuses on recent findings related to the Rpd3/Hda1 family with an emphasis on evolutionarily conserved domain organization, protein complex formation, regulation, and biological function.
Introduction
Acetylation of the ε-amino group of lysine residues was initially identified on histones four decades ago1 and is now known to occur in different organisms for regulating diverse cellular processes (Box 1). This modification is reversed by lysine deacetylases (KDACs, Box 1). Since histones were first identified as the substrates in 1969 (ref. 2), eukaryotic KDACs have been referred to as histone deacetylases (HDACs). In 1978, butyrate was discovered to be a non-specific HDAC inhibitor3–5, and then the fungal antibiotic Trichostatin A was identified in 1990 as the first specific inhibitor6. Using a similar inhibitor as an affinity tag, Taunton et al.7 purified in 1996 the first HDAC (now known as HDAC1) from cow protein extracts and discovered it to be an orthologue of yeast Rpd3 (reduced potassium dependency 3, Fig. 1a). Because yeast genetics had already established Rpd3 as a global gene regulator8, the unexpected sequence similarity strengthened the link of histone acetylation to gene regulation. Two independent studies confirmed the multiplicity of HDACs9,10 and, together with the identification of HDAC1, set the stage for the establishment of the general notion that these enzymes can function as transcriptional corepressors and spurred the search for additional members of the Rpd3/Hda1 (histone deacetylase 1) family11 (Fig. 1; see also Supplementary information S1 (figure)).
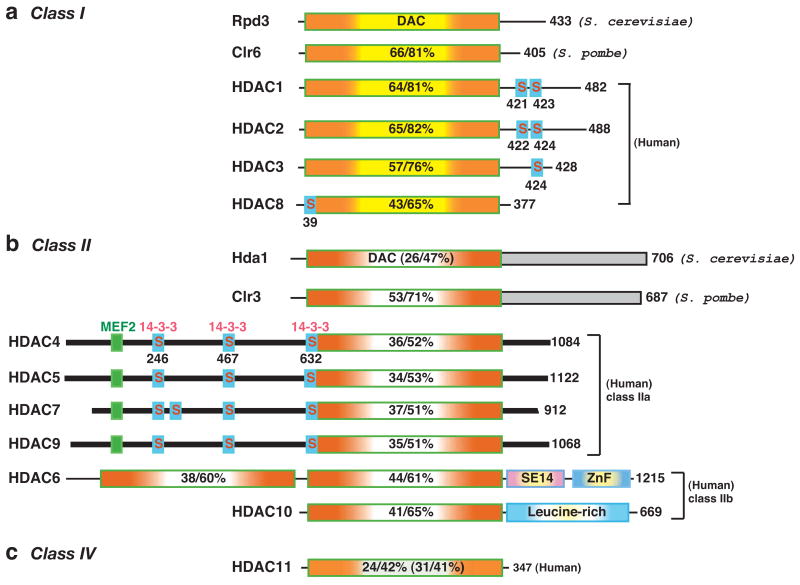
The deacetylases are grouped into different classes according to sequence similarity to yeast prototypes. In mammals, class I members (Rpd3-like) include HDAC1, 2, 3 and 8; class II members are HDAC4, 5, 6, 7, 9 and 10 (similar to Hda1); and class IV is made up of HDAC11 (Fig. 1). Class II is further divided into two subclasses, IIa (HDAC4, 5, 7 and 9) and IIb (HDAC6 and 10). The total number of amino acid residues in each deacetylase is shown to the right of the figure. Many of the deacetylases have isoforms that result from alternative splicing. For simplicity, the number refers to the longest isoform. The deacetylase (DAC) domain is depicted by a rectangle containing a yellow (class I, (a)), clear (class II, (b)), or light green (class IV, (c)) centre flanked by orange shading, and the percentage amino acid sequence identity/similarity to that of Rpd3 (for class I) or Hda1 (class II/IV) is shown. The sequence identity/similarity of Hda1 and HDAC11 to Rpd3 is given in brackets. The C-terminal domains (shaded rectangles) of Hda1 and Clr3 are homologous (identity/similarity, 26/57%). Dark bars represent similar N-terminal domains and C-terminal tails of class IIa histone deacetylases (HDACs). Myocyte enhancer factor 2 (MEF2)-binding motifs are depicted as small green boxes, whereas 14-3-3 binding motifs are indicated by small boxes labelled with the letter “S” (for serine). SE14, Ser-Glu-containing tetradecapeptide repeats; ZnF, ubiquitin-binding zinc finger.
In 2000, the Sir2 (silent information regulator 2) or sirtuin (Sir two (“tu”)-like protein) family of NAD+-dependent deacetylases was discovered12, which led to the designation of the zinc-dependent Rpd3/Hda1 family as the “classical family”13. HDACs can also be grouped according to phylogenetic analysis and sequence homology14–16. In this classification system, the sirtuin family constitutes class III, whereas the classical family is grouped into class I, II and IV (Fig. 1), with class II being further divided into two subclasses (IIa and IIb); such subclass division is also necessary to accommodate classification of members from C. elegans, D. melanogaster and zebrafish (Supplementary information S1 (figure)).
The HDAC field has been expanding at an amazing pace, owing largely to the following three factors. First, chromatin research has exploded over the past 10–15 years, and HDACs have a key role in deacetylating nucleosomes, interacting with other chromatin regulators and shaping epigenetic landscapes17,18. Second, in addition to histones, these enzymes deacetylate numerous transcription factors and many other proteins that are important for a multitude of cellular processes in eukaryotic and prokaryotic systems (Boxes 1 & 2). Finally, HDAC inhibitors have great potential for treating cancer and other diseases19,20. A majority of compounds that are currently under clinical evaluation target the Rpd3/Hda1 family.
This review covers mainly the recent findings relating to this family of deacetylases. We begin with an overview of domain organization and tertiary structure of the mammalian members and compare this organization to that of homologues in yeast and other model organisms. We then describe multisubunit complexes of class I/II HDACs, roles of class IIa members in signal transduction, and cytoplasmic functions of HDAC6. After that, we discuss how these molecular and cellular studies are related to genetic analyses in mice, C. elegans and D. melanogaster, as well as the implications for pathology and drug development. Two themes link different sections of this review: first, how amino acid sequences of the deacetylases determine the structure, function and regulation; and, second, how a simple catalytic domain co-evolves with different motifs and modules to connect functions in diverse cellular processes from various organisms. Both themes are fundamental issues relevant to many other protein families.
Domain organization of class I/II/IV HDACs
Domains and motifs of class I HDACs
As a founding member of class I, Rpd3 from budding yeast comprises an N-terminal deacetylase domain and a C-terminal tail8,10 (Fig. 1a). Both regions are conserved in Clr6 (cryptic locus regulator 6) from Schizosaccharomyces pombe21, but only the catalytic domain is conserved in the bacterial deacetylase AcuC (Box 2) and in class I members from higher eukaryotes13. Mammalian HDAC1 and HDAC2 are almost identical, and in addition to the catalytic domains, they are homologous in the C-terminal tail, which harbors tandem casein kinase 2 (CK2) phosphorylation sites22. The tail of HDAC1 also contains a sumoylation motif22. There are three orthologues in C. elegans, but only one in D. melanogaster or zebrafish (Supplementary information S1 (figure)). Unlike HDAC1, HDAC3 is only conserved from flies to mammals (Fig. 1; and Supplementary information S1 (figure)). Slightly different from HDAC1 and HDAC2, HDAC3 possesses one CK2 phosphorylation site23. HDAC8 is present in sea urchin, but not in C. elegans or D. melanogaster, suggesting that this deacetylase is deuterostome-specific. Instead of CK2 sites, this deacetylase has a conserved motif (Supplementary information S2a (figure)) for protein kinase A phophorylation and negative regulation of deacetylase activity24,25.
Domains and motifs of class II HDACs
Hda1 consists of an N-terminal deacetylase domain and a long C-terminal extension of unknown function (Fig. 1b). This domain organization is conserved in Clr3, a class II member from S. pombe. By contrast, only the deacetylase domain is shared by metazoan homologues (Fig. 1b)14,15,26. In addition to the deacetylase domain, a long N-terminal extension is conserved among the class IIa members in mammals (HDAC4, 5, 7, and 9). This N-terminal extension contains binding sites for myocyte enhancer factor 2 (MEF2) and 14-3-3 proteins. The MEF2-binding motif is conserved from C. elegans to mammals (Supplementary information S1 (figure))26 and displays resemblance to sites in other MEF2-binding proteins27. Only one or two of the 14-3-3 binding sites are conserved in C. elegans and D.melanogaster (Supplementary information S1 (figure)), but three are present in an orthologue from sea urchin (Supplementary information S2b (figure)). Consistent with the sequence conservation, MEF2 and 14-3-3 are major partners of class IIa HDACs.
HDAC3 was shown to be responsible for deacetylase activities associated with HDAC4 and HDAC7 (ref. 28). Consistent with this, mammalian class IIa members possess low deacetylase activity as a result of a Tyr-His substitution29. This substitution occurs in zebrafish, but not in C. elegans, D. melanogaster or sea urchin (Supplementary information S2b (figure)), which implies that it is vertebrate-specific. Interestingly, an equivalent tyrosine residue is dispensable for the esterase activity of a bacterial HDAC-like protein30. It will be important to determine how the substitution is reflected with regard to the function and regulation of class IIa deacetylases in vertebrates.
Unlike class IIa members, HDAC6 contains tandem deacetylase domains and a C-terminal zinc finger (Fig. 1b & Supplementary information S1 (figure)), and has emerged as a major cytoplasmic deacetylase. The zinc finger can bind ubiquitin, and its core sequence is similar to ZnF-UBP (ubiquitin carboxyl-terminal hydrolase-like zinc finger)26,31. An SE14-repeat domain that is found in human HDAC6 (ref. 26) is almost identical in macaques (XP_001101619) and highly conserved in horses (GenBank Accession No., XP_001493915), but is only partial in mice, implying that this domain is specific to high mammals.
The other class IIb member, HDAC10, has an N-terminal half that is highly similar to the first deacetylase domain of HDAC6, whereas the C-terminal half is leucine-rich (Fig. 1b). Although there are no invertebrate orthologues, a distant relative exists in zebrafish (Supplementary information S2c (figure)), which indicates that HDAC10 might have a vertebrate-specific role. However, very little is known about its real biological function.
HDAC11 as a highly conserved deacetylase
HDAC11 shows sequence similarity to class I and II HDACs (Fig. 1c)32. It is highly conserved from C. elegans and D. melanogaster to humans (Supplementary information S1 (figure)), and there are also related proteins in bacteria16 and plants (atHDA2, NP_568480). HDAC11 is sometimes referred to as a class I member, but phylogenetic analysis indicated that this deacetylase and its homologues belong to a separate class16. Little is known about its function or regulation, but its evolutionary conservation implies a fundamental role in diverse organisms.
Structures of Zn2+-dependent deacetylase domains
Crystal structures have been solved for the catalytic domains of HDAC8 (refs 25,33), HDAC7 (PDB code, 2NVR) and two bacterial deacetylases34,35. The structures are very similar: a single α/β domain composed of an eight-stranded β-sheet sandwiched between over a dozen α-helices (also see Supplementary information S3 (figure)). The catalytic centre contains a zinc ion chelated by side chains of histidine and aspartate residues. These structures indicate that the deacetylase domains from these family members share not only sequence similarity but also overall tertiary structure. Additional domains/motifs and sequence divergence within the deacetylase domains form the molecular basis for the unique subunit associations, and biological function, of each individual family member (see below).
Diversity of class I/II HDAC complexes
With the exception of HDAC8, all class I members can be catalytic subunits of multiprotein complexes (Table 1 and Supplementary information S4 (table)). In mammals, HDAC1 and HDAC2 interact with one another and form the catalytic core of several multisubunit complexes11. This HDAC1-2 core is present in mammalian Sin3, nucleosome remodeling deacetylase (NuRD) and corepressor of REST (repressor element 1 silencing transcription factor (CoREST)) complexes (Table 1 and Supplementary information S5 (table)). As discussed below, one emerging theme is that SANT domain-containing subunits stimulate deacetylase activities36. Another theme is that the subunits with which HDACs associate possess chromatin-remodelling or chromatin-binding ability, which provides an important platform for coordinating deacetylase functions with other chromatin-regulating mechanisms.
Table 1
Class I HDAC complexes from different eukaryotic organisms
| Complex | Mammal | Drosophila | C. elegans | S. cerevisiae | S. pombe | Arabidopsis | Protein domain | ||
|---|---|---|---|---|---|---|---|---|---|
| Rpd3L | Rpd3S | I | II | ||||||
| Sin3 | HDAC1/2 | dRpd3 | ceHda-1 | Rpd3 | Rpd3 | Clr6 | Clr6 | Rpd3 | Class I deacetylase |
| RbAp46/48 | p55 | Rba-1, Lin-53 | Ume1 | Ume1 | Prw1 | Prw1 | WD40 repeat | ||
| Sin3A | dSin3 | Sin3 | Sin3 | Sin3 | Pst1 | Pst2 | AtSin3 | PAH motifs | |
| Sds3/BRMS1 | Sds3 | Sds3 | Sds3 | Coiled coil | |||||
| RBP1 | |||||||||
| SAP30 | Sap30 | ||||||||
| SAP18 | AtSap18 | Ubiquitin-fold | |||||||
| ING1/2 | Pho23 | Png2 | PHD finger | ||||||
| Rco1 | PHD finger | ||||||||
| Eaf3 | Chromodomain | ||||||||
| Cph1 | PHD finger | ||||||||
| Cph2 | PHD finger | ||||||||
| Alp13 | Chromodomain | ||||||||
| Mi2/NuRD | HDAC1/2 | dRpd3 | ceHda-1 | Rpd3 | Rpd3 | Clr6 | Clr6 | Rpd3 | Class I deacetylase |
| RbAp46/48 | p55 | Rba-1, Lin-53 | WD40 repeat | ||||||
| Mi-2α/β | dMi-2 | Chd-3/-4 | Pickle | Helicase | |||||
| MTA1/2/3 | Egl-1, Egl-27 | SANT domain | |||||||
| MBD3 | Methyl CpG binding | ||||||||
| p66α/β | p66 | ||||||||
| CoREST | HDAC1/2 | dRpd3 | ceHda-1 | Rpd3 | Rpd3 | Clr6 | Clr6 | Rpd3 | Class I deacetylase |
| CoREST | dCoREST | Spr-1 | SANT domain | ||||||
| LSD1 | Su(Var)3-3 | Spr-5 | SWIRM, K-demethylase | ||||||
| BHC80 | PHD finger | ||||||||
| CtBP1 | Dehydrogenase | ||||||||
| N-CoR/SMRT | HDAC3 | dHDAC3 | Class I deacetylase | ||||||
| N-CoR/SMRT | SMRTER | SANT domain | |||||||
| TBL1/TBLR1 | WD40 repeat | ||||||||
| GPS2 | |||||||||
| JMJD2A | Jumonji demethylase | ||||||||
| Kaiso | Methyl CpG binding | ||||||||
Sin3 complexes from fungi and plants to mammals
S. cerevisiae Rpd3 forms large and small Sin3 complexes, known as Rpd3L and Rpd3S, respectively (Table 1 and Supplementary information S4 (table))10,37–40. The former typically possesses the subunits Ume1, Sds3, Sap30, and Pho23 (an ING (inhibitor of growth) protein). DNA-binding proteins, such as the zinc-finger repressor Ume6, target it to specific loci for transcriptional repression41. Rpd3S shares Sin3 and Ume1 with Rpd3L, but also contains two unique subunits, Eaf3 and Rco138,39. Eaf3 contains a chromodomain for recognizing K36-methylated histone H3 and recruits Rpd3S to nucleosomes that have been methylated by the Set2 histone methyltransferase38,39,42. The recognition efficiency depends on the plant homeodomain-linked (PHD) finger of Rco143. Whereas Rpd3L deacetylates histones at promoter regions, Rpd3S targets transcribed regions to suppress intragenic transcription initiation38,39.
S. pombe has three Sin3 orthologues, two of which have been shown to form deacetylase complexes. One associates with Clr6 to form two slightly different complexes21, both of which share two other subunits, Sds3 and Prw1; however, only one of the complexes contains the ING protein Png2. These complexes silence mating-type genes and are required for cell viability. The second Sin3 orthologue, Pst2, forms a third Clr6 complex with Prw1 and two PHD finger-containing proteins, one of which possesses a helicase domain. This complex reverses K9 acetylation of histone H3 and blocks antisense transcription21.
Mammals express two Sin3 proteins, Sin3A and Sin3B. The Sin3A complex contains HDAC1-2 catalytic core and RbAp46/48 (Table 1 and Supplementary information S5 (table))44, as well as RBP1 (Rb-binding protein 1), SAP180 (similar to RBP1), SDS3, BRMS1 (breast cancer metastasis suppressor 1, similar to SDS3), SAP30, SAP18, and ING1/2. As some of the subunits have different isoforms, there are several distinct Sin3A complexes. Interestingly, ING2 recognizes K4-methylated histone H3 and targets the Sin3A complex to deacetylate nucleosomes in a methylation-dependent manner45. The Sin3B complex shares some similarity to the Sin3A complex but might also contain distinct subunits46. Sin3 homologues are present in C. elegans, D. melanogaster and Arabidopsis thaliana (Table 1), which indicates that similar complexes might exist in various eukaryotes.
Metazoan and plant Mi-2 and NuRD
NuRD contains a catalytic core of HDAC1, HDAC2, RbAp46, and RbAp48 (Table 1 and Supplementary information S5 (table))44,47. In addition, this complex contains MTA (metastasis-associated) proteins, the nucleosome-remodelling ATPase Mi-2 (dermatomyositis-specific autoantigen), MBD3 (methyl CpG-binding domain 3), and p66. Through MBD3, this complex is recruited for DNA methylation-dependent gene silencing. It also associates with MeCP1 (methyl CpG-binding protein 1) and MeCP2 to provide an intimate connection with DNA methylation47,48. Three mammalian MTA isoforms (MTA1, 2 and 3) have been identified47. They possess SANT domains that are crucial for the integrity and deacetylase activity of NuRD. Mi-2 and p66 have α and β isoforms, enabling the formation of distinct complexes. NuRD subunits have been identified in C. elegans, D. melanogaster and A. thaliana (Table 1). Similar complexes probably exist, therefore, in animal and plant kingdoms.
CoREST complex
Initially identified as a corepressor of the transcriptional repressor REST49, CoREST is a component of an HDAC1-2 complex referred to as a neuronal corepressor complex (Table 1 and Supplementary information S5 (table))50–52. One subunit, KIAA0601/BHC110, possesses an FAD-binding domain with sequence homology to maize polyamine oxidase51. This subunit was recently shown to be a demethylase for K4-methylated histone H3 and therefore renamed lysine-specific demethylase 1 (LSD1)53. The CoREST complex promotes nucleosomal deacetylation through its SANT domains54. Interestingly, histone demethylation stimulates the deacetylase activity, which, in turn, is required for optimal demethylation, indicating that the two activities are interdependent55. CoREST subunits have been found in C. elegans and D. melanogaster (Table 1), but not in fungi or plants. This complex might, therefore, be metazoan-specific, which is consistent with its primary role in repressing neuronal gene expression.
HDAC3 complexes: from flies to men
Biochemical purification of HDAC3 uncovered an unexpected link to two homologous nuclear receptor corepressors, nuclear receptor co-repressor (N-CoR) and silencing mediator of retinoic acid and thyroid hormone receptors (SMRT) (Table 1 and Supplementary information S5 (table))56–58. Conversely, HDAC3 was identified through affinity purification of both corepressors59–61. The N-CoR and SMRT corepressors stimulate the deacetylase activity of HDAC3 through their SANT domains62, which are highly conserved in SMRTER, the D. melanogaster homologue of SMRT and N-CoR. An HDAC3 orthologue exists in D. melanogaster, suggesting that SMRTER might form a functional deacetylase complex. Similar proteins are absent from C. elegans and plants, so HDAC3 complexes might only be conserved from flies to mammals.
Class II HDAC complexes
Yeast Hda1 self-associates and forms a tetrameric complex with Hda2 and Hda3, in which both are important for the deacetylase activity63. Hda2 and Hda3 homologues are present in Candida albicans, but not in S. pombe or higher organisms. Instead, Clr3 forms a quartet core complex in S. pombe64. One subunit, Mit1, contains a PHD finger and a helicase domain, thus conferring both deacetylase and chromatin-remodelling activities within one complex64 (somewhat reminiscent of the NuRD complex in metazoans). The Clr3 complex is recruited to euchromatic and heterochromatic loci for transcriptional gene silencing through Mit1, a heterochromatin protein 1 (HP1) homologue and a telomere-binding protein64.
Non-catalytic subunits of Hda1 and Clr3 complexes are not conserved in metazoans, so these complexes might be fungi-specific. This is consistent with the fact that metazoan class II HDACs possess unique domains and motifs absent in Hda1 and Clr3 (Fig. 1 and Supplementary information S1 (figure)). Indeed, affinity purification of mammalian class IIa HDAC complexes identified 14-3-3 proteins65 and HSP70 (E.S. & X.J.Y., unpublished results), whereas HDAC6 was found to associate with cytoskeletal proteins and chaperones66,67. As discussed below, 14-3-3 proteins play a major role in regulating the trafficking of class IIa HDACs, and the association partners of HDAC6 shed important light on its important functions in the cytoplasm.
Class IIa HDACs as signal transducers
Nucleocytoplasmic trafficking mediated by 14-3-3
Depending on the cellular context, mammalian class IIa members (HDAC4, 5, 7, 9) can exist in both the nucleus and the cytoplasm14,15,26. They possess intrinsic nuclear import and export signals for dynamic nucleocytoplasmic trafficking, so the association of partners might alter this equilibrium and the subsequent subcellular distribution. MEF2 binds to class IIa HDACs and promotes their nuclear localization. By contrast, 14-3-3 proteins stimulate their cytoplasmic retention in a phosphorylation-dependent manner15,26. HDAC4 contains three 14-3-3-binding sites, S246, S467 and S632, all of which are conserved in HDAC5, 7 and 9 (Fig. 1b). Only the former two sites seem to be functional in HDAC5 and HDAC968,69, while a fourth site might be important in HDAC7 (Fig. 1b)70. Mutational analysis revealed that the S246 site of HDAC4 is the most important15,26. This site is conserved in both C. elegans and D. melanogaster (Fig. 1b and Supplementary information S1b (figure)).
Both 14-3-3 binding and the presence of a nuclear export signal are necessary for the cytoplasmic localization of HDAC4 and paralogues in mammals. The mechanistic impact of 14-3-3 binding is multifaceted (Fig. 2). First, the S246 site is adjacent to the nuclear localization signal (NLS, Fig. 2a), so 14-3-3 might impede access of the nuclear import receptor importin α/β to the NLS. Second, 14-3-3 binding might provide a signal in trans to promote the nuclear export of class IIa HDACs. Third, 14-3-3 binding affects the subnuclear distribution of class IIa HDACs. For example, upon binding to 14-3-3 proteins, an HDAC9 isoform lacking the deacetylase domain and nuclear export signal disperses from nuclear punctae into a uniformly nuclear pattern71. Similarly, 14-3-3 binding to C. elegans HDAC4 (Supplementary information S1a (figure)) does not seem to promote cytoplasmic localization72, perhaps due to the lack of a nuclear export signal in this deacetylase. Fourth, 14-3-3 association protects HDAC7 from proteasomal degradation73,74, which contributes to its cytoplasmic accumulation. Finally, 14-3-3 binding sites are subject to reversible phosphorylation. By binding to phosphorylated residues, 14-3-3 binding might occlude phosphatases, and, therefore, inhibit dephosphorylation, which might function as a feed-forward reinforcing mechanism70.

a Multiple kinases (calcium/calmodulin-dependent protein kinase (CaMK), protein kinase D (PKD), microtubule affinity-regulating kinase (MARK), salt-inducible kinase (SIK), checkpoint kinase 1 (CHK1) and other kinases) mediate specific phosphorylation of human HDAC4 on three 14-3-3-binding sites. Myosin phosphatase-targeting subunit-1 (MYPT1)/protein phosphatase (PP)1β and PP2A can act on these sites. The association of 14-3-3 proteins with HDAC4 retains it in the cytoplasm and prevents its interaction with transcription factors such as myocyte enhancer factor 2 (MEF2), thereby releasing them for transcriptional activation. This model can also be extended to other class IIa members, although some of the kinases and phosphatases have only been shown for HDAC4, 5, 7 and/or 9 (see text for references). At the bottom of the panel, the sequence surronding the major 14-3-3 binding site (S246) of HDAC4 is compared with the corresponding regions of its paralogues (HDAC5, 7 and 9) as well as orthologues from D. melanogaster and C. elegans. The key residues crucial for phosphorylation and 14-3-3 binding are highlighted. This 14-3-3 site constitutes a sequence conservation island140. The nuclear localization signal, which can be impeded by 14-3-3 binding, is also shown. b Signal-responsive transcriptional repression by HDAC4 (left) is analogous to transcriptional activation by transforming growth factor-β (TGFβ)-stimulated SMADs (middle) and cytokine-activated signal transducer and activator of transcription (STAT) proteins (right). 14-3-3 proteins bind to HDAC4 and retain it in the cytoplasm, thereby inhibiting its corepressor activity; upon dephosphorylation, HDAC4 translocates to the nucleus for transcriptional repression. The letter P circled in red denotes phosphorylation. For simplicity, only two 14-3-3 molecules are shown. This schematic also applies to HDAC5, 7 and 9.
Kinases and phosphatases for class IIa HDACs
Five groups of kinases have been shown to phosphorylate the 14-3-3-binding sites on HDACs: Ca2+/calmodulin-dependent kinases (CaMKs)68,75, protein kinase D76–78, microtubule affinity-regulating kinases70,79, salt-inducible kinases72,80, and checkpoint kinase-1 (CHK1) (Fig. 2a)81. Conversely, two phosphatases have been identified, myosin phosphatase-targeting subunit-1 (MYPT1)/protein phosphatase (PP)1β82 and PP2A70,83,84. Available data indicate that most of the kinases phosphorylate all class IIa members in mammals. The S246 site is conserved from C. elegans and D. melanogaster to mammals (Supplementary information S1 (figure)), and most of the kinases have orthologues in invertebrates, indicating that the regulatory mechanisms might be evolutionarily conserved. Consistent with this, the salt-inducible kinase orthologue Kin-29 in C. elegans phosphorylates HDAC4 (Supplementary information S1 (figure)) and promotes 14-3-3 binding72. The nuclear function of class IIa HDACs seems to be regulated by different signalling pathways through reversible phosphorylation at 14-3-3 binding sites. In this regard, these deacetylases function as novel signal transducers, very similar to the STAT and SMAD signal-responsive transcription factors (Fig. 2b). Mapping out events upstream of the kinases (Fig. 2a) will be necessary to determine how the regulation is integrated in cellular signalling networks under various physiological contexts.
Other regulatory mechanisms for class IIa HDACs
In addition to phosphorylation, class IIa HDACs are subject to ubiquitylation and sumoylation73,85,86. Ubiquitylation promotes protein degradation73,74, but the functional consequence of sumoylation is less clear85,87. HDAC4 sumoylation is very efficient, which makes it a potentially important modification. One possibility is that this modification generates a transcriptional repression domain and increases the transcriptional repression potential of HDAC4. In addition, this deacetylase is subject to non-reversible processing. For example, caspase cleavage produces an HDAC4 fragment corresponding to the N-terminal 289 residues88,89, within which there is a transcriptional repression domain26. Moreover, alternative splicing of HDAC9 mRNA produces isoforms that contain the N-terminal repression domain86. The HDAC4 fragment and some of the HDAC9 isoforms possess an intact MEF2-binding site and can still function as transcriptional corepressors, supporting that class IIa HDACs can repress transcription independent of their cryptic deacetylase activity28,29. Regulating the enzymatic activity is a potentially important mechanism. Furthermore, while HDAC4 expression is controlled by microRNA90, transcription of HDAC9 mRNA is regulated by a negative-feedback loop in which HDAC9 represses its own promoter91. Therefore, class IIa members are signal-responsive transcriptional corepressors subject to regulation by multiple mechanisms. Although the nuclear function of class IIa members is well established, their role in the cytoplasm (if any) is unclear. By comparison, the class IIb member HDAC6 is functionally important in this compartment.
HDAC6 as a key cytoplasmic regulator
Unlike many other HDACs, HDAC6 is predominantly (if not exclusively) cytoplasmic31. This finding has led to the simple hypothesis that HDAC6 possesses functions that are unrelated to histone modification, and available results convincingly support this theory. Indeed, multiple cytoplasmic processes seem to be regulated by HDAC6 (Fig. 3).
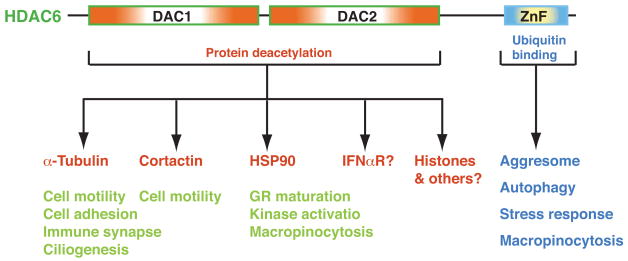
Through its tandem deacetylase (DAC) domains, HDAC6 deacetylates α-tubulin, cortactin and HSP90 to regulate cell motility, cilium assembly, maturation of the glucocorticoid receptor (GR), and activation of some protein kinases. HDAC6 may also deacetylate the type I interferon receptor (IFNαR)133. Additional cytoplasmic substrates are likely to be identified. Through its ubiquitin-binding zinc finger (ZnF), HDAC6 binds to ubiquitin and regulates aggresome formation, autophagy, activation of heat-shock factor 1 (HSF1), and epidermal growth factor receptor (EGFR)-induced macropinocytosis. It remains to be established whether HDAC6 has a direct role in the nucleus.
HDAC6 regulates cytoskeletal dynamics
HDAC6 has been shown to function as an α-tubulin deacetylase92–94, and it associates with microtubules and colocalizes with a microtubule motor complex92. Exogenous expression of HDAC6 leads to global deacetylation of α-tubulin and RNA interference-mediated knockdown of HDAC6 increases α-tubulin acetylation. Ectopic expression of HDAC6 also promotes chemotaxis92, which indicates that HDAC6-mediated deacetylation probably regulates microtubule-dependent cell mobility. In addition, HDAC6 might regulate cell adhesion95, the formation of the immune synapse96, and motor-mediated cargo transport along microtubules97,98. HDAC6 also has an unexpected role in the reabsorption of primary cilia of mammalian cells99. Like most HDACs, HDAC6 is a phosphoprotein100. Phosphorylation by Aurora A kinase activates the tubulin deacetylase activity of HDAC6 and promotes ciliary disassembly99. HDAC6 thus has roles in various microtubule-dependent cytoplasmic processes.
The filamentous (F)-actin-binding protein cortactin also interacts with HDAC6 and functions as its substrate67. As shown for α-tubulin, exogenous expression of HDAC6 leads to cortactin hypoacetylation, whereas inhibition of the deacetylase activity results in hyperacetylation. Independently, cortactin was identified as an acetylated protein in a proteomic study (M. Yoshida, personal communication). Interestingly, HDAC6 alters the ability of cortactin to bind F-actin by modulating the charge in the repeat region of cortactin and, in this way, modifies cell movement67. Therefore, in addition to its role in controlling microtubule-dependent cell motility, HDAC6 also seems to influence actin-dependent cell motility.
HDAC6 deacetylates HSP90
The heat shock chaperone protein HSP90 also associates with HDAC6 and is a bona fide cytoplasmic substrate101,102. HDAC6 is required for HSP90-dependent activation of the glucocorticoid receptor (GR). Treatment of cells with an HDAC6-sensitive deacetylase inhibitor or small interfering RNA-mediated knockdown of HDAC6 causes HSP90 to dissociate from p23 (a co-chaperone) and GR101. Studies in a cell-free system showed that HDAC6 promotes the assembly of stable HSP90–GR heterocomplexes but does not affect the intrinsic properties of GR103. Moreover, depletion of HDAC6 induces HSP90 acetylation and reduces its association with the leukemic tyrosine kinase Bcr-Abl102. HSP90 acetylation also regulates kinase activity of CHK1 (ref. 104). As a large number of other proteins, including many protein kinases involved in oncogenesis, require HSP90 for their maturation and function, it will be exciting to examine whether HSP90 deacetylation is important for their regulation. Although the list of cytoplasmic substrates of HDAC6 will probably continue to grow, the issue whether this deacetylase has a nuclear role awaits further investigation31.
HDAC6 and ubiquitin-dependent pathways
HDAC6 from mouse testes associates with the mammalian homologue of yeast UFD3 (ubiquitin fusion degradation protein 3) and p97/VCP31,66, a member of the AAA+ family of ATPases. Strikingly, the C-terminal zinc finger of HDAC6 efficiently interacts with ubiquitin, which causes HDAC6 to dissociate from the protein complex31. These results indicate an unexpected role for HDAC6 in ubiquitin-dependent pathways.
The expression of misfolded proteins in cells often leads to the formation of aggresomes105, a process that is facilitated by the delivery of misfolded proteins to these inclusion bodies via dynein-dependent transport on microtubules105. HDAC6 is a component of the aggresome, and treatment of cells with a proteasome inhibitor results in the accumulation of HDAC6 in polyubiquitin-containing aggresomes106. Mechanistically, HDAC6 associates with dynein motors and functions as an adaptor to link these motors to ubiquitylated misfolded proteins for delivery to aggresomes and degradation by proteasomes106. p97/VCP disrupts HDAC6–ubiquitin complexes and counteracts the ability of HDAC6 to promote the accumulation of polyubiquitylated proteins107. While the deacetylase activity of HDAC6 was found to be required for aggresome formation106, two recent studies showed that tubulin acetylation or HDAC6 inhibition accelerates dynein- and kinesin-mediated transport97,98.
Further support for the proposal that HDAC6 is essential for efficient management of cytotoxic proteins is provided by two recent studies suggesting that HDAC6 increases clearance of various substrates via autophagy108,109, a lysosome-mediated degradation pathway. Impairment of the ubiquitin–proteasome system induces autophagy, and this induction rescues neurodegeneration associated with such impairment108. Exogenous expression of HDAC6 alone compensates for an impaired ubiquitin–proteasome system and rescues neurodegeneration by autophagy108. Exactly how HDAC6 accelerates the turnover of misfolded proteins by autophagy is not known. Whatever the mechanism, it is clear that HDAC6 has a key role in the cellular clearance of misfolded proteins by different pathways and might function as a potential therapeutic target for neurodegenerative diseases. By associating with HSP90 and binding to ubiquitin, HDAC6 functions as a sensor for ubiquitylated protein aggregates to regulate the activation of heat-shock factor 1 (HSF1) during stress responses110. Moreover, HDAC6 is an essential component of stress granules111. This deacetylase, therefore, has key roles in different levels of cellular responses.
HDAC6 might also regulate cytoplasmic processes, such as macropinocytosis112. The mechanism by which the deacetylase and ubiquitin-binding activities are coordinated remains to be addressed. Together, recent studies have established HDAC6 as a major cytoplasmic regulator (Fig. 3) with properties that are not shared by other deacetylase, which is consistent with its unique domain organization (Fig. 1 and Supplementary information S1 (figure)).
Class I/II/IV HDACs in metazoan development
Genetic analyses of some class I/II members in C. elegans, D. melanogaster and mice have not only revealed important roles for HDAC1, 2 and 3, and class IIa members, in different developmental processes, but have also raised the important issue how to correlate these developmental functions with results from biochemical and cell-based assays. The findings from the genetic analyses are summarized in Table 2 and Supplementary information S6 (table), but some important points are noteworthy. First, HDAC1, 2 and 3 are catalytic subunits of multiprotein proteins (Table 1), so in addition to the deacetylases themselves, genetic analyses of associated subunits are very informative about functions of different deacetylase complexes (Table 2). Second, global deletion experiments have revealed that the functions of mouse HDAC1 and HDAC2 are not identical during development, and that they function differently in cultured tumour cells, suggesting that these two deacetylases do not always remain together as a catalytic core. Third, deletion of Sds3 or Sin3A affects K9 methylation of histone H3 and the subsequent distribution of HP1 nuclear foci113,114, indicating that K9 deacetylation is a prerequisite for subsequent methylation. This notion is well established in S. pombe21,64, and provides further support for an intimate relationship between histone deacetylation and methylation. And finally, all four members of class IIa have been subject to gene targeting in mice, with resultant phenotypes linked to MEF2 (Table 2), thereby indicating that the MEF2–HDAC axis (Fig. 2b) operates in many tissues aside from muscle115.
Table 2
Class I/II/IV HDACs and associated subunits in human disease and mouse development
| Class | Member | Associated subunit | Map position | Human disease | Effect of mouse gene deletion |
|---|---|---|---|---|---|
| I | HDAC1 | 1p34.1 | Embryonic lethal by E9.5 (refs. 141, 142) | ||
| Proliferation defects in ES cells141,143 | |||||
| I | HDAC2 | 6q21 | Colon cancer | Perinatal death from cardiac defects142 | |
| Cardiomyopathy from heart-specific deletion of both HDAC1 and HDAC2 (ref. 142) | |||||
| Sin3A | 15q24.2 | Embryonic lethal by E6.5; splenomegaly & glomerulopathy in heterozygotes114,144 | |||
| Sds3 | 12q24.23 | Embryonic lethal by E6.5 | |||
| Defects in heterochromatin formation & chromosome segregation113 | |||||
| Mi2β | 12p13 | Defects in T cell development & CD4 gene expression145 | |||
| MBD3 | 19p13.3 | Pre- or peri-natal death146 | |||
| Defects in ES cell pluripotency | |||||
| p66α | 19p13.11 | Embryonic lethal by E9.5 (ref. 147) | |||
| LSD1 | 1p36.12 | Embryonic lethal by E7.5 (ref. 148) | |||
| I | HDAC3 | 5q31.3 | Embryonic lethality with vascular defects (R. Mongomery and E. Olson, pers. commun.) | ||
| Liver hypertrophy (S. Hiebert, pers. commun.) | |||||
| N-CoR | 17p11.2 | Embryonic lethal by E15.5 & defects in neuronal and hematopoitic differentiation149 | |||
| I | HDAC8 | Xq13 | Craniofacial abnormalities (M. Haberland and E. Olson, pers. commun.) | ||
| IIa | HDAC4 | 2q37.2 | Osteodystrophy? | Small size, chondrocyte hypertrophy | |
| Exencephaly150 | |||||
| IIa | HDAC5 | 17q21 | Cardiac hypertrophy151 | ||
| IIa | HDAC7 | 12q13.1 | Cardiovascular defects152 | ||
| IIa | HDAC9 | 7p12.1 | Cardiac hypertrophy153 | ||
| IIb | HDAC6 | Xp11.22 | Neurodegeneration? | Viable with massive increase of α-tubulin acetylation (P. Matthias, pers. commun.) | |
| IIb | HDAC10 | 22q13.3 | ? | ||
| IV | HDAC11 | 3p25.2 | ? |
Note: For associated subunits, only those subject to gene inactivation analysis in mice are listed here.
Implications for disease and therapy
As discussed above, studies in cell-based and animal models shed important light on the potential involvement of the Rpd3/Hda1 family members in the pathophysiology of human diseases. Related to this, HDAC2 has been linked to tumourigenesis in colorectal cancers and increased expression of this deacetylase has been found in a majority of human colon cancer explants116. In addition, a frame shift mutation, which causes loss of HDAC2 and confers resistance to HDAC inhibition, has been detected in some human cancers117.
Members of the family have also been implicated in neuropathological conditions. Accumulating data indicate a key role for HDAC6 in neurodegeneration. It is required for aggresome formation and present in Lewy bodies associated with neurodegenerative disorders like Parkinson’s diseases and dementia106. Impairment of autophagy resulting from decreased HDAC6 activities might predispose individuals to neurodegeneration108. On the other hand, HDAC6 inhibition promotes tubulin acetylation and facilitates microtubule-based vesicle transport, thereby compensating transport deficit in Huntington’s disease98. Thus, functional roles of HDAC6 may vary in different pathological states. Overall, compared to the rapidly accumulating knowledge about the molecular and developmental roles of the Rpd3/Hda1family of deacetylases, much less is known about their direct and specific roles in human pathology. Additional studies are also required to extrapolate accurately results from animal studies to humans and to determine how different HDACs might be altered or mutated in different types or stages of diseases.
Classical HDAC inhibitors have the potential to be therapeutic and chemo-preventive agents for cancer19,118, neurodegenerative disorders20, heart disease69, muscular dystrophy119, spinal muscular atrophy120, and transplantation intolerance121. Attesting to this potential, one such inhibitor was recently approved for treating cutaneous T-cell lymphoma19. Most of the inhibitors currently available display limited selectivity towards Rpd3/Hda1 family members in humans, raising the important question about the identity of the deacetylase(s) actually targeted in vivo. It might not any of the class IIa members since knockout studies suggest that their activation may be beneficial (Table 2). HDAC6 plays an important role in the management of misfolded proteins, so its activation of HDAC6 may also be beneficial (Fig. 3). However, it positively regulates cell motility and the activation of some oncogenic kinases, and its inhibition accelerates microtubule-based vesicle transport, so this deacetylase could be one in vivo target. Other candidates include class I members. If HDAC1 or 2, then the question is which deacetylase complex is the authentic target in patients. Of relevance, RNA interference-mediated knockdown of HDAC1 in D. melanogaster and mammalian cells recapitulates some effects of trichostatin A on cell cycle progression122,123.
In terms of designing new HDAC inhibitors, are isoform-specific compounds preferable to pan-HDAC inhibitors? With regard to isoform-specific intervention, would it be more reasonable to target the signalling pathways that control HDAC functions? Each HDAC has multiple substrates (such as those for HDAC6; Fig. 3) or binding partners (such as MEF2 and 14-3-3 for class IIa HDACs; Fig. 2), so the interaction interface might be a suitable intervention point for designing selective inhibitors.
It is important to note that the real in vivo target may vary in various diseases and different pathological stages. As inactivation of some classical HDACs is detrimental under certain conditions, small-molecule activators may have therapeutic potential26. Because many of the deacetylase are evolutionarily conserved (Fig. 1 and Supplementary information S1 (figure)), different model organisms provide economical alternatives for screening HDAC modulators and testing novel strategies of therapy and prevention.
Conclusions and perspectives
Since identification of the first HDACs in 1996, additional members of the classical family have been found in various organisms (Fig. 1, Supplementary information S1 (figure) and Box 2). There are 11 members in humans, and with the exception of HDAC8, all class I members form multisubunit complexes (Table 1). These complexes show different patterns of evolutionary conservation and carry out key roles in deacetylating histones, interacting with other chromatin regulators and shaping epigenetic programmes. Genome-wide localization maps have been generated for classical HDACs and components of their multisubunit complexes in yeast18,43,64 and should function as valuable guides for further identification and characterization of similar complexes in multicellular organisms.
Class I deacetylases also target non-histone substrates (Box 1), but further characterization is needed to address how the deacetylase complexes are involved. Members of class IIa function as signal transducers to repress transcription in a phosphorylation-dependent manner (Fig. 2). They are at a central interface between signalling and gene regulatory networks. The class IIb member HDAC6 is a major deacetylase in the cytoplasm, using its ubiquitin-binding domain as a stress sensor (Fig. 3). HDAC8 and HDAC10 appear to be specific to deuterostomes and vertebrates, respectively, whereas HDAC11 is conserved even in invertebrates and plants. Compared to other mammalian HDACs, much less is known about functions of these three deacetylases.
Classical HDACs and some of their associated subunits have been subjected to genetic analyses in mice (Table 2), as well as in C. elegans, D. melanogaster and zebrafish (supplementary information S6 (table)). One challenge is to correlate the resulting phenotypes with results from biochemical, molecular and cellular studies. A coherent and integrated picture from a variety of studies will not only provide a valuable guide for identifying and characterizing related human diseases, but will also form an important basis for rational drug design. Global or tissue-specific gene knockout in mice tends to provide information related to early development, whereas human diseases often develop in the late stages of life. Therefore, time-specific gene inactivation or knockdown is expected to greatly extend our understanding of human pathology and to efficiently mimic effects of HDAC inhibition to identify the best in vivo targets for developing new generations of HDAC inhibitors.
Recent proteomic studies have identified a diverse array of Nε-acetylated proteins124–126, but the responsible enzymes are largely unknown. There are also acetylation-like modifications such as formylation127 and propionylation128,129. Moreover, protein O-acetylation has just been reported130, and classical HDACs display intrinsic esterase activity30. These exciting discoveries make us wonder whether we are still at the dawn of a new era for protein acetylation research. How the Rpd3/Hda1 family is involved in deacetylating newly-identified Nε-acetylated proteins, controlling O-acetylation and lysine acetylation-like modifications, and interacting with other post-translational modifications, such as methylation, phosphorylation, ubiquitylation and sumoylation, is an exciting question awaiting further investigation. Answers to this question will provide rich and novel insights into the fundamental issue how members of the Rpd3/Hda1 family are involved in shaping ‘histone code-like’ regulatory programmes that have evolved from bacteria and yeasts to mice and men.
Acknowledgments
Owing to strict space constraints and the wealth of literature, we have used seminal reviews and supplemental figures/tables to cover some of the primary findings from this field. Research in our laboratories has been supported by grants from the NIH, AHA, and the Kaul Foundation (to E.S.) and from the Canadian funding agencies NCIC, CIHR, NSERC and CFI (to X.J.Y.).
Glossary terms
- 14-3-3 proteins
- Members of a family of acidic proteins conserved from yeast and plants to humans. They are highly abundant, ~30 kDa in size and often recognize target proteins with the sequence motif RXXpS/TXP or RXXXpS/TXP, where X is any residue and pS/T denotes phosphoserine or phosphothreonine
- AAA+ ATPASES
- ATPases that are associated wit h a variety of cellular activities. A superfamily of proteins with one or two nucleotide-binding domains (‘AAA modules’), which often form ring-like oligomers and function as chaperones in diverse cellular processes
- ASTROCYTE
- A star-shaped glial cell that supports the tissue of the central nervous system
- AUTOPHAGY
- A pathway for the recycling of cellular contents, in which materials inside the cell are packaged into vesicles and are then targeted to the vacuole or lysosome for bulk turnover
- BASAL BODY
- The structure found at the base of eukaryotic cilia and flagella that consists of an array of nine microtubule triplets, as well as other proteins, and which is involved in the organization and assembly of the ciliary axoneme
- BROMODOMAIN
- A conserved protein module first described for the Drosophila melanogaster homeotic gene regulator Brahma (means ‘creator’ in Hindu). This domain is present in many gene and/or chromatin regulators and has the ability to recognize acetyllysine motifs
- CHAPERONE
- Protein that mediates assembly of another polypeptide-containing structure, but does not form part of the completed structure, or participate in its biological function
- CHEMOTAXIS
- A type of migration that is stimulated by a gradient of a chemical stimulant or chemoattractant
- CHONDROCYTE
- A differentiated cell of cartilage tissue
- CHROMODOMAIN
- Originally identified as a 37-residue chromobox shared by the heterochromatin protein HP1 and the polycomb protein Pc2. Subsequently found in many other chromatin regulators and recognizes methyllysine protein motifs
- DEUTEROSTOME
- An animal in which the anus develops from the first opening of the embryo, and the mouth is formed later. These include echinoderms and chordates
- DYNEIN
- Microtubule-based molecular motor that moves towards the minus end of microtubules
- EUCHROMATIC
- DNA that contains most of the structural genes, changes structure during the cell cycle and undergoes transcriptional regulation
- F-ACTIN
- (Filamentous actin). A flexible, helical polymer of G-actin (globular actin) monomers that is 5–9 nm in diameter
- HOMOLOGUES
- Genes (or their products) that are descended from a common ancestral gene
- IMMUNE SYNAPSE
- A junction that forms at the contact region between a T cell and its target cells. T-cell activation occurs here
- INCLUSION BODIES
- Insoluble aggregates of misfolded proteins; inclusion bodies are common in prokaryotes and the brains of patients affected by triplet-repeat diseases
- KINESIN
- Microtubule-based molecular motor, most often directed towards the plus end of microtubules
- MACROPINOCYTOSIS
- refers to a series of events initiated by extensive plasma membrane reorganization or ruffling to form an external macropinocytic structure that is then enclosed and internalized
- MATING TYPE GENES
- Genes in the yeast chromosome that control the sexual fate of the yeast cell
- microRNA
- A small RNA of ~21 nucleotides for regulating expression of mRNAs to which it is complementary in sequence
- MICROTUBULES
- Hollow tubes, 25 nm in diameter, formed by the lateral association of 13 protofilaments, which are themselves polymers of α- and β-tubulin subunits
- MYOCYTE ENHANCER FACTOR 2 (MEF2)
- An evolutionarily conserved transcription factor, the specific DNA-binding activity of which is mediated by an N-terminal MADS box and an adjacent MEF2-specific domain. Although initially identified as a muscle-specific transcription factor, different MEF2 isoforms are important in various tissues
- NUCLEOSOME
- The basic structural subunits of chromatin, which consist of ~200 base pairs of DNA wrapped around an octamer of histones
- ORTHOLOGUE
- product of a gene in two or more species that has evolved from a common ancestor
- PARALOGUES
- Sequences, or genes, that have originated from a common ancestral sequence, or gene, by a duplication event
- PHD FINGER
- A plant homeodomain-linked (PHD) zinc finger that chelates double zinc ions. This type of zinc finger is present in many chromatin regulators and was recently shown to bind the N-terminal tails of core histones in a methylation-dependent or -independent manner
- SANT DOMAIN
- A 60-residue module initially identified in Swi3, Ada2, N-CoR, and TFIIIB as a putative DNA-binding domain. Present in various transcriptional and chromatin regulators and now considered as a histone-binding module
- SCAFFOLD PROTEIN
- A protein that serves as a platform for the assembly of other proteins
- SE14-REPEAT
- Ser-Glu-containing tetradecapeptide repeats found in HDAC6 proteins from high mammals
- ZnF-UBP
- A ubiquitin-binding zinc finger present in HDAC6 and several ubiquitin-specific proteases
Footnotes
Weblinks
Rpd3: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=417699
Hda1: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=6324307
HDAC1: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=13128860
HDAC2: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=68068066
HDAC3: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=13128862
HDAC4: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=153085395
HDAC5: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=62750347
HDAC6: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=13128864
HDAC7: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=13259522
HDAC8: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=8132878
HDAC9: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=17158039
HDAC10: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=20070354
HDAC11: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=13376228
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrm2346
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2667380?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15(1):9496, 03 Nov 2024
Cited by: 0 articles | PMID: 39489725 | PMCID: PMC11532494
Exploring fatty acids from royal jelly as a source of histone deacetylase inhibitors: from the hive to applications in human well-being and health.
Epigenetics, 19(1):2400423, 10 Sep 2024
Cited by: 0 articles | PMID: 39255363 | PMCID: PMC11404605
Identification of HDAC10 as a candidate oncogene in clear cell renal carcinoma that facilitates tumor proliferation and metastasis.
Diagn Pathol, 19(1):120, 05 Sep 2024
Cited by: 0 articles | PMID: 39237939 | PMCID: PMC11378624
A key component Rxt3 in the Rpd3L histone deacetylase complex regulates development, stress tolerance, amylase production and kojic acid synthesis in Aspergillus oryzae.
Biotechnol Lett, 46(6):1121-1131, 31 Jul 2024
Cited by: 0 articles | PMID: 39083116
The RPD3L deacetylation complex is required for facultative heterochromatin repression in Neurospora crassa.
Proc Natl Acad Sci U S A, 121(32):e2404770121, 29 Jul 2024
Cited by: 1 article | PMID: 39074265
Go to all (764) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants.
BMC Plant Biol, 9:37, 28 Mar 2009
Cited by: 48 articles | PMID: 19327164 | PMCID: PMC2671507
Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo.
J Biol Chem, 276(38):35826-35835, 20 Jul 2001
Cited by: 140 articles | PMID: 11466315
The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans.
J Antimicrob Chemother, 70(7):1993-2003, 29 Mar 2015
Cited by: 29 articles | PMID: 25825380
Class IIa histone deacetylases: regulating the regulators.
Oncogene, 26(37):5450-5467, 01 Aug 2007
Cited by: 158 articles | PMID: 17694086
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA109699
Grant ID: R01 CA109699-04