Abstract
Free full text

The mode of Hedgehog binding to Ihog homologs is not conserved across different phyla
Associated Data
Abstract
Hedgehog (Hh) proteins specify tissue pattern in metazoan embryos by forming gradients that emanate from discrete sites of expression and elicit concentration-dependent cellular differentiation or proliferation responses1,2. Cellular responses to Hh and the movement of Hh through tissues are both precisely regulated, and abnormal Hh signaling has been implicated in human birth defects and cancer3-7. Hh signaling is mediated by its N-terminal domain (HhN), which is dually lipidated and secreted as part of a multivalent lipoprotein particle8-10. Reception of the HhN signal is modulated by several cell-surface proteins on responding cells, including Patched (Ptc), Smoothened (Smo), Ihog/CDO and the vertebrate-specific proteins Hip and Gas111. Drosophila Ihog and its vertebrate homologs CDO and BOC contain multiple immunoglobulin (Ig) and fibronectin type III (FNIII) repeats, and the first FNIII repeat of Ihog binds Drosophila HhN in a heparin-dependent manner12,13. Surprisingly, pull-down experiments suggest that mammalian Sonic hedgehog (ShhN) binds a nonorthologous FNIII repeat of CDO12,14. We report here biochemical, biophysical, and X-ray structural studies of a complex between ShhN and the third FNIII repeat of CDO. We show that the ShhN-CDO interaction is completely unlike the HhN-Ihog interaction and requires calcium, which binds at a previously undetected site on ShhN. This site is conserved in nearly all Hh proteins and is a hot spot for mediating interactions between ShhN and CDO, Ptc, Hip, and Gas1. Mutations in vertebrate Hh proteins causing holoprosencephaly and brachydactyly type A1 map to this calcium-binding site and disrupt interactions with these partners.
CDO is composed of 5 Ig domains followed by 3 FNIII repeats, a membrane-spanning region, and a cytoplasmic tail15. The second FNIII repeat (CDOFn2) is homologous to the Ihog repeat that binds Drosophila HhN and heparin, but the third FNIII repeat (CDOFn3) appears to be most important for binding Shh12,14 (Fig. 2a). Purified CDOFn2, CDOFn3, and a tandem repeat of these domains (CDOFn23) all failed to interact appreciably with ShhN in the presence or absence of heparin as judged by size-exclusion chromatography (SEC), however. Pull-down experiments showing an interaction between ShhN and CDOFn3 were performed in the presence of serum12,14, and a search for additional factors involved in ShhN-CDO binding led to the discovery that calcium ions promote high affinity interactions between ShhN and CDOFn3 (Supplementary Fig. 1). Analytical ultracentrifugation (AUC) and isothermal titration calorimetry (ITC) studies demonstrate that the interaction between ShhN and CDOFn3 is 1:1, requires calcium, is not heparin-dependent, and has a dissociation constant of ~1.3 μM (Figs. (Figs.1a,1a, ,2c2c and Supplementary Figure 2).
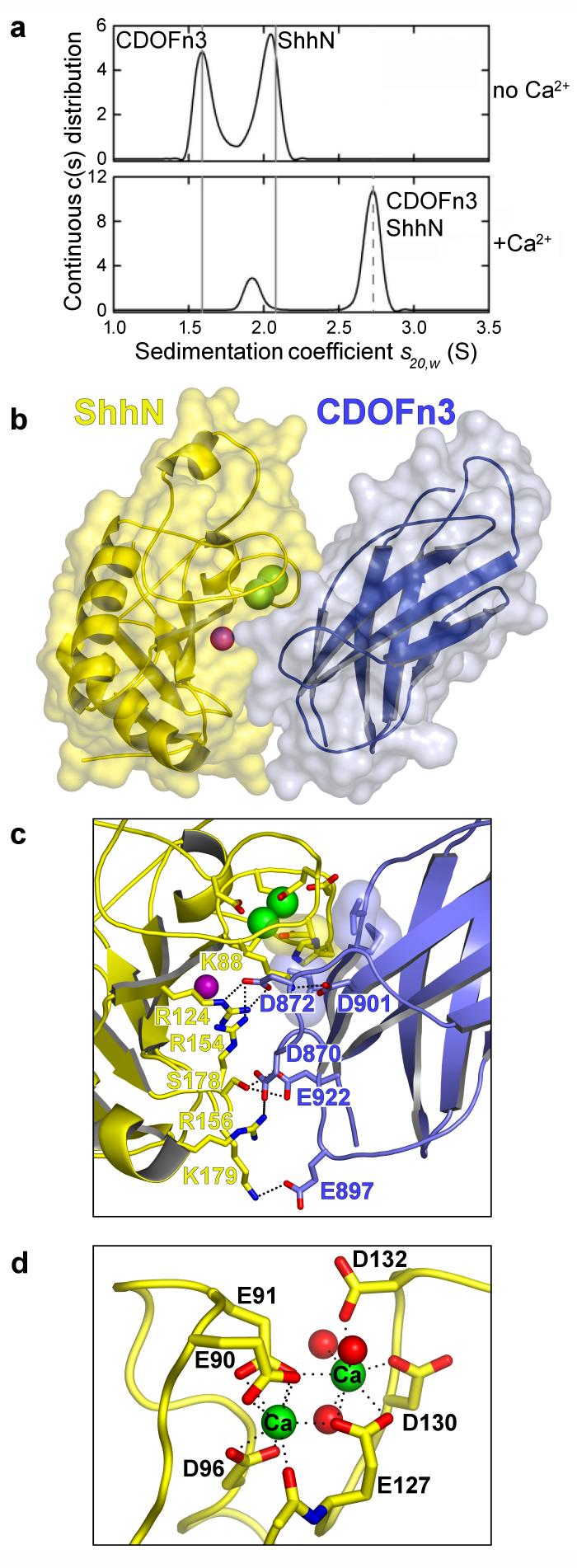
a, Sedimentation velocity results for a 1:1 mixture of 20.5 μM ShhN and CDOFn3 in the absence and presence of 1 mM CaCl2. Solid grey lines indicate the experimental sedimentation coefficients for monomeric CDOFn3 and ShhN. The dashed line indicates the sedimentation coefficient calculated in HYDROPRO for the 1:1 complex. b, Semi-transparent molecular surface over a ribbon diagram of ShhN (yellow) bound to CDOFn3 (blue). Calcium and zinc ions are depicted as green and purple spheres, respectively. c, The ShhN-CDOFn3 interface shown in the same orientation as b. The mostly acidic residues on CDOFn3 that interact with mostly basic residues on ShhN are labelled. Semi-transparent surface representations are shown for CDO V918, M919 and I920, which make van der Waal's contacts with Shh H134 and the methylene groups of E90. d, Sidechains in ShhN directly coordinating the two calcium ions are shown in stick representation. Calcium ions are green and three water molecules are shown as red spheres. Hydrogen bonds are depicted as dotted lines. All structure images were rendered with PyMOL (http://www.pymol.org).
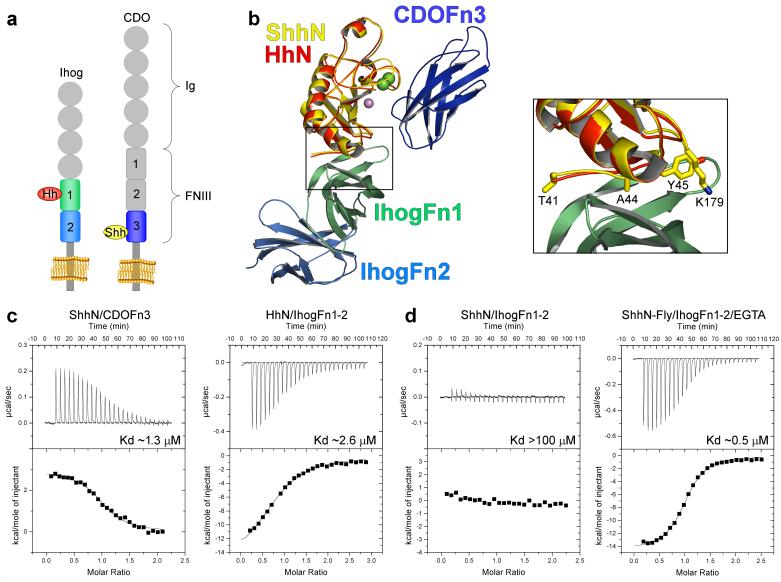
a, Schematic of Ihog and CDO. b, Ribbon diagram of the ShhN/CDOFn3 structure superimposed on the HhN/IhogFn1-2 structure by alignment of the Hh molecules. ShhN is colored yellow, CDOFn3 dark blue, HhN red, IhogFn1 light green, and IhogFn2 light blue. Calcium and zinc ions are depicted as green and purple spheres, respectively. The boxed area shows a close-up of the interface between IhogFn1 and the Hh proteins. The sidechains of four ShhN residues changed in the ShhN-Fly mutant are shown as sticks. c, ITC data for: ShhN and CDOFn3 with calcium (left), and HhN and IhogFn1-2 with heparin and calcium (right). d, ITC data for: ShhN and IhogFn1-2 with heparin and calcium (left), and ShhN-Fly and IhogFn1-2 with heparin and EGTA (right).
The calcium-dependent ShhN-CDOFn3 complex was crystallized, and its structure determined by molecular replacement and refined to R/Rfree of .186/.229 at 1.7 Å resolution (Supplementary Table 1). The ShhN-CDOFn3 interface buries 1325 Å2 with a shape complementarity of 0.61 (Figs. 1b and 1c), values consistent with a physiological interface16,17. This interface encompasses a hydrophilic region involving K88, R124, R154, R156, S178, and K179 on ShhN and D870, D872, E897, D901, and E922 on CDOFn3 and a hydrophobic region involving V918, M919, and I920 on CDOFn3 and H134 and the methylene groups of E90 on ShhN (Figure 1c and Supplementary Figs. 3-5). Binding studies with four previously created clusters of surface mutations in ShhN (surfaces A, B, C, and D)12 and five new mutations support identification of this interface as functionally relevant (Supplementary Fig. 3). Sequence analysis suggests that BOC, a receptor for Shh in axon guidance18, is likely to bind ShhN in the same manner as CDO (Supplementary Figs. 5 and 6).
The structure of the ShhN-CDOFn3 complex reveals a previously unknown binuclear calcium-binding site in ShhN buried at the interface with CDOFn3, which accounts for the calcium-dependence of this interaction (Fig. 1). The calcium ions are coordinated by 3 aspartate and 3 glutamate residues in ShhN (Asp 96, Asp 130, Asp 132, Glu 90, Glu 91, and Glu 127) in an arrangement similar to binuclear calcium sites in other proteins19. Each ion is coordinated by 8 oxygen atoms with an average distance of 2.5 Å, consistent with calcium coordination20. Identification of these ions as calcium was confirmed by anomalous difference Fourier analysis (Supplementary Fig. 7). The level of conservation of the six calcium-coordinating residues in Hh sequences suggests calcium binding is a general property of Hh proteins (Supplementary Fig. 8). Attempts to measure the dissociation constant of ShhN for calcium indicate a Kd > 100 μM. This weak affinity explains the failure to observe this calcium site in earlier HhN crystal structures, as calcium would have dissociated during purification13,21. 1 mM calcium is sufficient to restore calcium-dependent activity to ShhN in vitro, and the presence of ~1 mM calcium ions in the extracellular space suggests that calcium is likely to be a constitutive component of HhN proteins.
Strikingly, the ShhN-CDOFn3 interaction is completely unlike the interaction between Drosophila HhN and Ihog13 (Fig. 2b). The surfaces on ShhN and HhN that bind CDO and Ihog, respectively, overlap at only a single residue and interact with non-orthologous domains of CDO and Ihog (CDOFn3 vs. IhogFn1). This result is surprising given the 68% sequence identity between human ShhN and Drosophila HhN and sequence and functional data implicating CDO and BOC as the closest vertebrate homologs of Ihog12,14,22. To confirm the distinct binding modes, four simultaneous amino acid substitutions (T41Y, A44V, Y45L, and K179R) were introduced into ShhN to create a variant, ShhN-Fly, that contains all Ihog-contacting residues found at the Drosophila HhN-Ihog interface13. ShhN-Fly binds Ihog with >100-fold greater affinity than wild-type ShhN and has a slightly stronger affinity for Ihog (Kd ~0.5 μM) than does Drosophila HhN (Kd ~2.6 μM) (Fig. 2c, d). ShhN-Fly does not bind well to dPtc-expressing cells (Supplementary Fig. 9). As is the case for HhN, ShhN-Fly requires heparin but not calcium to bind Ihog, and the interaction is exothermic (Fig. 2c, d). In contrast, the interaction between ShhN or ShhN-Fly and CDO is endothermic (Fig. 2c, Supplementary Fig. 10). Expression of Shh-Fly within clones in the anterior compartment of Drosophila wing imaginal discs also induces expression of the Hh pathway target gene Ptc to a greater extent than wild-type Shh, and this expression extends several cells beyond the clone boundary (Supplementary Fig. 11).
Although modelling suggests that a complex containing ShhN-Fly, IhogFn12, and CDOFn3 is possible, this complex is not observed by SEC. The CDO binding site on ShhN overlaps extensively with a heparin-binding site required for interactions between HhN and Ihog13, and heparin may impede binding of CDOFn3 to ShhN-Fly. Analysis of Hh sequences suggests that the CDO-like binding mode is conserved in vertebrates and the Ihog-like mode in arthropods, but the binding mode for intermediate and earlier branchpoint clades is not clear.
Given the importance of this previously unappreciated calcium-binding site for Shh-CDO interactions, we examined its role in mediating interactions between ShhN and other ShhN binding partners. Four simultaneous amino-acid substitutions in the ShhN calcium binding site (E90Q, E91Q, E127Q, D132N) or increasing concentrations of EGTA, a calcium chelator, led to a >10-fold reduction in ShhN binding to Hip-transfected cells and a ~2-fold reduction in binding to Ptc-transfected cells when compared to wild-type ShhN (Fig. 3a, b). EGTA also reduces binding of Drosophila HhN to Ptc-transfected cells (Supplementary Fig. 12). The effects of EGTA appear stronger, possibly owing to effects of calcium chelation on the cell membrane or chelation of a previously identified zinc ion in ShhN21. The presence of increasing amounts of soluble CDOFn3 or CDOFn23, but not CDOFn2, also reduced the ability of ShhN to bind to Hip and Ptc-transfected cells of human or fly origin (Fig. 3c and Supplementary Figs. 13 and 14), suggesting that Hip and Ptc may directly compete with CDO for binding to ShhN. Although consistent with our results, direct competition would conflict with an earlier observation of synergy between CDO and Ptc for Shh binding23. Resolution of this issue awaits studies with purified proteins.
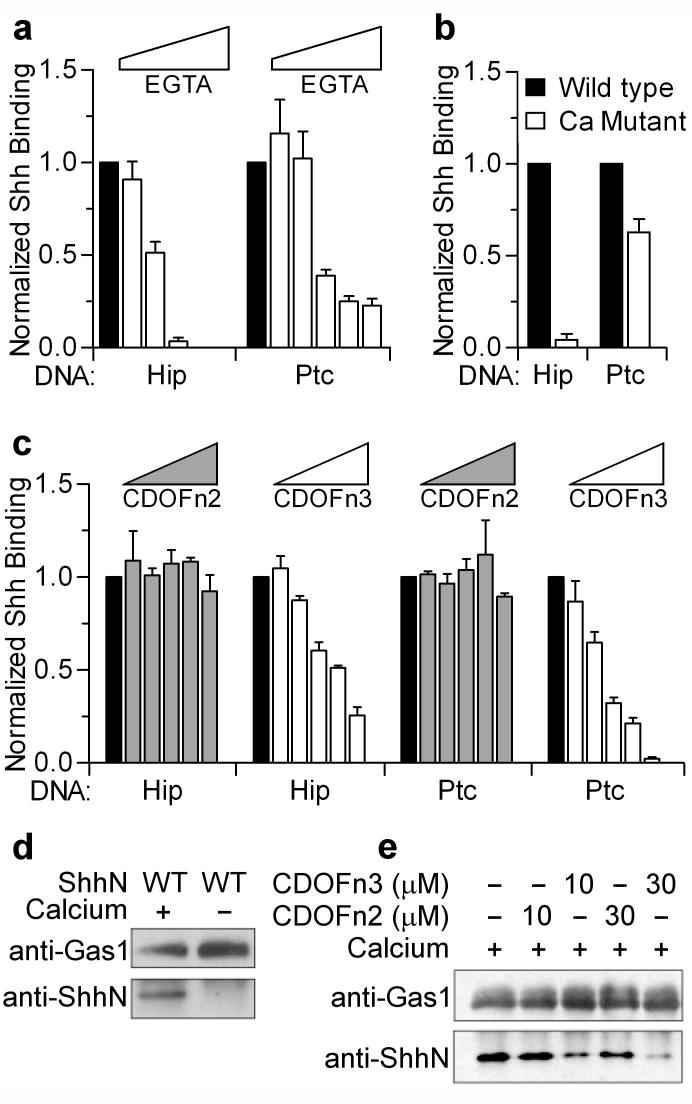
a, Cells expressing Hip or Ptc were assayed for Shh binding in the presence of 1.8 mM Ca2+ (black bars) and increasing EGTA concentrations (1.8-2.7 mM, white bars). b, Cell-based binding assays were performed with wild-type Shh and a Shh calcium-binding site mutant (Ca Mutant) in the presence of calcium. c, Cell-based binding assays with wild-type Shh were performed in the presence of calcium (black bars) and 0.5-30 μM soluble CDOFn2 (grey bars) or CDOFn3 (white bars). Error bars in a, b and c indicate standard deviation with n = 3. d, Gas1 Fc-fusion protein was immobilized to Protein G beads and binding to ShhN was assayed by a Western blot in the presence and absence of calcium. e, Gas1 pull-down experiments were repeated in the presence of CDOFn3 or CDOFn2.
Gas1 has also been reported to interact with Shh23, and a pull-down assay was developed in which Fc-tagged Gas1 immobilized on Protein G beads is used to precipitate ShhN. The absence of calcium in the precipitating medium or the presence of CDOFn3 (but not CDOFn2) greatly reduced the ability of Gas1 to precipitate ShhN (Fig. 3d, e). These results suggest that an intact and accessible calcium-binding site on ShhN is required to mediate interactions between ShhN and Gas1.
A missense substitution in human Shh (D88V) that causes holoprosencephaly (HPE)24, a developmental disorder involving abnormal formation of midline face and forebrain features, maps to a loop on ShhN that contains 3 of the 6 calcium-coordinating residues (Fig. 4a). D88 does not directly contact CDOFn3 or coordinate calcium but stabilizes the conformation of the calcium-coordinating loop by forming hydrogen bonds to two backbone amides. Introduction of the D88V substitution into ShhN results in loss of detectable binding to CDOFn3 (Fig. 4b) and reduces binding of ShhN to Hip expressing cells by ~4-fold and to Ptc expressing cells ~5-fold (Fig. 4d). D88V ShhN also binds less well to Gas1 (Supplementary Fig. 15). Although no molecular explanation for the effects of the D88V mutation was previously apparent, it is now clear that the D88V reduces the affinity of ShhN for multiple partners through disruption of the calcium-coordinating region.
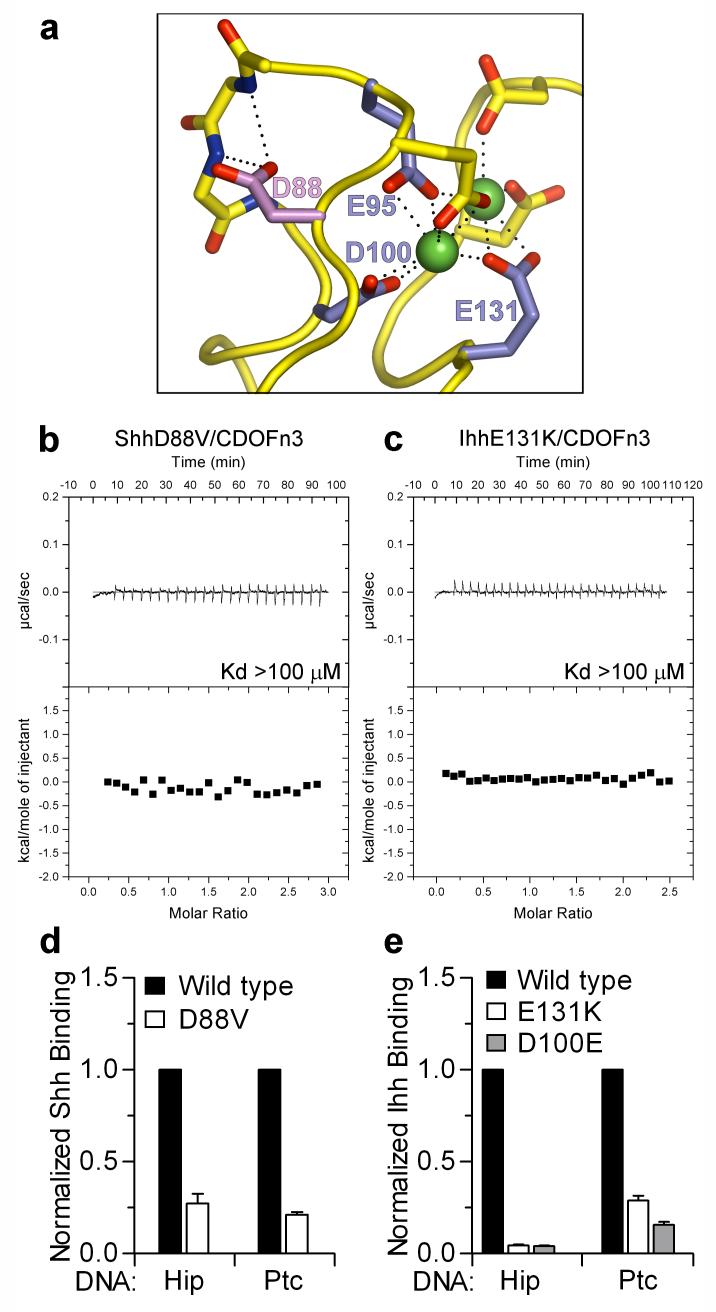
a, Loop representation of the mShhN calcium-binding site. Mutation of human Ihh residues E95, D100 and E131 (blue) cause BDA1. Mutation of D88 (purple) in human Shh causes HPE. Calcium ions are depicted as green spheres and hydrogen bonds are shown as dotted lines. b, c, ITC data showing loss of CDOFn3 binding to ShhD88V (b) and IhhE131K (c) in the presence of calcium. d, Normalized binding of Shh and ShhD88V to cells expressing Ptc and Hip. Error bars indicate standard deviation with n = 5. e, Normalized binding of Ihh, IhhE131K and IhhD100E to cells expressing Ptc and Hip. Error bars indicate standard deviation with n = 3.
Brachydactyly type A1 (BDA1) is characterized by absent or shortened middle phalanges5. Five missense substitutions in human Indian hedgehog (Ihh) that cause BDA1 (E95K, E95G, D100N, D100E, and E131K) affect residues that directly coordinate calcium (Fig. 4a). ITC experiments show that wild-type Ihh binds CDOFn3 with a Kd of ~2.7 μM in the presence of calcium (Supplementary Fig. 16), similar to the value of 1.3 μM for ShhN binding to CDOFn3. Introduction of the E131K mutation into IhhN results in loss of detectable CDOFn3 binding (Fig. 4c), and the E131K and D100E substitutions reduce binding of Ihh to cells expressing Ptc by ~4-fold and 10-fold, respectively (Fig. 4e). The E131K and D100E substitutions also reduced Ihh binding to Hip expressing cells >20-fold (Fig. 4e); the presence of EGTA decreased binding of wild-type Ihh to Ptc and Hip-expressing cells by similar amounts (Supplementary Fig. 17), implicating disruption of calcium-dependent interactions as a common mechanism for BDA1-causing substitutions. The E127K substitution in Shh (equivalent to E131K in human Ihh) reduced the binding of ShhN to Gas1 in pull-down experiments (Supplementary Fig. 15), and substitutions in Shh equivalent to each of the five Ihh BDA mutations reduce binding of Shh to Hip and Ptc in cell-based assays (Supplementary Fig. 18).
Ptc, CDO, and Gas1 are positive regulators of Hh signaling25, and Hip is a negative regulator26. The BDA1-causing Ihh mutations act in a dominant, gain-of-function fashion3, and produce a phenotype resembling a dominant mutation that produces ectopic expression of the Shh gene within the developing digits27. Among the complex effects of BDA1 mutations, disruption of interactions with Hip and an increased range of signaling seem most likely to cause gain-of-function phenotype. Indeed, introduction of the E95K substitution into mouse Ihh (E91 in mouse Shh) increases the range of Ihh action within the digit primordia of the limb, albeit with a reduced signaling potency (Chan et al. personal communication). In contrast, the loss-of-function phenotype of the D88V mutation in Shh suggests that effects of this mutation on interactions with Ptc, CDO, and/or Gas1 predominate. In general, the phenotype produced by calcium-site mutations is likely to be a composite of positive and negative effects depending on the importance or expression of various partners in affected tissues and processes.
We have shown that vertebrate Hh proteins utilize a previously unappreciated binuclear calcium-binding site to mediate interactions with multiple partners. Mutations causing the developmental disorders HPE and BDA1 map to this region in Shh and Ihh, respectively. Surprisingly, structural and biochemical studies demonstrate that vertebrate Hh proteins interact with the adhesion-like molecule CDO in a completely different manner than the homologous HhN-Ihog pair in Drosophila13. This difference may have arisen through convergent evolution, in which vertebrate and invertebrate ancestors independently evolved use of homologous molecules to modulate Hh signalling. We find this scenario unlikely and suggest the following alternative. Hh proteins are secreted as part of a multivalent lipoprotein complex9,10, which must result in high-density clustering of Hh binding partners at a localized patch on the surface of target cells. At sites of high local concentration and reduced dimensionality, weak interactions may become functionally relevant, and single mutations may have profound positive or negative effects28. We propose that following evolution of an Ihog/CDO precursor that mediated Hh binding, clustering may have led to a bi-modal interaction between Hh and Ihog/CDO. Additional substitutions could have resulted in the loss of one of the two binding modes in different species. Once different binding modes that supply the same function emerged, evolution of different regulatory mechanisms became possible.
METHODS SUMMARY
Sedimentation velocity experiments
Experiments were conducted at 4.0 °C on a Beckman Coulter XL-I analytical ultracentrifuge using multiple loading concentrations and rotor speeds of 55 or 60 krpm. Scans were analyzed in SEDFIT 11.3 to obtain a sedimentation coefficient, s or a c(s) distribution29. Values were corrected to obtain s20,w or c(s20,w) using solvent parameters calculated in SEDNTERP 1.09.
Crystallization
mShhN (residues 26-189) and hCDOFn3 (826-924) were mixed in the presence of 2 mM CaCl2, incubated for 1 hour at room temperature, and the mShhN/hCDOFn3 complex was purified by SEC and concentrated to ~6 mg/ml total protein. The complex was crystallized by vapor diffusion in hanging drops at 20 °C by mixing 1 μL of protein solution with 1 μL of reservoir buffer (28% PEG 3350, 200 mM MgCl2, 100 mM Tris pH 8.3). The complex crystallized in space group C2221 with unit cell dimensions a=71.2, b=98.6 and c=144.0 Å.
ITC experiments
Titrations were performed using a VP-ITC MicroCalorimeter (MicroCal Inc.). When IhogFn1-2 binding was assayed, 300 μM LMW heparin (Sigma) was included. CDO or Ihog proteins (143-432 μM) were added to a 1.38 ml cell containing a specific HhN protein (10-38 μM) while stirring at 260 rpm. Data were processed with ORIGIN software and best fit by a single binding site model.
Cell-based binding assays
Cos1 cells plated in 6-well-plates were transfected with Fugene 6 (Roche) for 48 hr followed by incubation with conditioned medium containing ShhN-Ren or IhhN-Ren proteins on ice for 1 hr. Cells were washed three times with cold PBS or DMEM, lysed, and 50 μl of lysate was used to measure luciferase activities.
METHODS
Cloning, protein expression and purification
DNA fragments encoding hCDOFn2-3 (amino acids 722-924), hCDOFn3 (826-924), hBOCFn3 (710-817), D. melanogaster IhogFn1-2 (466-679) and IhogFn1 (466-577) were PCR amplified and cloned into the bacterial expression plasmid pT7HMT30. DNA fragments encoding mShhN (26-189), mIhhN (67-231), and D. melanogaster HhN (86-248) were PCR amplified and cloned into a modified pMAL-c2X (NEB) bacterial expression vector. Megaprimer PCR mutagenesis was used to create the mShhD89V (hShhD88V) mutant by introducing the desired base substitutions during two rounds of PCR and subcloning the product into the expression plasmid. A similar approach was used to create the ShhN-Fly (T41Y, A44V, Y45L, K179R) and IhhE131K mutants.
Proteins were expressed in BL21(DE3) cells during an overnight induction. After cell lysis, proteins were partially purified using Ni-NTA resin (Qiagen) and then digested with either TEV or HRV14-3C protease to remove N-terminal tags. Proteins were then further purified by ion-exchange and size-exclusion chromatography and stored frozen at -80 °C in 20 mM Tris-HCl pH 8.0, 200 mM NaCl.
Expression in Drosophila and mammalian cell cultures was driven by the Actin 5C promoter and CMV promoter, respectively. Renilla luciferase tagged HhN or ShhN expression vectors, and mPTCH1 expression vectors, have been described previously12,31.
Data collection and structure determination
Native ShhN/CDOFn3 crystals in reservoir solution were frozen directly in the cryostream. Diffraction data were collected at beamline X4C of the National Synchrotron Light Source at Brookhaven National Laboratory and processed with HKL200032. A molecular replacement solution was found with MOLREP33 using the mShhN20 and IhogFn213 structures as search models. Model building was performed using COOT34, and refinement was carried out by CNS35 and REFMAC33. Data collection and refinement statistics are presented in Supplementary Table 1. There are two complexes in the asymmetric unit and the final model consists of 3,911 protein atoms, 410 water molecules, four calcium ions, and two previously described zinc ions20.
Sedimentation equilibrium
Experiments were conducted at 4.0°C on a Beckman Optima XL-A analytical ultracentrifuge. Each sample (loading volume of 130 μl) in 20 mM Tris pH 8.0, 200 mM NaCl was studied at three loading concentrations corresponding to a loading A280 of approximately 0.4, 0.8 and 1.2, and experiments were carried out at rotor speeds of 20 to 36 krpm. Data were acquired as an average of 4 absorbance measurements at a wavelength of 280 nm using a radial spacing of 0.001 cm. Sedimentation equilibrium was achieved within 48 hours and data were analyzed globally in SEDPHAT 4.3 in terms of various models36. Solution densities ρ were calculated using SEDNTERP 1.09, as were values for the partial specific volumes ν of the proteins studied. Data collected for each of the ShhN and CDOFn3 samples at different speeds and different loading concentrations were best fit in terms of a single ideal solute, returning molecular masses consistent with monomeric, monodisperse species (Supplementary Table 2). 2:1, 1:1 and 1:2 loading mixtures of ShhN and CDOFn3 containing 1 mM CaCl2, each at three loading A280, were also studied and data were analyzed in terms of a reversible A + B ↔ AB or A + A + B ↔ AAB equilibrium using mass conservation. Analysis in terms of an A + B ↔ AB model returned excellent fits and an averaged Kd of 4 ± 2 μM. 1:1 loading mixtures of ShhN and CDOFn3 in the absence of calcium chloride, as well as ShhD88V and CDOFn3 in the presence of calcium chloride were studied in a similar manner. In both these instances no evidence for an interaction was noted and data were best fit in terms of free ShhN and CDOFn3.
Sedimentation velocity and molecular modeling
Structural data for CDOFn3, ShhN and their 1:1 complex were used to calculate the sedimentation coefficient based on a shell model with the program HYDROPRO37 (Supplementary Table 2). Sedimentation velocity c(s) analyses were carried out using an s-value range of 0.05 to 4.0 with a linear resolution of 100 and a confidence level (F-ratio) of 0.68; root mean square deviation values for the best fits ranged from 0.0035 to 0.0065 absorbance units.
Pull-down binding assays
mGas1 human-Fc fusion proteins were expressed in Cos1 cells and bound to Protein G beads overnight at 4 °C. The Protein G beads were then incubated with either wild type or mutant ShhN-conditioned media overnight at 4 °C and Shh proteins associated with the matrices detected by western blotting.
Western blot analysis
Primary antibodies were rabbit anti-Shh (1:1000) and rabbit anti-mGas1 (1:1000)38.
Generation of Shh/Shh-Fly expressing clones
hsp70-FLP, UAS-GFP; Pin/Cyo X actP<y<Gal4/Cyo; UAS-Shh (or Shh-Fly)/Tm6B, 2-3 instar larvae were heat shocked for 5 min at 37 °C to induce clones.
Acknowledgements
We thank R. Abramowitz and J. Schwanof of beamline X4C at the National Synchrotron Light Source for assistance with X-ray data collection, C.W. Vander Kooi for suggesting ions may promote Shh-CDO interactions, and W. Yang, J. Nathans, W.I. Weis, K.C. Garcia, P.A. Cole and L. M. Amzel for comments on the manuscript. We thank A.P. McMahon and C.M. Fan for the mHip1 and mGas1-Fc cDNAs, respectively. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (R.G.). D.J.L. is supported by R01 HD055545 and P.A.B. is an HHMI investigator. J.S.M. is supported by a National Science Foundation Graduate Research Fellowship. X.Z. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1915-06).
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature07358
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2679680?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature07358
Article citations
Short stature with brachydactyly caused by a novel mutation in the IHH gene and response to 4-year growth hormone therapy: a case report.
Transl Pediatr, 13(5):856-863, 24 May 2024
Cited by: 0 articles | PMID: 38840672
Suppression of apoptosis impairs phalangeal joint formation in the pathogenesis of brachydactyly type A1.
Nat Commun, 15(1):2229, 12 Mar 2024
Cited by: 1 article | PMID: 38472182 | PMCID: PMC10933404
Hedgehog on the Move: Glypican-Regulated Transport and Gradient Formation in <i>Drosophila</i>.
Cells, 13(5):418, 27 Feb 2024
Cited by: 1 article | PMID: 38474382 | PMCID: PMC10930589
Review Free full text in Europe PMC
Coordination of canonical and noncanonical Hedgehog signalling pathways mediated by WDR11 during primordial germ cell development.
Sci Rep, 13(1):12309, 29 Jul 2023
Cited by: 2 articles | PMID: 37516749 | PMCID: PMC10387110
Cellular and molecular mechanisms of Hedgehog signalling.
Nat Rev Mol Cell Biol, 24(9):668-687, 17 Mar 2023
Cited by: 42 articles | PMID: 36932157
Review
Go to all (111) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J Biol Chem, 285(32):24584-24590, 01 Jun 2010
Cited by: 42 articles | PMID: 20519495 | PMCID: PMC2915694
Structure of a heparin-dependent complex of Hedgehog and Ihog.
Proc Natl Acad Sci U S A, 103(46):17208-17213, 31 Oct 2006
Cited by: 62 articles | PMID: 17077139 | PMCID: PMC1859911
The ihog cell-surface proteins bind Hedgehog and mediate pathway activation.
Cell, 125(2):343-357, 01 Apr 2006
Cited by: 152 articles | PMID: 16630821
Cdon and Boc: Two transmembrane proteins implicated in cell-cell communication.
Int J Biochem Cell Biol, 44(5):698-702, 03 Feb 2012
Cited by: 27 articles | PMID: 22326621
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
Intramural NIH HHS (1)
Grant ID: Z99 DK999999
NICHD NIH HHS (1)
Grant ID: R01 HD055545




