Abstract
Free full text

microRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes
Abstract
microRNA-34a (miR-34a) is a transcriptional target of p53 that is down-regulated in some cancer cell lines. We studied the expression, targets and functional effects of miR-34a in brain tumor cells and human gliomas. Transfection of miR-34a down-regulated c-Met in human glioma and medulloblastoma cells and Notch-1, Notch-2 and CDK6 protein expressions in glioma cells. miR-34a expression inhibited c-Met reporter activities in glioma and medulloblastoma cells and Notch-1 and Notch-2 3′UTR reporter activities in glioma cells and stem cells. Analysis of human specimens showed that miR-34a expression is down-regulated in glioblastoma tissues as compared to normal brain and in mutant p53 gliomas as compared to wild-type p53 gliomas. miR-34a levels in human gliomas inversely correlated to c-Met levels measured in the same tumors. Transient transfection of miR-34a into glioma and medulloblastoma cell lines strongly inhibited cell proliferation, cell cycle progression, cell survival and cell invasion, but transfection of miR-34a into human astrocytes did not affect cell survival and cell cycle status. Forced expression of c-Met or Notch1/Notch2 transcripts lacking the 3′UTR sequences partially reversed the effects of miR-34a on cell cycle arrest and cell death in glioma cells and stem cells, respectively. Also, transient expression of miR-34a in glioblastoma cells strongly inhibited in vivo glioma xenograft growth. Together, these findings represent the first comprehensive analysis of the role of miR-34a in gliomas. They show that miR-34a suppresses brain tumor growth by targeting c-Met and Notch. The results also suggest that miR-34a could serve as a potential therapeutic agent for brain tumors.
Introduction
Each year more than 20,000 people in the United States are diagnosed with a primary malignant brain tumor. Gliomas are the most common and deadly brain tumors in adults and medulloblastoma is the most common brain tumor in children. Multiple molecular dysfunctions have been associated with glioma and medulloblastoma formation and growth. Among these, the HGF/c-Met pathway, the Notch pathway and CDK6 are recognized to play important roles. We and others have shown that overexpression of c-Met or HGF is frequently found in brain tumors including glioma and medulloblastoma, and elevated HGF and/or c-Met levels as well as co-expression of HGF and c-Met correlate with poor prognosis (1-7). The Notch pathway has also been implicated in brain tumor formation and growth. Notch-1 and Notch-2 play critical roles in glioma cell and stem cell survival and proliferation (8-12). The cell cycle regulator protein CDK6 is another adverse prognostic indicator and contributor to cell proliferation, differentiation and transformation of human brain tumors. The expression of CDK6 in brain tumors is often elevated relative to matched normal brain tissue. Overexpression of CDK6 in glioma and medulloblastoma significantly correlates with poor prognosis (13-15). The mechanisms of c-Met, Notch and CDK6 expression deregulation in brain tumors are not very well understood.
microRNAs are small noncoding regulatory RNA molecules, with profound impact on a wide array of biological processes (16, 17). microRNAs modulate protein expression by binding to the 3′ untranslated region (3′UTR) of mRNA and promoting RNA degradation and inhibiting transcription. microRNAs are thought to play important roles in cancer by regulating the expression of various oncogenes and tumor suppressors (18-20). Expression profiling identified microRNA-34a (miR-34a) as one of several microRNAs that are downregulated in some cancer cells (21). miR-34a expression was recently shown to be transcriptionally regulated by p53, but a direct correlation between miR-34a levels and the p53 status in human tumors has not been demonstrated to date (22-25). A few studies have shown that miR-34a expression in cancer cells inhibits c-Met expression (25-27). However, a correlation between miR-34a levels and c-Met levels in human tumors has not been established.
We studied the role of miR-34a in human brain tumors with a focus on gliomas. We found that miR-34a potently inhibits c-Met protein expression and c-Met 3′UTR-reporter activity in glioma and medulloblastoma cells. Furthermore, we found for the first time that miR-34a also inhibits Notch-1 and Notch-2 protein expression and 3′UTR reporter activities as well as CDK6 protein expression in glioma cells. Using quantitative reverse transcription-PCR analysis, we showed for the first time that average pre-miR-34a expression is down-regulated in human glioblastoma tissues as compared to normal human brain. Moreover, miR-34a expression was higher in wild-type p53 glioblastoma tissues as compared to mutant p53 glioblastoma tissues, and miR-34a levels in human gliomas inversely correlated with c-Met levels in the same tumors. Transient transfection of miR-34a into brain tumor cell lines inhibited cell proliferation, cell cycle progression, cell survival and cell invasion but did not affect human astrocyte cell survival and cell cycle. Moreover, transient expression of miR-34a strongly inhibited in vivo glioma xenograft growth. Forced c-Met expression partially rescued the effects of miR-34a on the cell cycle and forced Notch1 and Notch2 expressions partially rescued the effects of miR34a on cell death in glioma cells and stem cells. We have therefore shown that miR-34a is downregulated in human gliomas and that it suppresses tumor growth by affecting several malignancy endpoints via downregulation of multiple oncogenes.
Materials and Methods
Reagents
Eagle's minimum essential medium (MEM), Dulbecco's modified essential medium (DMEM) with 4.5 g/L glucose, DMEM with 1 g/L glucose, 0.15% sodium bicarbonate, 1 mmol/L sodium pyruvate, 0.1 mol/L nonessential amino acids and HEPES buffer were purchased from Cellgro Mediatech (Washington, DC). Improved MEM Zinc Option, neurobasal media, N2, B27, penicillin-streptomycin, Trizol, Oligofectamine and pcDNA3.1/Zeo vector were purchased from Invitrogen (Carlsbad, CA). Human recombinant bFGF and EGF were purchased from R&D Systems (Minneapolis, MN). Fetal bovine serum (FBS) was purchased from GEMINI Bio-Products (West Sacramento, CA). Pre-miR-34a or control pre-miR (miR-con), and pmiR-REPORT vector were purchased from Ambion (Austin, Texas). pGL3-promoter plasmids and Luciferase System Kit were purchased from Promega Corp (Madison, WI). Crystal violet was purchased from Sigma (Saint Louis, Missouri). Fugene 6 reagent was purchased from Roche (Indianapolis, IN). Bio-Rad's iScript cDNA Synthesis Kit was purchased from Bio-Rad Laboratories (Hercules, CA). Taqman MicroRNA Reverse Transcription Kit, MultiScribe reverse transcriptase and a human 18S rRNA Taqman probe were purchased from Applied Biosystems (Foster City, CA). Propidium iodide, Annexin V-PE and 7AAD were purchased from BD Pharmingen (San Diego, CA).
Cell culture
U87 human glioblastoma cells were grown in Eagle's MEM supplemented with 10% FBS, 0.15% sodium bicarbonate, 1 mmol/L sodium pyruvate, 0.1 mol/L nonessential amino acids, and 500 μg/mL penicillin-streptomycin. A172 human glioblastoma cells and LN-Z308 cells (a kind gift from Dr. Erwin Van Meir, Emory University) were grown in DMEM with 4.5 g/L glucose and 10% FBS. U373 and T98G human glioblastoma cells were grown in DMEM with 1 g/L glucose supplemented with HEPES buffer and 10% fetal calf serum. DAOY human medulloblastoma cells were grown in Improved MEM Zinc Option supplemented with 10% FBS. Glioma stem cells 0308 (a kind gift from Dr. Howard Fine, NIH) were grown in Neurobasal Media, N2 and B27 supplements (0.5 × each) and human recombinant bFGF and EGF (50 ng/ml each). Immortalized human astrocytes (a kind gift of Dr. Russ Pieper, UCSF) were grown in DMEM with 4.5 g/L glucose supplemented with 10% FBS. All cells were grown at 37 °C in 5% CO2-95% O2.
Vectors
The pcDNA-c-Met, pcDNA-Notch1 and pcDNA-Notch2 plasmids were constructed via respective insertions of the full-length human c-Met, Notch1 and Notch2 cDNAs that do not contain the 3′UTR regions into the pcDNA3.1/Zeo vector. The Notch-1 3′UTR reporter plasmid was constructed via insertion of Notch-1 3′UTR into the pGL3-promoter plasmid. The Notch-2 3′UTR reporter plasmid was constructed via insertion of Notch-2 3′UTR into the pmiR-REPORT vector. The c-Met 3′-UTR reporter plasmid was a kind gift of Dr. Lin He (Cold Spring Harbor Laboratory) (25). The miR-34a reporter plasmid was constructed via insertion of two copies of full site complementary to miR-34a into the pGL3-promoter plasmid. The p53 and mutant-p53 expression plasmids were kind gifts of Dr. Bert Vogelstein (Johns Hopkins University).
Quantitative Real-Time RT-PCR
Patient glioblastoma specimens and normal brain samples were obtained from University of Virginia Health System under an approved IRB protocol. Total RNA was extracted with Trizol according to the manufacturer's instructions. For endogenous controls, cDNA was synthesized using Bio-Rad's iScript cDNA Synthesis Kit. For microRNA expression analysis, 10 ng total RNA was used along with miR-34a–specific primers supplied with miR-34a Taqman MicroRNA Assay. cDNA was synthesized using Taqman MicroRNA Reverse Transcription kit and quantitative real-time PCR analysis was performed using the 7500 Real-Time PCR System. A human 18S rRNA Taqman probe was used as endogenous control. For c-Met expression analysis 50 ng total RNA was used and cDNA was synthesized using Multiscribe reverse transcriptase and c-Met expression was analyzed using c-Met specific primers. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were used as endogenous controls.
Cell transfections
Transfections of microRNAs were performed using Oligofectamine and 10 nM pre-miR-34a or 10 nM pre-miR-con according to the manufacturer's instructions. Plasmid transfections were performed with Fugene 6 reagent according to the manufacturer's instructions.
Luciferase assays
To determine if miR-34 can bind to c-Met, Notch-1 or Notch-2 3′-UTR, brain tumor cells or stem cells were transfected with pre-miR-34a or pre-miR-con for 24 hrs and subsequently transfected with either 3′UTR-control, 3′UTR-Met, 3′UTR-Notch-1, or 3′UTR-Notch-2 in addition to control cytomegalovirus-β-galactosidase reporter plasmids for 48 hrs. To determine if wild-type p53 but not mutant p53 activates miR-34a, p53-null LN-Z308 cells were transfected with pre-miR-34a or pre-miR-con reporter plasmids for 6 hrs prior to transfection with either wild-type p53, mutant p53 (R273H) or control vectors for 48 hrs. Luciferase assays were performed using the Luciferase System Kit and luminescence was measured on a Promega GloMax 20/20 luminometer and normalized as described previously (28, 29).
Immunoblotting
Immunoblotting was performed as previously described using antibodies specific for c-Met and CDK6 (Cell Signaling Technologies, Danvers, MA), Notch-1, Notch-2 (Santa Cruz Biotechnologies, Santa Cruz, CA). All blots were stripped and re-probed with β-actin or α-tubulin antibodies (Santa Cruz Biotechnologies, Santa Cruz, CA) as loading controls.
Cell proliferation assays
U87, A172 and DAOY cells (30,000/well) were transfected with pre-miR-34a or pre-miR-con as described above. After 72 hrs, the cells were collected every day for five subsequent days and counted with a hemocytometer.
Propidium Iodide Flow Cytometry
The effects of miR-34a expression on cell cycle progression were assessed using propidium iodide (PI) flow cytometry as previously described 28, 29. Briefly, U87 and A172 cells or astrocytes were transfected with pre-miR-34a or pre-miR-con for 72 hrs. The cells were washed with PBS, harvested and fixed in 70% (v/v) ethanol. The cells were then treated with 20 μg of DNase-free RNase and stained with propidium iodide. Cell samples were analyzed on a FACscan (Becton-Dickinson, Fullerton, CA) and G0/G1, S and G2/M fractions were determined.
Annexin V-PE and 7AAD flow cytometry
The effects of miR-34a expression on cell death were assessed by Annexin V-PE and 7AAD flow cytometry as previously described 1. Briefly, A172, 0308, DAOY and astrocytes were transfected with pre-miR-34a or pre-miR-con for 96 hrs. The cells were harvested and stained with Annexin V-PE and 7AAD according to the manufacturer's instructions. Cell samples were analyzed on a FACsan and apoptotic fractions were determined.
Cell invasion assays
The effects of miR-34a expression on cell invasion were assessed using a transwell invasion assay as previously described (28). Briefly, A172 cells were transfected with pre-miR-34a or pre-miR-con for 72 hrs. The transfected cells (1×105) were resuspended in 300 μL 0.1% FBS medium and placed in the upper chamber of the wells. Six hundred μL 10% FBS medium were placed in the lower chamber. After incubation for 6 hrs at 37°C in 5% CO2, the cells on the upper membrane surface were mechanically removed. Cells that had migrated to the lower side of the collagen IV-coated membrane were fixed and stained with 0.1% crystal violet. Migrated cells were counted under a microscope in five randomly chosen fields and photographs were taken.
c-Met and Notch rescue experiments
To determine if c-Met or Notch can rescue miR-34a-induced cell cycle arrest or cell death, the effects of miR-34a on cell cycle and apoptosis were tested in the setting of forced expression of plasmids encoding either c-Met or Notch1 and Notch2 transcripts lacking the respective 3′-UTR regions and that therefore cannot be inhibited by miR-34a. The plasmids were transfected in U87 or 0308 cells for 6 hrs prior to transfection with pre-miR-34a or pre-miR-con for 48 hrs. The cells were collected and analyzed for cell cycle with propidium iodide flow cytometry or for cell death with Annexin V-PE/7AAD flow cytometry as described above. c-Met and Notch expression changes were verified by immunoblotting.
Tumor formation in vivo
The effects of miR-34a on in vivo tumor growth were tested in an intracranial glioma xenograft model. U87 cells were transfected with pre-miR-34a or pre-miR-con for 24 hrs. Transfected cells (3 × 105) were stereotactically implanted into the striata of immunodeficient mice (n=10). The animals were sacrificed after 4 weeks of tumor implantation. The brains were removed, sectioned, and stained with H&E. Maximal tumor cross-sectional areas were measured by computer-assisted image analysis and tumor volumes were calculated.
Statistics
When appropriate, two group comparisons were analyzed with a t-test and multiple group comparisons were analyzed with a Dunnett test and p values were calculated. P < 0.05 was considered significant and symbolized by an asterisk in the graphs.
Results
miR-34a inhibits the expression of multiple oncogenes and binds to their 3′UTR
Based on the Sanger microRNA database, miR-34a has several predicted seed matches in the 3′UTR of multiple oncogenes including c-Met, Notch1, Notch-2, CDK6 and PDGFRA. Therefore, we tested the effects of miR-34a on c-Met, Notch1, Notch-2, CDK6 and PDGFRA expressions in brain tumor cells and stem cells. These oncogenes were selected because they are frequently upregulated in human brain tumors and because they are subject to research in our laboratories. We tested the effects of miR-34a transfection on protein levels of all the above oncogenes and the 3′UTR reporter activities of c-Met, Notch-1 and Notch-2. Transfection of miR-34a reduced the protein levels of c-Met protein in glioma and medulloblastoma cells and astrocytes. Transfection of miR-34a also reduced the protein levels of Notch-1, Notch-2 and CDK6 in glioma cells and stem cells (Figure 1A). miR-34a transfection did not affect the protein levels of PDGFRA in any tested cell line (not shown). To determine if miR-34a can directly inhibit c-Met, Notch-1 and Notch-2 protein expressions by binding to their 3′UTR, glioma, medulloblastoma and glioma stem cells were transfected with pre-miR-34a or pre-miR-control. The 3′UTR reporter activities of c-Met, Notch-1 and Notch-2 were assessed by luciferase assays. miR-34a inhibited c-Met, Notch-1 and Notch-2 luciferase activities in all cell lines tested (Figure 1B). miR-34a inhibited normalized c-Met 3′UTR luciferase activity by 90.4 ± 5.6% in U87 cells, 87.6 ± 2.4% in A172 cells and 74.6 ± 4.0% in DAOY cells (n=3; P<0.05 for all). miR-34a also inhibited normalized activity of the Notch-1 3′UTR reporter by 40.1 ± 10.3% in T98G cells and 24.0 ± 4.1% in the glioma stem cell line 0308 and activity of Notch-2 3′UTR reporter by 50.1 ± 3.4% in U373 cells and by 35.0 ± 2.1% in 0308 glioma stem cells (n=3; P<0.05 for all). Together, these data show that miR-34a binds to c-Met 3′UTR, Notch-1 3′UTR and Notch-2 3′UTR and downregulates c-Met, Notch-1, Notch-2 and CDK6 protein expressions.
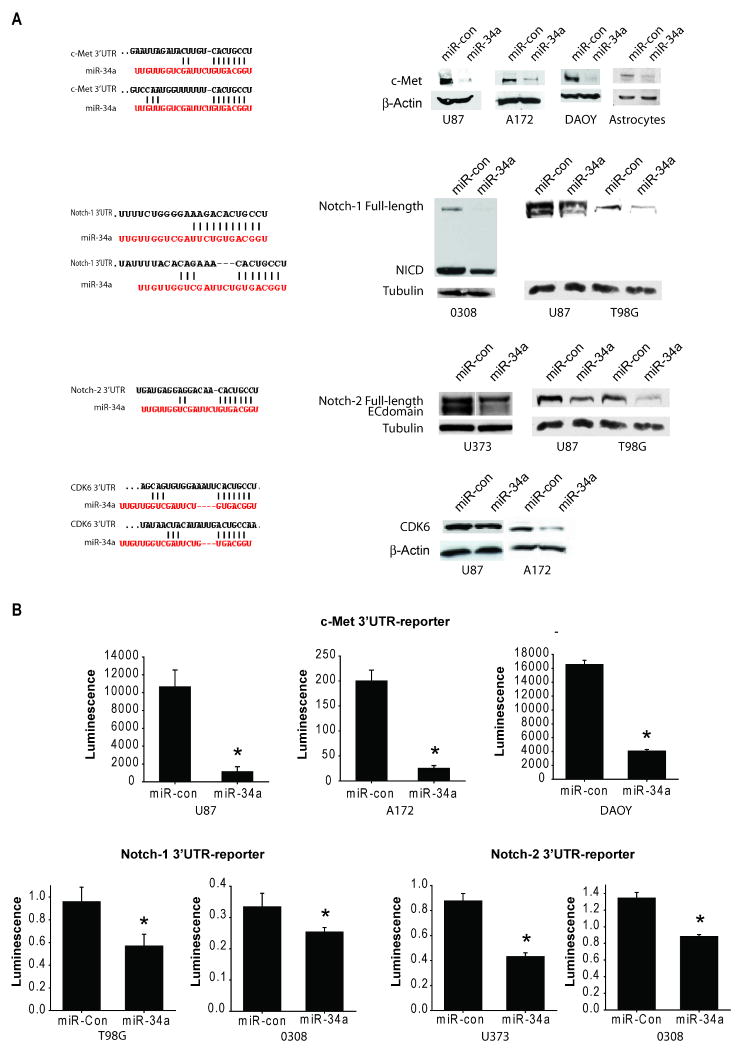
A) Glioma cells and stem cells, medulloblastoma cells or astrocytes were transfected with either pre-miR-34a or pre-miR-con for 72 hrs. c-Met, Notch-1, Notch-2 and CDK6 protein levels were measured by immunoblotting. The results show that miR-34a expression downregulates c-Met, Notch-1, Notch-2 and CDK6 protein levels (right panels). The predicted seed matching between miR-34a and the oncogenes' 3′UTR sequences is shown in the left panels. B) Glioma cells were transfected with pre-miR-34a or pre-miR-con for 24 hrs prior to transfection with either c-Met 3′UTR, Notch-1 3′UTR, Notch-2 3′UTR or control reporter plasmids together with β-Gal plasmids for 48 hrs and 3′UTR reporter activity was measured by a luciferase assay and normalized to β-Gal. The results show that miR-34a expression downregulates c-Met, Notch-1 and Notch-2 luciferase activities in the cells. NICD = Notch intracellular domain. * = p<0.05.
miR-34a expression in human glioblastoma tissues
A previous study screened for microRNA expressions in the NCI-60 tumor cells and found that miR-34a is downregulated in brain tumor cell lines (21). In this study, we analyzed for the first time miR-34a expression in human glioblastoma surgical specimens and normal brain tissue by quantitative RT-PCR. We found that the average level of miR34 expression is lower in glioblastoma tissues (n=11) than in normal brain tissue (n=6) (Figure 2A). Importantly, we also measured c-Met mRNA expression levels in the same specimens described above and found a significant inverse correlation between the levels of c-Met and miR-34a expression in human glioblastoma and normal tissues (r = 0.84) (Figure 2C). To confirm that miR-34a affects c-Met mRNA levels, we transfected U87 glioma cells with pre-miR-34a and measured the effects on c-Met mRNA with quantitative RT-PCR. miR-34a expression reduced c-Met mRNA levels, albeit less than it reduced protein levels (Figure 2D and Figure 1A). This suggests that miR-34a affects both c-Met transcription and c-Met mRNA degradation.
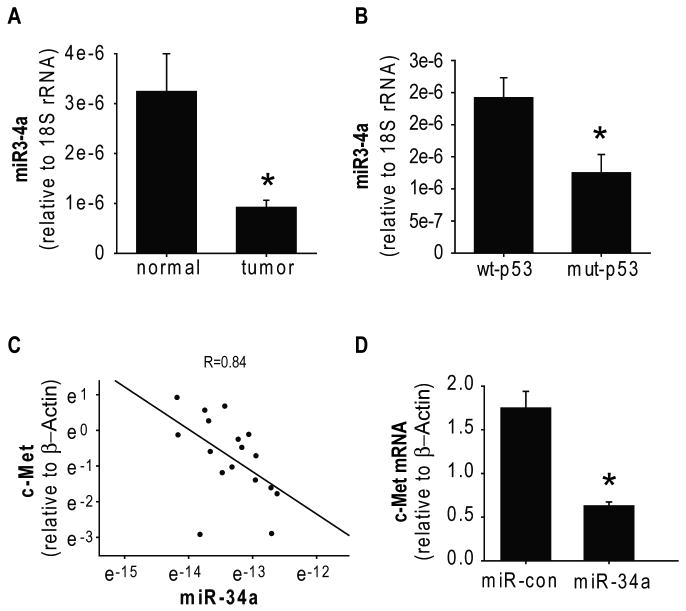
A) miR-34a levels were measured by qRT-PCR in 11 glioblastoma surgical specimens and 6 normal brain samples and normalized to 18S rRNA measured in the same samples (arbitrary units). The results show that average levels of miR-34a in gliomas are lower than in normal brain. B) The p53 status of the glioma specimens described in (A) were determined. The blots show that average expression of miR-34a in wild-type p53 glioblastoma tumors (n=7) is significantly higher than miR-34a expression in mutant p53 tumors. C) c-Met expression in the same tissues described in (A) was measured by RT-PCR and normalized to β-Actin. Plotting of miR-34a vs. c-Met expressions shows an inverse correlation between them. D) Glioma cells were transfected with miR-34a for 48 hrs prior to measurement of c-Met mRNA levels by RT-PCR. The results show that miR-34a reduces c-Met mRNA levels. * = p<0.05.
p53 has been recently shown to transcriptionally regulate miR-34a (22-24). We therefore assessed the levels of miR-34a in wild-type-p53 tumors as compared to mutant-p53 tumors. We found that the average level of miR-34a expression was higher in wild-type p53 glioblastoma (n=7) as compared to mutant p53 glioblastoma (n=4) (p<0.05) (Figure 2B). Previous studies that linked p53 to miR-34a transcription compared p53-null to wt-p53 cells and tissues. The effects of mutant-p53 protein, which is highly expressed in many tumors and which can possess gain-of-function activities, on miR-34a expression have not been examined to date. To determine the effects of mutant p53 on miR-34a activity in brain tumor cells, we constructed a miR-34a reporter plasmid and miR-con reporter plasmid. We transfected the reporters together with either wild-type p53 or mutant p53 in p53-null LN-Z308 glioma cells and assessed the effects of p53 on miR-34a activity. Luciferase assays showed that wild-type p53 but not mutant p53 downregulates normalized miR-34a luciferase activity (not shown), indicating that mutant p53 does regulate miR-34a transcription.
miR-34a inhibits brain tumor malignancy
We assessed the effects of miR-34a on brain tumor malignancy parameters including cell proliferation, cell cycle, cell death and cell invasion in human glioblastoma, medulloblastoma and astrocytes. Pre-miR-34a or pre-miR-con were transiently transfected into the cells and cell proliferation, cell cycle, cell death and cell invasion were analyzed by cell counting, propidium iodide flow cytometry, Annexin V-PE 7AAD flow cytometry, and transwell invasion assays, respectively. miR-34a expression significantly inhibited cell proliferation in U87 and A172 glioblastoma and DAOY medulloblastoma cells (n=6, p<0.05). miR-34a restoration inhibited cell proliferation by 78.4 ± 2.4% in U87, by 93.9 ± 3.8% in A172, and by 70.5 ± 4.9% in DAOY after seven days of expression (Fig 3A). miR-34a also significantly induced cell cycle arrest and increased the G0/G1 fraction from 73.9 ± 4.0% to 91.7 ± 2.1% in U87 cells (n=3, p=<0.05) and from 68.8 ± 1.1% to 86.4 ± 1.4% in A172 cells (n=5, p=<0.05), respectively but did not significantly change the cell cycle status in human astrocytes (Figure 3B). miR-34a also inhibited brain tumor cell but not astrocyte survival. miR34a expression induced cell death by 24.3 ± 0.3% in A172 cells and 30.7 ± 1.5% in DAOY cells but no significant cell death induction was observed in human astrocytes (Figure 3C). Moreover, miR34a increased the levels of cleaved PARP in A172 cells. miR34a also markedly inhibited transwell invasion of A172 cells by 61.7 ± 7.7% (Figure 3D). The above data show that miR-34a suppresses several malignancy parameters in human brain tumor cells but not in human astrocytes.
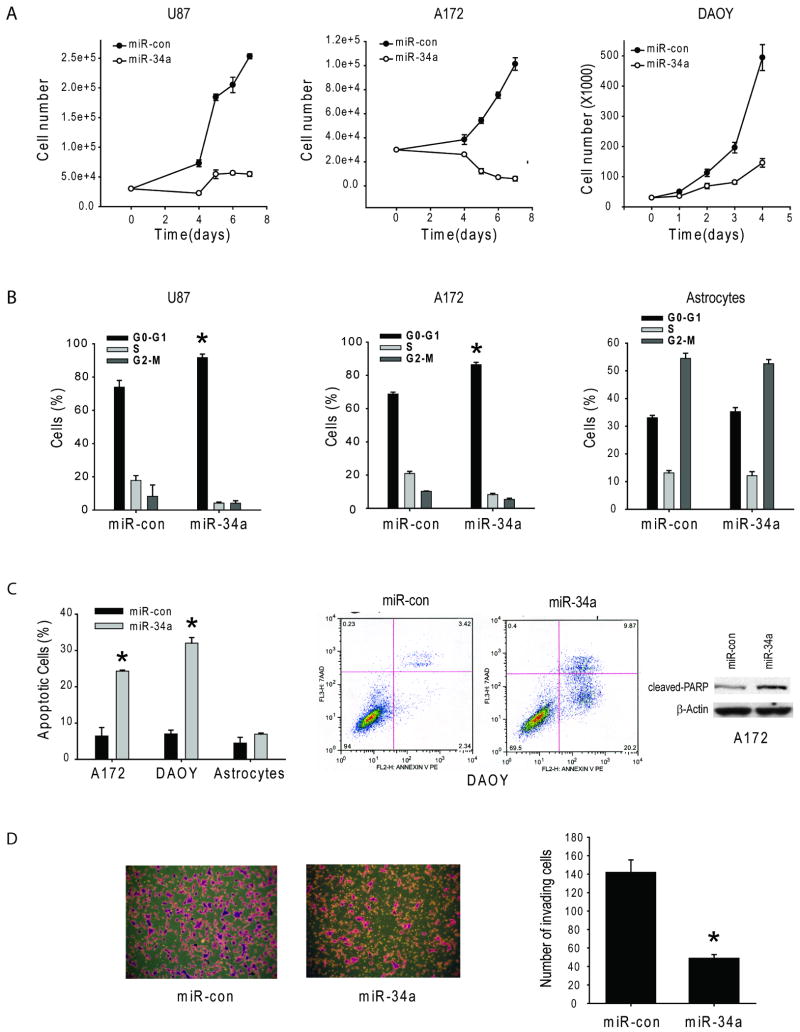
Glioma and medulloblastoma cells were transfected with pre-miR-34a or pre-miR-con and subsequently assessed for cell proliferation by cell counting (A), for cell cycle by propidium iodide flow cytometry (B), for apoptosis by Annexin V flow cytometry and cleaved PARP immunoblotting (C) and for invasion by a transwell invasion assay (D). The results show that miR-34a strongly inhibits cell proliferation (A), induces cell cycle arrest (B), induces apoptosis (C) and inhibits transwell cell invasion (D). * = p<0.05.
Forced c-Met or Notch1 and Notch2 expressions partially rescue cell cycle arrest and cell death induced by miR-34a in glioma cells or stem cells
To determine if the effects of miR-34a on the cell cycle are mediated by c-Met and/or Notch, we overexpressed c-Met cDNA or Notch1 and Notch2 cDNAs that lack the respective 3′-UTR regions and that therefore cannot be inhibited by miR-34a in U87 cells (c-Met) or 0308 stem cells (Notch) prior to transfection with miR-34a and testing for cell cycle status and apoptosis. The results show that forced c-Met expression partially but significantly rescues miR-34a-induced cell cycle arrest (n=3 ; p<0.05) in U87 cells (Figure 4A) and that forced Notch1 and Notch2 expressions partially but significantly rescue miR-34a-induced cell death (n=3 ; p<0.05) in 0308 stem cells (Figure 4B). c-Met and Notch1 expression levels were verified by immunoblotting which confirmed that miR-34a only partially inhibits forced c-Met or Notch1 expressions in the cells. Altogether, these data suggest that the effects of miR-34a on the cell cycle are partially mediated by c-Met and Notch.
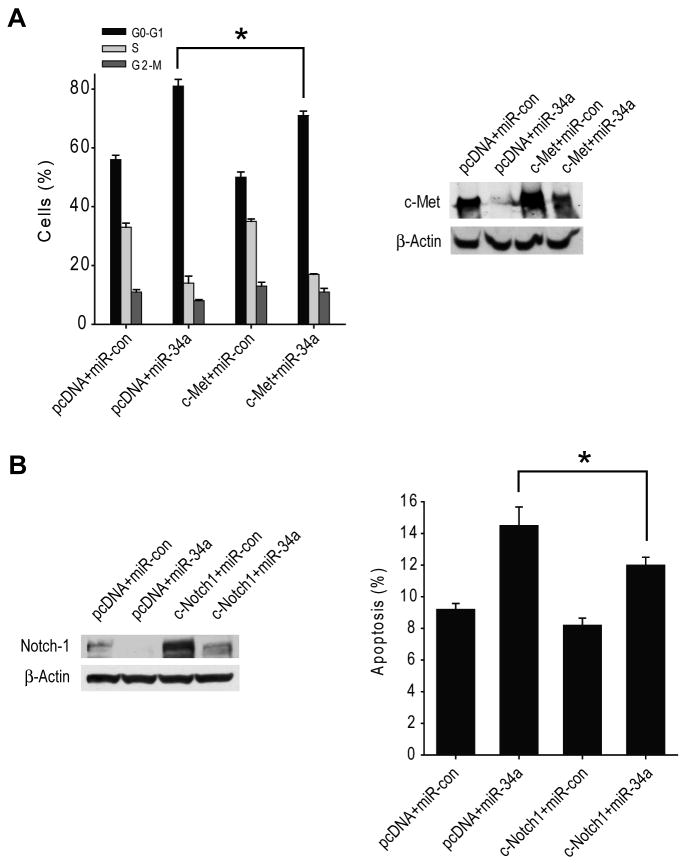
A) U87 cells were transfected with 5 μg pcDNA-Met or pcDNA control for 6 hrs prior to transfection with pre-miR-34a or pre-miR-con for 48 hrs. The cell cycle status of the transfected cells was analyzed by propidium iodide flow cytometry. c-Met expression changes were analyzed by immunoblotting. The results show that overexpression of 3′UTR-deleted c-Met partially rescues the cells from miR-34a-induced cell cycle arrest. B) 0308 stem cells were transfected with 2.5 μg pcDNA-Notch1 and 2.5 μg pcDNA-Notch2 or 5 μg pcDNA-control for 6 hrs prior to transfection with pre-miR-34a or pre-miR-con for 48 hrs. Apoptosis of the transfected cells was analyzed by Annexin V flow cytometry. Notch1 expression changes were analyzed by immunoblotting. The results show that overexpression of 3′UTR-deleted Notch1 and Notch2 partially rescues the cells from miR-34a-induced cell death. * = p<0.05.
miR-34a expression inhibits in vivo glioblastoma xenograft growth
We assessed the effects of miR-34a expression on in vivo glioblastoma xenograft growth. Pre-miR-34a or pre-miR-con were transfected ex vivo into U87 cells. The cells were subsequently implanted in the striata of immunodeficient mice (n=10) and tumor sizes were measured after 4 weeks. miR-34a-transfected cells generated xenografts (3.4 ± 1.0 μm2) that were statistically significantly smaller than control miR-transfected xenografts (22.1 ± 1.0 μm2) (p<0.05) (Figure 5). U87 cells transfected with miR-34a as described above were also tested for viability with anchorage-independent growth in soft agar. The miR-34a-transfected cells formed colonies in soft agar, albeit fewer and smaller colonies than control-transfected cells (not shown). Therefore, miR-34a inhibits the in vivo growth of human glioma experimental tumors.
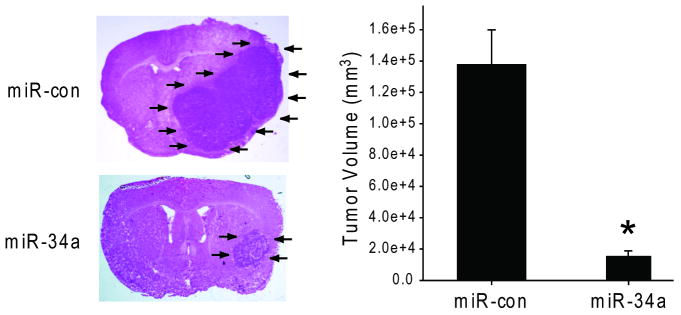
U87 cells were transfected with pre-miR-34a or pre-miR-con for 24 hrs. The transfected cells were implanted into the brains of immunodeficient mice (n=10). After 4 weeks, the mice were euthanized and the brains were cryosectioned and H&E stained. Tumor sizes were measured with computer-assisted image analysis. The results show that miR-34a inhibits in vivo glioblastoma xenograft growth. * = p<0.05.
Discussion
We studied the role of miR-34a in brain tumors with a focus on glioblastoma. We found that miR-34a inhibits c-Met in brain tumor cells and Notch-1/ Notch-2 in glioma cells and stem cells via binding to the 3′UTRs. We also found that pre-miR-34a expression is lower in glioblastoma tissues than in normal brain tissues and in mutant p53 tumors as compared to wild type p53 glioblastomas. Moreover, we found a significant inverse correlation between miR-34a expression and c-Met expression in these tissues. We showed that forced miR-34a expression in brain tumor cells strongly inhibits cell proliferation, cell cycle progression, cell survival, cell invasion and in vivo glioma xenograft growth but does not affect astrocyte survival and cell cycle. We also showed that the effects of miR-34a on glioma cells are partially mediated by c-Met and Notch downregulations. We therefore present the first comprehensive study of the role of miR-34a in human gliomas.
In the last two years, a few studies have investigated the role of miR-34a in cancer. Our study provides several new findings on this microRNA. We show for the first time that miR-34a expression is reduced in human gliomas and that miR-34a levels inversely correlate with c-Met levels in the same tumors. We also show for the first time that miR-34a targets Notch. We show that miR-34a inhibits in vivo tumor growth and that it has no inhibitory effects on human astrocyte growth. Our findings also show for the first time partial rescue of miR-34a-induced cell cycle and cell death by forced c-Met and Notch expressions.
miR-34a was reported to be down-regulated in human cancer cell lines (22, 30-32). In this paper, we show that miR-34a expression is lower in human glioblastoma tissues than in normal brain. The downregulation of miR-34a expression in glioblastoma could be associated with the frequent deletion of chromosome 1p36, p53 mutations and CpG methylation of the miR-34a promoter in these tumors. Allelic loss at 1p is seen in 70% to 85% of oligodendrogliomas and 20-30% of astrocytomas. Most 1p deletions in gliomas involve almost the entire chromosome arm (33). miR-34a was reported to locate within chromosome 1p36 (34). It was also recently shown that miR-34a was inactivated by aberrant CpG methylation in multiple types of cancer, but brain tumors were not investigated (32). In addition, several groups have reported that p53 binds to and transactivates the miR-34a promoter (22-24). However, a correlation between downregulation of miR-34a expression and p53 mutations in tumor tissues was not described before. We show that the level of miR-34a expression in wild-type p53 glioblastoma tissues is higher compared with mutant p53 glioblastoma tissues. Moreover, we find that wild-type p53 but not mutant p53 (R275H) can increase miR-34a luciferase activity. p53 mutations are found in approximately 30% of all gliomas irrespective of tumor grade (35, 36).
miR-34a has multiple predicted targets among which are many oncogenes. We chose to focus on c-Met and Notch pathways because they are dysregulated in brain tumors and because they are the subject of intensive research in our laboratories. c-Met, Notch-1 and Notch-2 are frequently overexpressed in glioblastoma and medullloblastoma but the cause of their overexpressions is not well understood (1, 6, 8, 9, 12, 37). Our data which show that miR-34a is downregulated in gliomas, that miR-34a inhibits c-Met and Notch expressions, and that miR-34a levels inversely correlate with c-Met levels in human gliomas provide one potential explanation for the overexpression of these oncogenes in gliomas.
We found that miR-34a expression strongly suppresses multiple malignancy endpoints in glioblastoma and medulloblastoma. These effects are likely mediated by changes in a number of miR-34 target mRNAs. Recent work indicates that miR-34a-induced cell cycle arrest could be mediated by the inhibitory effects of miR-34a on E2F3, MYCN, CDK4, Cyclin E2, CyclinD1 and BCL2 (30, 31, 34, 38, 39). miR-34a also directly binds to SIRT1 and regulates cell proliferation and cell survival via SIRT-1-p53 (40). Here, we show that c-Met and Notch inhibitions contribute to miR-34a tumor suppressive effects, as c-Met and Notch expressions partially rescue the effects of miR-34a on cell cycle and cell death. The strong inhibitory effects of miR-34a on malignancy are further demonstrated by the effects on in vivo xenograft growth. The anti-tumor effects of miR-34a are probably achieved via targeting of multiple oncogenes. Interestingly, miR-34a transfection into human astrocytes only marginally affected cell death and cell cycle. This suggests that while restoration of miR-34a to cells in which it is downregulated will inhibit malignancy via inhibition of multiple oncogenes, transfection into cells in which miR-34a has normal expression levels and in which oncogene expression is low will not affect cell death. Consequently, miR-34a might serve as a potential cancer and glioma therapeutic agent.
Altogether, this study provides new insights into the role of miR-34a in human brain tumors. It shows that miR-34a is deregulated in gliomas and that miR-34a potently inhibits brain tumor growth by targeting multiple oncogenes. The study also suggests that miR-34a might serve as a brain tumor therapeutic agent.
Acknowledgments
Supported by NIH grant RO1 NS045209 (RA) and a University of Virginia Cancer Center Pilot Project Grant (RA).
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-09-0529
Read article for free, from open access legal sources, via Unpaywall:
https://aacrjournals.org/cancerres/article-pdf/69/19/7569/2615268/7569.pdf
Free after 12 months at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/content/full/69/19/7569
Free to read at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/content/abstract/69/19/7569
Free after 12 months at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/reprint/69/19/7569.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/0008-5472.can-09-0529
Article citations
Exploring the clinical implications and applications of exosomal miRNAs in gliomas: a comprehensive study.
Cancer Cell Int, 24(1):323, 27 Sep 2024
Cited by: 0 articles | PMID: 39334350 | PMCID: PMC11437892
Review Free full text in Europe PMC
Combination of microRNA and suicide gene for targeting Glioblastoma: Inducing apoptosis and significantly suppressing tumor growth in vivo.
Heliyon, 10(17):e37041, 29 Aug 2024
Cited by: 0 articles | PMID: 39286083 | PMCID: PMC11403485
Genomic, epigenomic and transcriptomic landscape of glioblastoma.
Metab Brain Dis, 39(8):1591-1611, 24 Aug 2024
Cited by: 0 articles | PMID: 39180605
Review
Multifunctional elastin-like polypeptide nanocarriers for efficient miRNA delivery in cancer therapy.
J Nanobiotechnology, 22(1):293, 27 May 2024
Cited by: 0 articles | PMID: 38802812 | PMCID: PMC11131307
Mechanism of Notch Signaling Pathway in Malignant Progression of Glioblastoma and Targeted Therapy.
Biomolecules, 14(4):480, 15 Apr 2024
Cited by: 1 article | PMID: 38672496 | PMCID: PMC11048644
Review Free full text in Europe PMC
Go to all (405) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
microRNA-34a is tumor suppressive in brain tumors and glioma stem cells.
Cell Cycle, 9(6):1031-1036, 15 Mar 2010
Cited by: 204 articles | PMID: 20190569 | PMCID: PMC3278213
P53-induced microRNA-107 inhibits proliferation of glioma cells and down-regulates the expression of CDK6 and Notch-2.
Neurosci Lett, 534:327-332, 07 Dec 2012
Cited by: 43 articles | PMID: 23220650
MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme.
Cancer Biol Ther, 12(6):477-483, 15 Sep 2011
Cited by: 58 articles | PMID: 21743299
Targeting strategies on miRNA-21 and PDCD4 for glioblastoma.
Arch Biochem Biophys, 580:64-74, 02 Jul 2015
Cited by: 31 articles | PMID: 26142886
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: R01 CA134843
NINDS NIH HHS (2)
Grant ID: R01 NS045209
Grant ID: R01 NS045209-06A2





