Abstract
Free full text

A putative viral defence mechanism in archaeal cells
Abstract
Clusters of regularly spaced direct repeats, separated by unconserved spacer sequences, are ubiquitous in archaeal chromosomes and occur in some plasmids. Some clusters constitute around 1% of chromosomal DNA. Similarly structured clusters, generally smaller, also occur in some bacterial chromosomes. Although early studies implicated these clusters in segregation/partition functions, recent evidence suggests that the spacer sequences derive from extrachromosomal elements, and, primarily, viruses. This has led to the proposal that the clusters provide a defence against viral propagation in cells, and that both the mode of inhibition of viral propagation and the mechanism of adding spacer-repeat units to clusters, are dependent on RNAs transcribed from the clusters. Moreover, the putative inhibitory apparatus (piRNA-based) may be evolutionarily related to the interference RNA systems (siRNA and miRNA), which are common in eukarya. Here, we analyze all the current data on archaeal repeat clusters and provide some new insights into their diverse structures, transcriptional properties and mode of structural development. The results are consistent with larger cluster transcripts being processed at the centers of the repeat sequences and being further trimmed by exonucleases to yield a dominant, intracellular RNA species, which corresponds approximately to the size of a spacer. Furthermore, analysis of the extensive clusters of Sulfolobus solfataricus strains P1 and P2B provides support for the presence of a flanking sequence adjoining a cluster being a prerequisite for the incorporation of new spacer-repeat units, which occurs between the flanking sequence and the cluster. An archaeal database summarizing the data will be maintained at http://dac.molbio.ku.dk/dbs/SRSR/.
Introduction
Clusters of regularly spaced repeats were first detected in Escherichia coli (Ishino et al. 1987), and were later found in archaea, both in chromosomes of the Haloferax genus of the kingdom Euryarchaeota (Mojica et al. 1993, 1995) and in conjugative plasmids of the Sulfolobus genus of the kingdom Crenarchaeota (She et al. 1998, Greve et al. 2004). The repeat clusters consist of a number of identical repeats interspaced with generally unique spacer sequences and have been assigned various acronyms in the literature including TREP, DVR, LCTR, SPIDR, SRSR and CRISPR. With the rapid progress in whole genome sequencing, it is now clear that repeat clusters are ubiquitous in the sequenced archaeal chromosomes, except that of Halobacterium sp. NRC-1,and they are present in 55% of the sequenced bacterial chromosomes. Striking for the Archaea, is that the chromosomal clusters are often both multiple and very large, such that in some Sulfolobus species, they constitute > 1% of the genome (Kawarabayasi et al. 2001, She et al. 2001).
Results from early experiments involving the transformation of repeat cluster-containing plasmids into species of the Haloferax genus implicated the clusters in chromosomal segregation (Mojica et al. 1995). Consistent with this view, it was demonstrated that, for both Euryarchaeota and Crenarchaeota, clusters tend to be replicated at the end of the replication cycle prior to chromosomal segregation (Zivanovic et al. 2002, Lundgren et al. 2004). Moreover, circumstantial evidence from studies of Sulfolobus conjugative plasmids supports the idea that plasmids carrying repeat clusters are more stably maintained in host cells (Greve et al. 2004).
Early attention focused on the structure and function of the repeat sequences, which generally carry a low level of nonpalindromic dyad symmetry. For Sulfolobus, a protein was isolated that bound specifically to the repeat and induced a distortion at its center, suggesting that it might have a role in folding the repeat clusters (Peng et al. 2003).
More recently, evidence has been presented that some spacer sequences in archaeal and bacterial chromosomes correspond closely to sequences occurring in extrachromosomal elements (Bolotin et al. 2005, Mojica et al. 2005, Pourcel et al. 2005). For archaea, sequence similarities were found with fuselloviruses and rudiviruses, as well as with a conjugative plasmid of Sulfolobus, and many matches lie within annotated open reading frames (ORFs) (Mojica et al. 2005). Moreover, for bacteria, cluster spacers of diverse strains of Streptococcus thermophilus were shown to produce matches to different bacteriophage or plasmid sequences (Bolotin et al. 2005). Furthermore, for Yersinia strains, spacers of the three small repeat clusters yielded multiple sequence matches with a chromosomal region containing a defective lambdoid prophage (Pourcel et al. 2005). These results are all consistent with the hypothesis that the spacer DNA derives from intracellular extrachromosomal elements.
Furthermore, evidence from comparative studies of Mycobacterium tuberculosis and Yersinia strains suggests that the sizes of repeat clusters can change by addition or deletion of one or more repeat-spacer units. In Yersinia, new repeat-spacer units can be added at the end of clusters, where a flanking or leader sequence is located (van Embden et al. 2000, Jansen et al. 2002, Tang et al. 2002, Pourcel et al. 2005). The mechanism by which this occurs could involve the products of a superoperon of genes that were originally implicated in DNA repair primarily in archaea and thermophilic bacteria (Makarova et al. 2002). These genes, some of which were later labeled cas genes, were considered to be co-functional with the repeat clusters because they are located close to chromosomal repeat clusters and are absent from bacterial chromosomes lacking repeat clusters (Jansen et al. 2002, Bolotin et al. 2005, Haft et al. 2005).
These results led to a common hypothesis for both archaea and bacteria that the cluster spacers are relics of an earlier, intracellular presence of extrachromosomal elements. The incorporation of their DNA into the repeat clusters then provides immunity against subsequent cellular invasion and propagation by identical, or closely related, genetic elements (Bolotin et al. 2005, Mojica et al. 2005). The hypothesis is strongly underpinned, at least for the archaea Archaeoglobus fulgidus and Sulfolobus solfataricus P1, by the finding that repeat clusters produce transcripts from one DNA strand, which may target and inactivate either gene transcripts or genes of the invading genetic elements (Tang et al. 2002, 2005). Moreover, such a mechanism is consistent with the finding of double-strand-specific endoribonucleases in both euryarchaeal and crenarchaeal species, which could be involved in the degradation of double helical RNA regions generated by the annealing of antisense-RNA and mRNAs (Stolt and Zillig 1993, Ohtani et al. 2004). Such a process is also reinforced by the structural characterization of euryarchaeal argonaute proteins which, in eukaryotes, have been implicated in the processing of interference RNAs (Parker et al. 2004, Song et al. 2004).
Recently, Makarova et al. (2006) extended the abovementioned knowledge by reassigning the cluster-associated cas genes as primarily encoding an RNA/DNA regulatory-processing system related to the eukaryal interference RNA (siRNA and miRNA) systems, which they defined as a prokaryotic interference RNA (piRNA).
In this article we summarize what is currently known about the structure and function of the repeat clusters of archaeal genomes and include some new data relevant to their structural and functional properties.
Materials and methods
Sequencing of clusters in S. solfataricus P1
Long range PCR products were obtained across the chromosomal cluster regions of strain P1, which differ in sequence from those of strain P2, using the Herculase II kit (Stratagene, La Jolla, CA) according to the protocol, with 150 ng genomic DNA in a 25 µl reaction. Similar regions were amplified in 1–2 kb sections with Taq DNA polymerase (New England Biolabs, Ipswich, MA) and 2 ng genomic DNA in a 15 µl reaction. The PCR products were purified using QIAquick PCR purification kit (Qiagen, Westburg, Germany). Sequencing was performed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA), where each 10 µl sequencing reaction consisted of 1.4 µl purified PCR product, 1.6 pM primer and 2 µl Big-Dye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems). The reaction was run on a TRIO-thermoblock (Biometra, Goettingen, Germany) (30 s at 96 °C, 15 s at 50 °C, 4 min at 60 °C) × 25 and then maintained at 4 °C, whereupon it was ethanol-precipitated and redissolved in 12.5 µl Template Suppressing Reagent (Applied Biosystems). The sequences were analyzed with Sequencher (Gene Codes, Ann Arbor, MI), BLAST searches were performed against the Sulfolobus Database (http://dac.molbio.ku.dk/dbs/Sulfolobus/cbin/mutagen.pl).
Preparation of total RNA and Northern blotting
Sulfolobus acidocaldarius cells were grown at 78 °C in complex medium containing 2% tryptone (Schleper et al. 1995). Total RNA was prepared from exponentially growing and stationary phasecells by the phenol-guanidium-thiocyanate-chloroform method (Sambrook and Russell 2001) with DNase I treatment. Twenty µg of total RNA was fractionated in an 8% polyacrylamide gel with 7 M urea, 90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA, pH 8.3, together with a 10–100 nt RNA ladder (Decade Marker System, Ambion, Huntingdon, U.K.) or a 0.1–2.0 kb RNA ladder (Invitrogen, Paisley, U.K.). The RNA was transferred onto nylon membranes (Hybond N+, Amersham Biosciences, Amersham, U.K.) using the Bio-Rad semi-dry blotting apparatus (Trans-blot SD, Bio-Rad, Hercules, CA). After immobilizing the RNAs using a Crosslinker (Stratagene), the nylon membranes were prehybridized for 1 h in 6 × SSPE (0.9 M NaCl, 60 mM NaH2PO4, 4.6 mM EDTA and pH 7.4), 0.5% SDS and 5 × Denhardt’s solution at 59 °C. Oligonucleotide primers (26-mers), complementary to each strand of a terminal spacer in saci-4 (5′-GATACGTTGCAGGCAGATGATGAGAG-3′, 5′-CTCTCATCATCTGCCTGCAACGTATC-3′), were end-labeled with [32P]ATP and T4 polynucleotide kinase. Hybridization was carried out at 59 °C in 6 × SSPE, 0.5% SDS, 3 × Denhardt’s solution and 100 µg ml–1 tRNA for 18 h. The sample was washed three times at room temperature in 6 × SSPE and 0.1% SDS for 15 min each and subsequently at 59 °C in the same buffer. Membranes were exposed to Ultra UV-G X-ray film (Dupharma, Kastrup, Denmark) for 3 days.
Genome sequence analyses
Genome sequences were downloaded from National Center for Biotechnology Information (NCBI), except that of Haloferax volcanii, which was obtained from The Institute for Genomic Research (http://www.tigr.org), and the Hyperthermus butylicus sequence (H.-P. Klenk, eGene Biotechnologie, Feldafing, Germany, unpublished data). All sequences were searched using the program LUNA obtainable from http://dac.molbio.ku.dk/bioinformatics/luna/. Short perfect repeats were identified with LUNA and the sequences were then extracted with a perl-script (available on request from this web-site). All clusters were further analyzed by BLAST searches against the Genbank databases. Matches to extrachromosomal elements were considered to be significant if they contained > 20 identical nucleotides. If the matches were 25–40 bp, a few mismatches were allowed. A cut-off at 20 bp was selected because a shorter sequence would be expected to occur randomly once in a 1.1 Mbp genome.
Results and discussion
The archaea for which genome sequences are available fall into the major kingdoms Crenarchaeota and Euryarchaeota, and cover a wide range of optimal growth conditions. The crenarchaea are all extreme- or hyper-thermophiles, whereas the euryarchaea, including haloarchaea, methanoarchaea and thermophiles, grow optimally over a wide range of temperatures. The full names of the organisms, their optimal growth temperatures and the Accession numbers of their genome sequences are listed in Table 1.
Table 1.
Summary of properties of the archaeal chromosomal and plasmid clusters.
| Organis m/plasmid | Strain no. | Optimal growth | No. of temp. (°C) | Total no. clusters | Accession of repeats |
| Crenarchaea | |||||
| Hyperthermus butylicus | 95–106 | 2 | 93 | – | |
| Pyrobaculum aerophilum | IM2 | 100 | 5 | 136 | AE009441 |
| Aeropyrum pernix | K1 | 95 | 4 | 89 | BA000002 |
| Sulfolobus solfataricus | P2B | 80 | 7 | 425 | AE006641 |
| Sulfolobus tokodaii | 7 | 80 | 6 | 462 | BA000023 |
| Sulfolobus acidocaldarius | DSM639 | 75 | 5 | 227 | CP000077 |
| plasmid pNOB8 | 80 | 1 | 6 | AJ010405 | |
| plasmid pKEF9 | 80 | 1 | 6 | AJ748321 | |
| Nanoarchaea | |||||
| Nanoarchaeum equitans | Kin4-M | 100 | 2 | 42 | AE017199 |
| Euryarchaea | |||||
| Pyrococcus abyssi | GE5 | 103 | 5 | 62 | AL096836 |
| Pyrococcus furiosus | DSM3638 | 100 | 8 | 208 | AE009950 |
| Pyrococcus horikoshii | OT3 | 98 | 7 | 153 | BA000001 |
| Thermococcus kodakaraensis | KOD1 | 85 | 3 | 77 | AP006878 |
| Picrophilus torridus | DSM9790 | 60 | 3 | 120 | AE017261 |
| Archaeoglobus fulgidus | DSM4304 | 83 | 3 | 152 | AE000782 |
| Thermoplasma acidophilum | DSM1728 | 59 | 2 | 48 | AL139299 |
| Thermoplasma volcanium | GSS1 | 60 | 3 | 36 | BA000011 |
| Methanopyrus kandleri | AV19 | 98 | 5 | 41 | AE009439 |
| Methanocaldococcus jannaschii | DSM2661 | 85 | 20 | 200 | L77117 |
| Methanothermobacter thermoautotrophicus | DH | 65 | 2 | 171 | AE000666 |
| Methanosarcina barkeri | fusaro | 37 | 6 | 101 | CP000099/CP000098 |
| Methanosarcina acetivorans | C2A | 37 | 8 | 79 | AE010299 |
| Methanosarcina mazei | Go1 | 37 | 8 | 136 | AE008384 |
| Methanospirillum hungatei | JF-1 | 37 | 6 | 266 | CP000254 |
| Methanosphaera stadtmanae | DSM3091 | 37 | 2 | 119 | CP000102 |
| Methanococcoides burtonii | DSM6242 | 23 | 2 | 87 | CP000300 |
| Haloarcula marismortui plasmids pNG300, pNG400 | ATCC 43049 | 45 | 3 | 129 | AY596297/AY596298 |
| Haloferax volcanii | 45 | 3 | 76 | – | |
| Natronomonas pharaonis | DSM2160 | 43–45 | 4 | 25 | CR936257 |
The numbers of clusters and repeat-spacer units that occur in each archaeal chromosome or plasmid are presented in Table 1. Most chromosomes contain two to eight clusters, the exception being M. jannaschii, which contains 20. Furthermore, the chromosomes carry a large number of repeat-spacer units extending from 25 for N. pharaonis to 462 for S. tokodaii. On average, the thermophilic organisms carry more repeat-spacer units, but this is not a strict rule (Table 1).
Clusters also occur in extrachromosomal elements (Tables 1 and and2).2). Both conjugative plasmids of Sulfolobus, pNOB8 and pKEF9, exhibit a single cluster with six repeat-spacer units (She et al. 1998, Greve et al. 2004). Moreover, two plasmids of H. marismortui, pNG300 and pNG400, and two plasmids of N. pharaonis contain single clusters (Baliga et al. 2004, Falb et al. 2005). For the N. pharaonis plasmid, one cluster is identical to a chromosomal cluster except that the latter contains an additional repeat-spacer unit (Table 2).
Table 2.
Properties of the repeat clusters. Organisms are arranged in the same order as in Table 1. Symbols: * = cluster containing an inserted transposable element; † = cluster located on a plasmid; and # = single repeats that lack a flanking sequence.
| Organism/plasmid (conserved bp) | Repeat sequence | Repeats per cluster (altered in sequence) | Flanking sequence |
| H. butylicus | CTTGCAATTCTCTTTTGAGTTGTTC | 47(1), 46 | 2 (394) |
| P. aerophilum | GTTTCAACTATCTTTTGATTTCTGG | 15(2), 18, 14(2) | 3 (257) |
| CTTTCAATCCTCTTTTTGAGATTC | 81 | ||
| GTTTCAATTCTTTTGTAGATTCTTC | 8 | ||
| A. pernix | CTTGCAATTCTATCTCGAAGATTC | 1, 27(8), 19(2) | 3 (476) |
| CTTTCTATTCCCTTTAGGGATATGC | 42 | ||
| S. solfataricus | CTTTCAATTCCTTTTGGGATTAATC | 1, 103, 95(47) | 3 (502) |
| CTTTCAATTCTATAAGAGATTATC | 32, 96(1), | 2 (266) | |
| CTTTCAATTCTATAGTAGATTAGC | 7, 91(2) | ||
| S. tokodaii | CTTTCAATTCCTTTTGGGATTCATC | 74(2), 112(48) | 2 (482) |
| CTTTCAATTCCATTAAGGATTATC | 48(1), 104(1), 121(4) | 3 (269) | |
| CTTTATTCATAATGCTAATTCCGT | 3 | ||
| S. acidocaldarius | GTTTTAGTTTCTTGTCGTTATTAC | 133(1), 78(5) | 2 (239) |
| CTTTCAATCCCTTTTGGGATTCATC | 4(1), 11(1), 1 | 3 (506) | |
| pNOB8 | CTTTCAATTCTATAGTAGATTATC | 6† | |
| pKEF9 | GTTGCAATTCCCTAAATGTGCGGG | 6† | |
| N. equitans | CTTTCAATATTTCTAATATATTAGAAAC | 13, 29(1) | 2 (190) |
| P. abyssi | CTTTCAATTCTATTTTAGTCTTATTGGAAC | 23(3), 4(2), 27 | |
| CTTTCCACACTACTAAGTTCTACGGAAAC | 7(2), 1 | 2 (401) | |
| P. furiosus | CTTTCAATTCTATTTTAGTCTTATTGGAAC | 52(3), 21(1), 23(2), 31, 46(2), 1#, 22, 12(5) | 7 (524) |
| P. horikoshii | CTTTCCACACTATTTAGTTCTACGGAAAC | 18(1), 25(3), 66(14), 1# | 3 (526) |
| CTTTCAATTCTATTTTAGTCTTATTGGAAC | 18(1), 7(2), 18(1) | 2 (258) | |
| T. kodakaraensis | CTTTCAATTCTCTTAGAGTCTTATTGCAAC | 16(7), 24(5), 33(7) | 3 (437) |
| P. torridus | CTTCCATACTATCTAGTAATTCTTAAAC | 15(1), 17(1) | 2 (322) |
| CTTTCAATCCTATTTAGGTTATTATTTAAC | 88(2) | ||
| A. fulgidus | CTTTCAATCCCATTTTGGTCTGATTTCAAC | 60, 48(1) | 2 (347) |
| CTTTCAATCTCCATTTTCAGGAGCCTCCCTTTCTTAC | 44(4) | ||
| T. acidophilum | CTTTCAATCCTATTAAGGTTCTATTTTAC | 47(1), 1# | |
| T. volcanium | CTTCCATACTAACTAGTACATCTTAAAC | 19(1), 16(1), 1 | 3 (287) |
| M. kandleri | GTTTCATTACCCGTATTATTACGGGTTAATTGCGAG | 12(2), 5, 8(1), 4(2), 12(3) | 5 (564) |
| M. jannaschii | TTTCCATTCCGAAACGGTCTGATTTTAAT/ | 26(1), 3(1), 4(2), 1, 16(2), 24(4), | 19 (440) |
| TTTCCATCCTCCAAGAGGTCTGATTTTAAC | 12(2),15, 7(2), 13(2), 2, 3(1), 14(4), 14(7), 5(1), 10(1), 1, 9(2), 9(3), 12(1) | ||
| M. thermoautotrophicus | ATTTCAATCCCATTTTGGTCTGATTTTAAC | 124, 47 | 2 (460) |
| M. barkeri | GTTTCAATCCCTCTAAGGCCTGATTTTAAC | 51(2) | |
| GTTTCAATCCTTGTTTTAGTGGATCTTGCTCACGAAT | 1#, 4(2), 1#, 19(1) | 1 (171) | |
| GTTTCCATAACCGAAAGGTTGTGGCAGAATTGAAGC | 25(5) | ||
| M. acetivorans | GTTTCAATCCTTGTTTTAGTGGATCTTGCTCGCGAAT | 1#, 7, 33(2)*, 2, 1#, 1#, 2(1) | 2 (171) |
| GTTTCAATCCCTCTAAGGTCTGATTTTAAC | 31(1) | ||
| M. mazei | GTTTCAATCCTTGTTTTAGTGGATCTTGCTCACGAAT | 1#, 2(1), 2(1), 1#, 1#, 47(7), 1#, 81(8) | 2 (171) |
| Methanospirillum hungatei | GTTGCAAGTGACCCGAAAATAGAAGGGTATGGCAAC | 31, 8, 37(2) | 2 (210) |
| GTTTCAATCCCTATCGGGTTTTCTTTTCCATTGTGAC | 44(1), 66 | ||
| GGTTCATCCCCATACACACGGGGAACTC | 80 | ||
| Methanosphaera stadtmanae | GTTTAAAATAGACTTAATAGTATGAAAAC | 62(2) | |
| CTTTCAATTTCATTATGATCTTATTCTATT | 57 | ||
| Methanococcoides burtonii | GAGTTCCCCATGCATGTGGGGATAAACCG | 65 | |
| GTTTCAATCCCTCTAAGGTCTGATTTTAAC | 22(2) | ||
| H. marismortui | GCTTCAACCCCACAAGGGTCCGTCTGAAAC | 48(13)† | |
| GCTTCAACCCCACGAGGGTCCGTCTGTAAC | 24(1)†, 57(1) *† | 2 (132) | |
| H. volcanii | CTTCAATCCCACAAGGGTTCGTCTGAAAC | 25(1), 11(1), 40 | 2 (169) |
| N. pharaonis | GCTTCAACCCCACAAGGGTTCGTCTGAAAC | 5(1), 3(2)† | |
| GTCGAGACGGACTGAAAACCCAGAACGGGATTGAAAC | 9(1), 8(1)† | ||
Properties of the sequence repeats
Repeat sequences vary in both length and sequence and are presented for each cluster in Table 2. The crenarchaeal genomes range in size from 24 to 26 bp, whereas the euryarchaea and the nanoarchaeon genomes vary from 26 to 37 bp. There is only limited conservation of many repeat sequences, with the left half showing the higher conservation, as was noted earlier (Peng et al. 2003). Nevertheless, some repeat sequences show major differences, especially in the right half of the sequence; compare, for example, the right halves of the repeats of plasmids pNOB8 and pKEF9 (Table 2). Most repeat sequences show some kind of weak, dyad symmetry generally in the form of interrupted and imperfect short inverted repeats and it has been shown, at least for the genus Sulfolobus, that this provides a recognition site for a repeat binding protein (Peng et al. 2003).
Within some clusters, the repeat sequence exhibits a little variation. For example, in the S. solfataricus cluster, ssol-95, the repeat sequence changes at the center of the cluster to another sequence (albeit with a single nucleotide change) (She et al. 2001). Therefore, we examined the constancy of repeat sequences within each genomic cluster and the results demonstrate that most clusters are not homogeneous in their repeat sequence (Table 2). Many carry one to four altered repeat sequences and two, the aforementioned ssol-95 cluster of S. solfataricus and stok-112 of S. tokodaii, show more dramatic changes. In addition, six of the 20 clusters in M. jannaschii carry two repeat sequences differing in their central regions (Table 2).
Uniqueness of the spacer sequences?
Spacers vary in size from 35 to 44 bp between clusters as well as between organisms, and tend to be conserved within a cluster (Table 2). However, occasional exceptions were detected. In the S. solfataricus cluster ssol-91, a half spacer precedes two atypical repeat sequences, the second of which is followed by a regular repeat sequence and not a spacer. In S. tokodaii cluster stok-121, two atypical repeats are 18 bp longer than the other repeats, but are followed by shorter spacers such that the repeat-spacer unit is conserved in size. In N. equitans, two spacers are 25/26 bp longer than the others. The 56 bp spacers of M. kandleri are more than 10 bp longer than those found in any other archaea.
All spacer sequences within a cluster and within a chromosome are generally different, but a systematic search revealed several exceptions. Spacers are occasionally repeated, sometimes more than once within a cluster, and can appear in different clusters within the same chromosome. The results demonstrate that 12 of the 28 chromosomes investigated carry one or more repeated spacers, which tend to be located in the larger clusters. The distributions of duplicated spacers are indicated in Table 3. There are no clear patterns for the arrangement of duplicated spacers, as some are arranged consecutively, others are located in different parts of a given cluster and a few occur in different clusters. The greatest number of duplicated spacers (36) occurs in the two clusters of M. thermautotrophicus (Table 3) and their distribution is illustrated for the larger mthe-124 cluster in Figure 1A. Although identical groups of spacer-repeat units have been observed in closely related strains, they have not been detected in different species.
Table 3.
Spacer sequences repeated either within a cluster or between different clusters of the same organism.
| Organism/ plasmid | Repeated once in a cluster | Repeated more than once in a cluster | Present in different clusters | Total no. of repeated spacers |
| H. butylicus | 2 | 4 | ||
| S. solfataricus | 1 | 3 | 8 | |
| S. tokodaii | 2 | 1 | 5 | 17 |
| S. acidocaldarius | 2 | 1 | 9 | |
| P. furiosus | 1 | 3 | 8 | |
| P. torridus | 10 | 20 | ||
| A. fulgidus | 1 | 2 | ||
| M. jannaschii | 2 | 2 | 14 | |
| M. thermautotrophicus | 18 | 36 | ||
| M. barkeri | 1 | 2 | ||
| M. mazei | 2 | 4 | ||
| H. marismortui pNG300 | 1 | 2 | ||
| H. marismortui pNG400 | 2 | 4 | ||
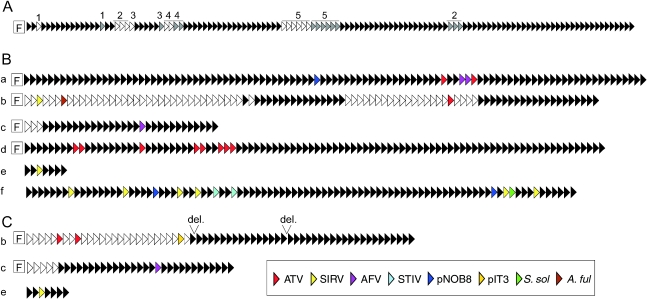
(A) A map of mthe-124 from M. thermoautotrophicus showing the locations of duplicated spacer-repeat units, or groups of units, labeled 1–5. Each triangle represents a spacer-repeat unit. Duplicated spacer-repeat units are shaded in gray. (B) Schematic representation of the six clusters which occur in the genome of S. solfataricus P2B (She et al. 2001). Each triangle represents a spacer-repeat unit. The colored triangles are coded to indicate the archaeal viruses, plasmids or chromosomes (S. solfataricus or A. fulgidus) that yield good matches with the spacer sequence. (C) A corresponding scheme is presented for three of the six clusters (ssol-95, ssol-32 and ssol-7) that are present in the chromosome of the closely related strain S. solfataricus P1 (Accession numbers: DQ831675, DQ831676 and DQ831677). Open triangles denote those spacer-repeat units that differ in sequence between the strains P1 and P2A. Abbreviation: del. indicates the two sites where deletion of several spacer-repeat units has occurred in strain P1 (or where insertions have occurred in strain P2B).
Integrity of the clusters
Repeat clusters are generally highly conserved in their repeat sequences and in the sizes of their repeats and spacers. Such integrity extends to an almost complete lack of insertion sequences (ISs) or other mobile elements. This is surprising, given the large number of mobile elements in some archaeal genomes and the large variety of spacer sequences that could provide potential target sites for the insertion of mobile elements (Brügger et al. 2002). Nevertheless, one IS element (ISH4) was identified within the repeat cluster, hmar-57, of the H. marismortui megaplasmid, pNG400 (Table 2). Moreover, a miniature inverted-repeat transposable element (MITE) occurs in the mace-33 cluster of M. acetivorans (Table 2), located at the center of a repeat sequence close to the end carrying the flanking sequence, but not in any of the other identical repeat sequences. The 132 bp MITE shares a 14 bp inverted terminal repeat and a 3 bp direct repeat with the IS element, ISMac11, which exists in the same chromosome and encodes a transposase likely responsible for the MITE transposition (Brügger et al. 2002).
Properties of the flanking sequence
Many chromosomal clusters carry a flanking sequence at one end, which is sometimes referred to as a “leader,” although its function is unknown (see below). These sequences tend to be rich in short homopolynucleotide sequences and AT-rich regions, and they lack open reading frames (Jansen et al. 2002, Tang et al. 2002). Archaeal flanking sequences range in size from 132 bp for H. marismortui to 564 bp for M. kandleri (Table 2), and they directly adjoin the first repeat sequence of the cluster. Moreover, they invariably occur at the same end of the cluster, with respect to the strand orientation of the repeat sequence. There is an approximate direct correlation between the sequence length and the optimal growth temperature of the organism (Tables 1 and and2). 2).
For chromosomes carrying multiple clusters with identical repeats, the flanking sequence (if present) is often conserved (Table 2). However, there is generally no conservation of the flanking sequence between organisms, with the exception of three Methanosarcina species, which share five highly similar 171 bp sequences with > 87% sequence identity (Figure 2A).

(A) Alignment of the five conserved flanking sequences adjoining the first repeat (in bold type) of five clusters of the three Methanosarcina strains. A putative TATA-like box is outlined. (B) Alignment of two flanking regions of the P. abyssi genome where paby-1 appears to exhibit a defective start site.
Several clusters lack a flanking sequence, including those with different repeat sequences found in P. aerophilum and A. pernix (Table 2) and some with identical repeats found in S. solfataricus P2B (see ssol-7 and ssol-91 in Figure 1B).
Protein genes associated with clusters
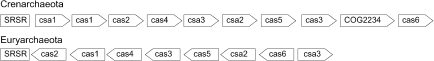
So far, the only protein that has been shown to interact directly with a cluster is the genus-specific SSO0454 from S. solfataricus P2, which binds and distorts the repeat sequence (Peng et al. 2003). However, superoperons containing > 20 genes often flank one or more repeat clusters within archaeal and bacterial chromosomes (Jansen et al. 2002). Because they are absent from bacterial genomes that lack repeat clusters, they were inferred to be co-functional with the repeat clusters (Jansen et al. 2002). Although some of these genes (now denoted cas or csa genes) were earlier annotated as unusual DNA repair enzymes (Makarova et al. 2002), they have recently been reassigned to the regulation and processing of the repeat clusters and to a putative role in piRNA function (Makarova et al. 2006). Predicted functions of the more common gene products are listed in Table 4. Some of the genes show a degree of specificity for different archaeal or bacterial phyla including five archaea-specific genes (denoted csa1 to csa5) (Bolotin et al. 2005, Haft et al. 2005), although we find csa2 homologs in some bacterial genomes. Cas5 and cas6 should not be confused with identically named genes exclusive to bacterial genomes lacking cas2, cas3 and cas4 (Bolotin et al. 2005). An overview of the genes commonly found in archaeal genomes is presented in Table 5.
Table 4.
Predicted functions of common cas-genes in archaea. Data are summarized from Makarova et al. 2006 and earlier papers (Jansen et al. 2002, Makarova et al. 2002, Haft et al. 2005). Symbols: * = formerly COG3578; and ** = formerly COG3574.
| Family/cas-name | Predicted function | Comments |
| COG1518/cas1 | Nuclease/integrase | Possibly involved in inserting new DNA sequences |
| COG1343/cas2 | Nuclease | Possibly involved in inserting new DNA sequences |
| COG1203/cas3 | DNA helicase | Often fused to HD-nuclease domain/ related to COG2254 |
| COG2254 | HD-like nuclease | |
| COG1468/cas4 | RecB-like nuclease | |
| COG1688/cas5 | RNA-binding | “Ramp” superfamily |
| COG1583/cas6 | RNA-binding | “Ramp” superfamily |
| COG4343*/csa1 | RecB-like nuclease | |
| COG1857/csa2 | Nuclease | |
| COG2462/csa3 | HTH-type transcriptional regulator | |
| COG1353 | RNA polymerase | “Loosely” associated with repeat clusters |
| AF0070/csa4 | None | Crenarchaea-specific |
| AF1870**/csa5 | None | Crenarchaea-specific |
Table 5.
Summary of the occurrence of common cluster-associated genes in archaea. COG2254 typically adjoins a cas3 gene (in A. fulgidus, M. jannaschii and several Crenarchaea) and they are sometimes fused. Symbols: * = cas1/cas4 are fused; and ** = A cas gene operon links two clusters with different repeat sequences.
| Organism | Repeat sequence | Flank | No. of | COG | COG | COG | COG | COG | COG | COG | COG | COG | COG | COG | ||
| clusters | 1518/ | 1343/ | 1203/ | 1468/ | 1688/ | 1583/ | 2254 | 4343/ | 1857/ | 2462/ | 1353/ | |||||
| cas1 | cas2 | cas3 | cas4 | cas5 | cas6 | csa1 | csa2 | csa3 | pol | |||||||
| H. butylicus | CTTGCAATTCTCTTTTGAGTTGTTC | + | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| P. aerophilum | GTTTCAACTATCTTTTGATTTCTGG | + | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| CTTTCAATCCTCTTTTTGAGATTC | – | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| GTTTCAATTCTTTTGTAGATTCTTC | – | 1 | ||||||||||||||
| A. pernix | CTTGCAATTCTATCTCGAAGATTC | + | 3 | |||||||||||||
| CTTTCTATTCCCTTTAGGGATATGC | – | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| S. solfataricus | CTTTCAATTCCTTTTGGGATTAATC | + | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| CTTTCAATTCTATAAGAGATTATC | + | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | |||
| CTTTCAATTCTATAGTAGATTAGC | – | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| S. tokodaii | CTTTCAATTCCTTTTGGGATTCATC | + | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| CTTTCAATTCCATTAAGGATTATC | + | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| CTTTATTCATAATGCTAATTCCGT | – | 1 | ||||||||||||||
| S. acidocaldarius | GTTTTAGTTTCTTGTCGTTATTAC | + | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||
| CTTTCAATCCCTTTTGGGATTCATC | + | 3 | 1 | 1 | 1 | 1 | ||||||||||
| N. equitans | CTTTCAATATTTCTAATATATTAGAAAC | + | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| P. abyssi | CTTTCAATTCTATTTTAGTCTTATTGGAAC | – | 3 | |||||||||||||
| P. furiosus | CTTTCCACACTACTAAGTTCTACGGAAAC | – | 2 | 1 | 1 | |||||||||||
| P. horikoshii | CTTTCCACACTATTTAGTTCTACGGAAAC | + | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| CTTTCAATTCTATTTTAGTCTTATTGGAAC | + | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||
| T. kodakaraensis | CTTTCAATTCTCTTAGAGTCTTATTGCAAC | + | 3 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | ||||
| P. torridus | CTTCCATACTATCTAGTAATTCTTAAAC | + | 2 | 1 | 1 | 1 | 1 | |||||||||
| CTTTCAATCCTATTTAGGTTATTATTTAAC | – | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| A. fulgidus | CTTTCAATCCCATTTTGGTCTGATTTCAAC | + | 2 | 1 | 1 | 1 | ||||||||||
| CTTTCAATCTCCATTTTCAGGAGCCTCCCTTTCTTAC | – | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| T. acidophilum | CTTTCAATCCTATTAAGGTTCTATTTTAC | – | 2 | |||||||||||||
| T. volcanium | CTTCCATACTAACTAGTACATCTTAAAC | + | 3 | 1 | 1 | 1 | 1 | |||||||||
| M. kandleri | GTTTCATTACCCGTATTATTACGGGTTAATTGCGAG | + | 5 | 1 | 1 | 2 | ||||||||||
| M. jannaschii | ATTTCCATTCCCCGAGGGATCTGATTTTAC | + | 20 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| M. thermoautotroficus | ATTTCAATCCCATTTTGGTCTGATTTTAAC | + | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| M. barkeri** | GTTTCAATCCCTCTAAGGCCTGATTTTAAC | – | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| GTTTCAATCCTTGTTTTAGTGGATCTTGCTCACGAAT | + | 3 | ||||||||||||||
| GTTTCCATAACCGAAAGGTTGTGGCAGAATTGAAGC | – | 1 | 1* | 1 | 1 | 1* | ||||||||||
| M. acetivorans** | GTTTCAATCCTTGTTTTAGTGGATCTTGCTCGCGAAT | + | 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| GTTTCAATCCCTCTAAGGTCTGATTTTAAC | – | 1 | ||||||||||||||
| M. mazei | GTTTCAATCCTTGTTTTAGTGGATCTTGCTCACGAAT | + | 8 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||
| M. hungatei | GTTGCAAGTGACCCGAAAATAGAAGGGTATGGCAAC | + | 3 | 1 | 1 | 1 | 1 | |||||||||
| GTTTCAATCCCTATCGGGTTTTCTTTTCCATTGTGAC | – | 2 | 1 | 1 | 1 | 1 | 1 | |||||||||
| GGTTCATCCCCATACACACGGGGAACTC | – | 1 | 1 | 1 | 1 | |||||||||||
| M. stadtmanae | GTTTAAAATAGACTTAATAGTATGAAAAC | – | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||
| CTTTCAATTTCATTATGATCTTATTCTATT | – | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| M. burtonii | GAGTTCCCCATGCATGTGGGGATAAACCG | – | 1 | 1 | 1 | 1 | ||||||||||
| GTTTCAATCCCTCTAAGGTCTGATTTTAAC | – | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| H. marismortui, | GCTTCAACCCCACGAGGGTCCGTCTGTAAC | + | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| pNG400 | ||||||||||||||||
| N. pharaonis | GCTTCAACCCCACAAGGGTTCGTCTGAAAC | – | 2 | |||||||||||||
| GTCGAGACGGACTGAAAACCCAGAACGGGATTGAAAC | – | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
Typical examples of fairly conserved gene orders present in superoperons of crenarchaea and euryarchaea are presented in Figure 3 for the most commonly occurring genes. All euryarchaeal genomes lack Csa1, except for A. fulgidus. Each gene generally occurs once per superoperon, although duplicate copiesoccur in M. jannaschii and T. kodakaraensis. A few genes, including cas3, cas5, cas6, csa2, csa3 and COG2254, are sometimes located distantly from the repeat clusters, most commonly inthe genomes of the three Pyrococcus species and A. fulgidus.
Organisms with multiple clusters carrying the same repeat sequence exhibit only a single superoperon adjacent to one cluster (Jansen et al. 2002). In some genomes, the superoperons physically link two, or even three, clusters with identical repeat sequences (e.g., in P. aerophilum, S. solfataricus, S. tokodaii and H. marismortui). In contrast, organisms containing two or more clusters with different repeat sequences generally have two or more superoperons (Jansen et al. 2002), except for P. aerophilum, A. pernix and S. tokodaii, which each have one. Exceptionally, superoperons are absent from the chromosomes of T. acidophilum and P. abyssi and plasmids pNOB8 and pKEF9, which all carry repeat clusters (Table 5).
In M. acetivorans and M. barkeri, a superoperon links clusters with dissimilar repeats and both the gene order (cas6-cas4-cas1-cas2-Cluster1-csa2-cas5-cas3-Cluster2) and sequence (> 94% identity) are conserved. Since the repeat sequences of the corresponding pairs of clusters are identical between these two organisms, this suggests that a lateral transfer event of both superoperon and clusters has occurred. However, such an event must have happened at an early stage of cluster development, because no similarities were detected between the spacer sequences of the two organisms.
Development of the clusters
The only relevant study of cluster development was performed on related bacterial strains of M. tuberculosis (van Embden et al. 2000) and of Yersinia (Pourcel et al. 2005). Examination of 26 strains of M. tuberculosis revealed several differences in a large repeat cluster consistent with insertions/deletions of internal repeat-spacer units having occurred. For the related Yersinia strains, a similar phenomenon was observed, but it was also inferred that repeat-spacer units could have been added at the cluster end adjoining the conserved flanking sequence.
In order to shed some insight on how clusters may develop and change in archaea, we completely sequenced three of the six large cluster regions of S. solfataricus P1 and compared the spacer content with those of strain P2B. The two highly similar strains were originally sampled about one meter apart from a small stream at Pisciarelli, Naples (W. Zillig, deceased, personal communication). A comparative repeat-spacer alignment of the two strains (Figures 1B and and1C)1C) demonstrates the following: (1) new repeat-spacer units are added at, or near, the end of clusters b and c, which both exhibit an adjoining flanking sequence; (2) the large b clusters are more active in adding new repeat-spacer units than the smaller c clusters; (3) no new repeat-spacer units are added to the e clusters, which lack a flanking sequence, but the clusters remain highly conserved in sequence; (4) deletion and/or insertion of single, or multiple, repeat-spacer units can occur; and (5) deletion/insertion of repeat-spacer units occurs precisely, reinforcing the idea that the structural integrity of the clusters is important for their function.
Both data found in the literature (Pourcel et al. 2005) and the results presented in Figures 1B and and1C1C are consistent with new spacer-repeat units being added exclusively adjacent to the flanking sequence. Moreover, since no new units were added to the repeat clusters lacking a flanking sequence (cluster ssol-32 in Figures 1B and and1C),1C), this region is likely to provide a binding site for the cas genes involved in the copying-insertion events. Although there is no insight into the mechanism by which repeat-spacer units are added to a cluster, the process is likely to involve reverse transcription of mRNAs and recombination (Makarova et al. 2006).
There are several instances of a flanking sequence being followed by a single repeat or repeat-spacer unit (Table 2), consistent with the clusters developing from one end, but possibly also indicating a defective start site. One such region from P. abyssi is shown where the flanking sequence adjoins a one half repeat and truncated spacer, followed by a full repeat, and it is aligned with the start of paby-23 from the same genome (Figure 2B). There are also a few examples of single repeats that lack flanking sequences, mainly from the genera Pyrococcus and Methanosarcina (Table 2).
The preceding data suggest that development of clusters is primarily dependent on a combination of cas genes and the flanking sequence. An alternative hypothesis is that repeat clusters are spread intercellularly by plasmids (Godde and Bickerton 2006), and, at least for the Methanosarcina species, there is clear sequence evidence for the lateral transfer of a superoperon of cas genes and all its flanking sequences (Figure 2). Moreover, the presence of repeat clusters in conjugative plasmids provides a possible mechanism for intercellular transfer. However, the repeat cluster of pNOB8 does not appear in the S. tokodaii chromosome, where a closely similar plasmid sequence is encaptured (Kawarabayasi et al. 2001).
The role of RNA
Experimental studies have demonstrated that RNA is transcribed from the repeat-clusters of the euryarchaeon, A. fulgidus, and the crenarchaeon, S. solfataricus (Tang et al. 2002, 2005). Examination of clone libraries of reverse transcripts prepared from total cellular RNA (< 500 nt) yielded sequences of a series of cluster-encoded small RNAs (22 from A. fulgidus and one from Sulfolobus). Since the 5′-terminal sequence of each RNA was lacking (by about 10–25 nt), the start position could only be assigned approximately within a repeat sequence, while the 3′-terminus was located at or near the center of the following repeat, yielding an estimated average size of 50–70 nt (Tang et al. 2002, 2005).
Northern blotting, using a probe against the repeat sequence of the clusters, revealed a series of discrete RNA products that were multiples of ~68 nt (68, 136, 204, 272, 340 and 408 nt) for A. fulgidus (Tang et al. 2002) and of ~60 nt (60, 180, 360 and 540 nt) for S. solfataricus (Tang et al. 2005). In both studies, the smallest RNA detected corresponded approximately to the estimated sizes of most of the cloned RNAs. The RNA size distribution is consistent with the processing of larger transcripts at regular spatial intervals and the sequencing data support, but do not establish, that processing occurs at or near the center of each repeat.
To gain further insight into the RNA products, transcription was examined from a small cluster of S. acidocaldarius (saci-4) (Chen et al. 2005) using a Northern blotting approach. Probes were prepared against complementary sequences of the terminal spacer sequence adjacent to the flanking sequence. Total RNA was extracted from exponentially growing and stationary phase cell cultures. The results revealed a series of bands that correspond to transcription from the direction of the flanking sequence (Figure 4). The pattern of larger discrete bands is quite similar to that obtained earlier for S. solfataricus P1 (Tang et al. 2005). What is different, however, is that diffuse bands range in size from 52 to 35 nt for the exponentially growing sample, and from 52 to 30 nt for the stationary phase sample (Figure 4A). This suggests that progressive trimming of the smallest discrete band of 58 nt has occurred, possibly by exonucleases. Clearly, the yields of these smaller products increase, and their average size decreases, on going from the exponential to stationary growth phase. The largest band (~420 nt) exceeds the total size of the cluster (maximum 260 bp), which is consistent with the transcript extending beyond the cluster limits. Evidence is also provided for larger transcripts being produced by the complementary DNA strand in stationary phase cells (Figure 4B).
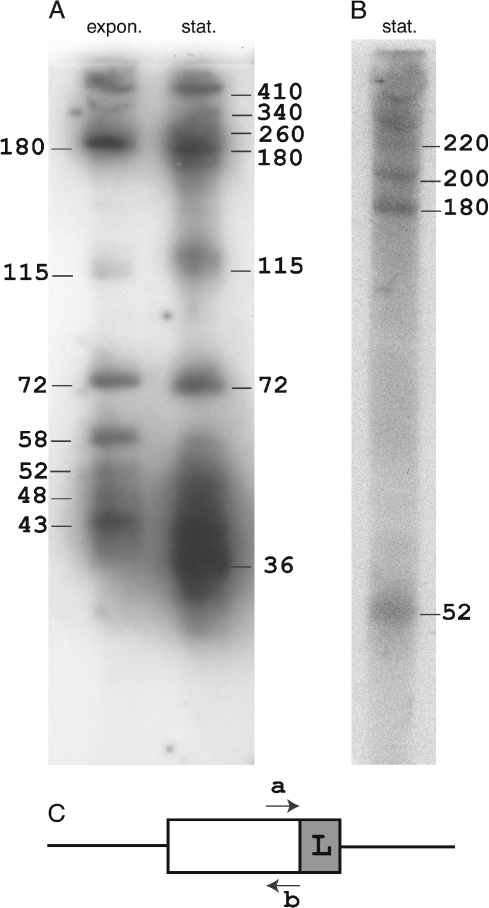
Northern blotting analyses of RNA transcripts obtained from the cluster saci-4 of S. acidocaldarius. (A) Transcripts from one strand were detected in total RNA extracted from cells grown to exponential (expon.) or stationary (stat.) phase. (B) Transcripts were observed from the complementary DNA strand at stationary phase. The approximate estimated nucleotide lengths of the RNA products are given. The minimal detection limit, using 26-nt. probes, was estimated at 15–16 nucleotides. (C) Denotes the location of the primers “a” and “b,” and “L” indicates the location of the putative transcriptional leader within the flanking sequence.
The observed sizes of the larger transcripts obtained from different clusters are consistent with transcription being initiated in a leader sequence within the flanking sequence adjoining the first repeat (Figure 2). The majority of the flanking sequences (20 out of 26) carry a putative TATA-like motif immediately upstream from the first repeat which, if active, would produce an initation start at the center of this repeat. However, this property is also shared by repeat clusters lacking a flanking sequence and one of these, from the plasmid pKEF, produces transcripts (R.K. Lillestøl, unpublished data), suggesting that the flanking sequence is not primarily involved in transcriptional initiation.
Derivation of the spacer sequences from extrachromosomal elements
Recently, evidence has been presented for both archaea and bacteria that some spacer sequences correspond closely to sequences occurring in extrachromosomal elements some of which lie within ORFs. For the archaea, sequence similarities were found with Sulfolobus fuselloviruses and rudiviruses as well as with the conjugative plasmid pNOB8 (Mojica et al. 2005), whereas, for the bacteria, they were found for bacteriophages of Streptococcus and a prophage of Yersinia (Bolotin et al. 2005, Pourcel et al. 2005).
We tested all the archaeal spacer sequences for matches against the GenBank database and against our in-house archaeal genome database, which contains all the available archaeal viral, plasmid and chromosomal sequences (Brügger et al. 2003). Several sequence matches were found, showing 88–100% identity with viruses, plasmids and chromosomes. They are listed in Table 6 and include those reported earlier (Mojica et al. 2005). Most matches were to ORFs; the few exceptions (Table 6) mainly correspond to predicted noncoding regions of chromosomes.
Table 6.
Spacer sequence matches to viruses, plasmids and chromosomes. Lengths of the sequence matches are given in columns 3–5 where sequence identity ranged from 88 to 100%. For transposase genes only the best match is given. For M. thermoautotrophicus, there were three matches to phage ΨM2 ORF6 and two to prophage ΨM100 ORF31.
| Organism/plasmid | Spacer matches | Sense | Antisense | Noncoding regions |
| P. aerophilum | P. aerophilum | 39 | ||
| A. pernix | A. pernix | 32 | ||
| S. solfataricus | ATV | 38, 35, 38, 40, 35, 34, 28 | 39, 39, 37, 39 | |
| SIRV | 31, 23, 41 | 40, 31, 29 | ||
| AFV6 | 29 | |||
| AFV7 | 25 | |||
| AFV8 | 38 | |||
| STIV | 27 | 23 | ||
| pNOB8 | 25, 38 | 40 | ||
| A. fulgidus | 32 | |||
| S. solfataricus | 40 | 40, 35, 38, 32 | ||
| S. tokodaii | 40, 40 | |||
| S. tokodaii | SIRV | 41 | ||
| SSV4 | 39 | |||
| pNOB8 | 38 | |||
| pKEF9 | 42 | |||
| S. tokodaii | 37 | |||
| S. acidocaldarius | pKEF9 | 24 | ||
| S. acidocaldarius | 35 | |||
| pKEF9 | SSV5 | 27 | ||
| SIRV | 41 | |||
| M. thermo-autotrophicus | M. wolfeii | |||
| prophage yM100 | 38 | 35, 36 | 36 | |
| M. marburgensis | ||||
| phage ΨM2 | 37, 35, 37 | 36, 36 | 38 | |
| M. acetivorans | M. acetivorans | 38 | ||
| M. mazei | M. barkeri | |||
| M. acetivorans | 36 | |||
| N. pharaonis | N. pharaonis | 35, 35 | ||
The majority of the positive archaeal matches are between spacer sequences of S. solfataricus and extrachromosomal elements of the related genera Sulfolobus and Acidianus. This strong bias probably reflects: (1) that Sulfolobus species are especially rich in repeat-spacer units (Table 1); and (2) that most sequenced archaeal viruses and plasmids derive from the related genera, Sulfolobus and Acidianus. Of the other positive matches, four were crenarchaeal (all to chromosomal sequences), eight were methanoarchaeal (mainly to phages or prophages), one was haloarchaeal and none were found to the haloarchaeal viruses, including SH1, His1 and His2 (Bamford et al. 2005, Bath et al. 2006) (Table 6).
The positions of the matching spacers for S. solfataricus are indicated in Figures 1B and and1C1C and the identities and predicted functions of the matching ORFs are presented in Table 7. Most positive matches are to viruses, in particular to ORFs of the Acidianus bicaudavirus ATV (62,730 bp) (Häring et al. 2005, Prangishvili et al. 2006). Matches also occurred to the similar rudiviruses SIRV1 and SIRV2 (Peng et al. 2001), the betalipothrixviruses AFV6, AFV7 and AFV8 (G. Vestergaard, University of Copenhagen, Denmark, unpublished data), the fusellovirus SSV4 (X. Peng, University of Copenhagen, Denmark, unpublished data) and the icosahedral virus STIV (Rice et al. 2004). Intriguingly, the cluster within the plasmid pKEF9 yielded matches with the rudivirus SIRV (Peng et al. 2001) and fusellovirus SSV5 (B. Greve, University of Copenhagen, Denmark, unpublished data; Table 7).
Table 7.
Spacer sequence matches between S. solfataricus strains P2 and P1 and viruses, plasmids and chromosomes. Matches of ATV and pIT3 to strain P1 are additional to those found for strain P2B, as illustrated in Figure 1. Symbol: * = an exclusive match to SIRV2. Literature references to the viral and plasmid sequences are given in the text for strain P2, and see Prato et al. 2006 regarding the pIT3 sequence.
| Virus/plasmid/ chromosome matches | Spacer match (mismatch) | Sense | Function |
| S. solfataricus P2 | |||
| ATV | |||
| ORF61 | 35 (1) | S | |
| ORF127 | 38 (0) | S | |
| ORF145 | 39 (0) | A | Virionprotein |
| ORF192 | 37 (0) | A | ORF198, pING1 |
| ORF326b | 39 (1) | A | ParBc |
| ORF529 | 34 (0) | S | AAA-ATPase |
| ORF545 | 35 (2) | S | Membrane protein |
| ORF618 | 40 (0) | S | AAA-ATPase/virionprotein |
| ORF710 | 39 (0) | A | |
| ORF892 | 28 (1) | S | VWA-domainprotein |
| ORF892 | 38 (0) | S | VWA-domainprotein |
| SIRV1/2 | |||
| ORF98 | 23 (0) | S | |
| ORF121* | 31 (0) | A | Hollidayjunctionresolvase |
| ORF134 | 31 (0) | S | Virionprotein |
| ORF268 | 41 (5) | S | |
| ORF356 | 29 (1) | A | Glycosyltransferase |
| ORF510 | 40 (2) | A | |
| STIV | |||
| A109 | 27 (0) | S | |
| C557 | 23 (0) | A | |
| AFVs | |||
| ORF267 AFV6 | 29 (2) | S | |
| ORF96 AFV7 | 25 (1) | S | |
| ORF593 AFV8 | 36 (0) | A | AFV-typehelicase |
| pNOB8 | |||
| ORF315 | 40 (3) | A | ParA |
| ORF406 | 25 (1) | S | IS element |
| ORF1025 | 38 (2) | S | TrbE family |
| Chromosome | |||
| SSO1736 | 40 (5) | A | IS element |
| (S. solfataricus) | |||
| AF1948 | 32 (3) | A | |
| (A fulgidus) | |||
| S. solfataricus P1 | |||
| ATV | |||
| ORF653 | 36 (2) | S | Virionprotein |
| pIT3 | |||
| ORF80 | 35 (0) | S | CopG |
A mechanism of defense
Earlier evidence was provided for the presence of several RNAs in S. solfataricus cells that were antisense to transposase mRNAs, and it was inferred that these regulate the transpositional events of the numerous IS elements and MITEs present in the S. solfataricus chromosome, either by facilitating degradation of the transposase mRNAs, or by inhibiting their translation (Tang et al. 2005). However, sense fragments corresponding to transposase mRNA fragments were also present, which remain unexplained (Tang et al. 2005). Similarly, the spacer transcripts that match extrachromosomal ORF sequences also occur in both sense and antisense orientations, given that the clusters seem to be transcribed primarily in one direction (Tang et al. 2002, 2005; Figure 4). Thus, of the 29 ORF matches listed for S. solfataricus, 17 correspond to a mRNA and 12 are antisense (Table 7). This has led to some speculation as to whether the putative inhibitory mechanism acts at an RNA level (piRNA), as in the eukaryal interference RNA systems (siRNA and miRNA), or whether it occurs at the gene level with either sense or antisense transcripts annealing directly to a gene, thereby facilitating degradation of the viral genome (Makarova et al. 2006). These ideas are consistent with the presence of double-strand-specific ribonucleases in both crenarchaea and euryarchaea (Stolt and Zillig 1993, Ohtani et al. 2004) and the discovery of argonaute family proteins in the euryarchaea P. furiosus and A. fulgidus, which are an essential part of the RNA-induced silencing complex (RISC) in Eukarya (Parker et al. 2004, Song et al. 2004). The proposals are also reinforced by the prediction that some cas genes may have RNA-related polymerase or processing functions (Makarova et al. 2006).
Conclusions
Strong circumstantial evidence has been accrued over the past year for repeat clusters being involved in an antiviral cellular defense mechanism for almost all archaea and about 50% of the bacteria investigated. The putative defence apparatus is shared by chromosomes and plasmids and is directed primarily against viruses. The genetic apparatus is complex and dynamic, undergoing rapid evolutionary change. It is likely to involve a large number of Cas proteins, including an essential core group, and some with more peripheral functions, which appear to be involved in the development of clusters and in the production and processing of the transcripts produced therefrom. The clusters appear to be extended by DNA spacers derived, directly or indirectly, from the genes of invading viruses. Transcripts from the spacer sequences are thought to inhibit, or possibly regulate, viral propagation by hybridizing at an mRNA or gene level.
Judging by its genetic complexity, this system must be important for survival of the archaeal cell in natural environments. This supposition is reinforced by the observation that large repeat clusters are present in all sequenced archaeal genomes, except that of Halobacterium sp. NRC-1 (Ng et al. 2000). Some insight into the latter exception may have been provided by a genomic mutation recently observed in S. solfataricus P2 (strain P2A in Redder and Garrett 2006). Strain P2 has been a laboratory strain for many years and, recently, a culture grown from a single colony was shown to exhibit a 124 kbp deletion constituting 4% of the chromosome. The deletion included each of the four repeat clusters which carry a flanking sequence (ssol-103, ssol-95, ssol-32 and ssol-96 in Figure 1B) and all the cluster-related genes. Halobacterium NRC1 is, similarly, a common laboratory strain and it is likely that when these organisms are grown free from invading extra-chromosomal elements over a longer time period, there is a tendency to lose this complex and energy consuming genetic apparatus. This could also explain why many bacteria, particularly endosymbionts (Jansen et al. 2002), lack such a system.
Acknowledgments
We thank Jan Christiansen, Elfar Torarinsson, Qunxin She, Xu Peng and Gisle Vestergaard for helpful discussions and Hien Phan for help with DNA sequencing. The research was supported by grants from the Danish Research Council for Natural Science. Grants from Copenhagen University supported K. Brügger and P. Redder, with K. Brügger also receiving grants from the Danish Science Research Council.
References
Articles from Archaea are provided here courtesy of Wiley
Full text links
Read article at publisher's site: https://doi.org/10.1155/2006/542818
Read article for free, from open access legal sources, via Unpaywall:
https://downloads.hindawi.com/journals/archaea/2006/542818.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1155/2006/542818
Article citations
Unity among the diverse RNA-guided CRISPR-Cas interference mechanisms.
J Biol Chem, 300(6):107295, 18 Apr 2024
Cited by: 1 article | PMID: 38641067 | PMCID: PMC11127173
Review Free full text in Europe PMC
Rational design of unrestricted pRN1 derivatives and their application in the construction of a dual plasmid vector system for Saccharolobus islandicus.
mLife, 3(1):119-128, 20 Mar 2024
Cited by: 0 articles | PMID: 38827506 | PMCID: PMC11139203
Genetic and technological diversity of Streptococcus thermophilus isolated from the Saint-Nectaire PDO cheese-producing area.
Front Microbiol, 14:1245510, 14 Nov 2023
Cited by: 2 articles | PMID: 38487210
Reconstruction of Archaeal Genomes from Short-Read Metagenomes.
Methods Mol Biol, 2522:487-527, 01 Jan 2022
Cited by: 0 articles | PMID: 36125772
Tools for Efficient Genome Editing; ZFN, TALEN, and CRISPR.
Methods Mol Biol, 2495:29-46, 01 Jan 2022
Cited by: 15 articles | PMID: 35696026
Review
Go to all (159) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 32 of 32)
- (1 citation) ENA - AE006641
- (1 citation) ENA - AE009439
- (1 citation) ENA - CP000098
- (1 citation) ENA - CP000077
- (1 citation) ENA - CP000099
- (1 citation) ENA - AL139299
- (1 citation) ENA - CP000254
- (1 citation) ENA - AJ748321
- (1 citation) ENA - DQ831677
- (1 citation) ENA - DQ831676
- (1 citation) ENA - AY596297
- (1 citation) ENA - AY596298
- (1 citation) ENA - AE017261
- (1 citation) ENA - DQ831675
- (1 citation) ENA - AP006878
- (1 citation) ENA - AJ010405
- (1 citation) ENA - AE009441
- (1 citation) ENA - BA000023
- (1 citation) ENA - BA000001
- (1 citation) ENA - BA000002
- (1 citation) ENA - AE010299
- (1 citation) ENA - AE017199
- (1 citation) ENA - AL096836
- (1 citation) ENA - AE000666
- (1 citation) ENA - CP000102
- (1 citation) ENA - CP000300
- (1 citation) ENA - BA000011
- (1 citation) ENA - L77117
- (1 citation) ENA - AE008384
- (1 citation) ENA - AE000782
- (1 citation) ENA - AE009950
- (1 citation) ENA - CR936257
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes.
J Bacteriol, 185(8):2410-2417, 01 Apr 2003
Cited by: 41 articles | PMID: 12670964 | PMCID: PMC152625
CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties.
Mol Microbiol, 72(1):259-272, 23 Feb 2009
Cited by: 140 articles | PMID: 19239620
Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism.
Biochem Soc Trans, 37(pt 1):23-28, 01 Feb 2009
Cited by: 61 articles | PMID: 19143596
CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea.
Nat Rev Microbiol, 6(3):181-186, 01 Mar 2008
Cited by: 495 articles | PMID: 18157154
Review
Funding
Funders who supported this work.