Abstract
Free full text

Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts
Abstract
To generate efficient vaccines and cures for Mycobacterium tuberculosis, we need a far better understanding of modes of infection, persistence and spreading. Host cell entry and establishment of a replication niche are well understood, but little is known about how tubercular mycobacteria exit host cells and disseminate the infection. Using the social amoeba Dictyostelium as a genetically tractable host for pathogenic mycobacteria, we discovered that M. tuberculosis and M. marinum but not M. avium are ejected from the cell through an actin-based structure, the ejectosome. This conserved nonlytic spreading mechanism requires a cytoskeleton regulator from the host and an intact mycobacterial ESX-1 secretion system. This insight offers new directions for research into the spreading of tubercular mycobacteria infections in mammalian cells.
Intracellular bacterial pathogens have evolved strategies to exploit host cell resources and replicate inside a variety of cell types, staying out of reach of the host’s immune system. The concept is emerging that pathogenic bacteria evolved from environmental species by adapting to an intracellular lifestyle within free-living, bacteria-eating protozoans. Consequently, cellular defense mechanisms active in animal immune phagocytes may have originated in amoebae (1, 2). For example, during differentiation of the social amoeba Dictyostelium to form a multicellular structure, Sentinel cells are deployed to combat pathogens (1).
Generally, infection follows entry of a bacterium inside a host cell, giving rise to a pathogen-containing vacuole usually called a phagosome. From this common starting point, pathogens subvert or resist the mechanisms that usually transform the phagosome into a bactericidal environment. Understanding of the passive or triggered uptake mechanisms and of the subsequent hijacking of host cell processes is increasing steadily, but little is known about how the pathogens exit their primary host cell and spread the infection.
Mycobacterium tuberculosis causes tuberculosis and other granulomatous lesions and is a major threat to human health. M. marinum is a close relative (3) responsible for fish and amphibian tuberculosis, in which it causes almost indistinguishable pathologies and lesions [reviewed in (4)]. Several elegant cross-species complementation studies between these two pathogens highlight their common mechanisms of pathogenicity [reviewed in (4)].
After passive uptake by immune phagocytes, M. tuberculosis and M. marinum arrest or bypass phagolysosome maturation and replicate inside a compartment of endosomal nature (57). Both species can escape from this vacuole into the host cytosol (8–11), though at varying frequencies. Efficient translocation into the cytosol depends on an intact region of difference (RD) 1 locus (10, 11), which encodes components of a type seven secretion system and essential secreted effectors (12). This ESX-1 secretion system has been implicated in arrest of phagosome maturation (13), granuloma formation and the spread of infection (14, 15); however, it is not essential for replication inside macrophages (14). It has also been directly associated with the secretion of a membranolytic activity (15). Specifically, the secreted effector ESAT-6 has recently been linked to niche breakage and pore-forming activity in macrophages (11).
M. marinum and M. tuberculosis can disseminate through release of bacilli following host cell lysis via necrotic or apoptotic cell death (16–18), but studies also document cell-to-cell, antibiotic-insensitive spreading inside an epithelial monolayer (19, 20). M. marinum induces plasma membrane protrusions (2, 9) suggested to participate in dissemination between macrophages in culture (21) and inside zebrafish embryos (22). Hence, escape into the cytosol may be a necessary precursor for cell-to-cell spread and may involve a direct nonlytic transmission process occuring within the granuloma (5, 6).
Here, we adopted the genetically tractable Dictyostelium-M. marinum model system to unravel basic mechanisms of intercellular dissemination. The course of M. marinum infection in Dictyostelium amoebae is very similar to that in macrophages (8). The mycobacteria replication vacuole accumulates a flotillin-like raft protein, then ruptures and releases M. marinum into the cytosol (see fig. S1). A Dictyostelium mutant lacking the RacH GTPase, which is involved in regulation of the actin cytoskeleton and endosomal membrane trafficking and acidification, was more permissive for M. marinum proliferation (8). Detailed FACS analysis (fig. S2) of these infected cells suggested an interference with intercellular dissemination (fig. S3).
To test this hypothesis, we designed a quantitative dissemination assay (Fig. 1A). Briefly, an infected Dictyostelium donor strain (either wild type or racH-) is mixed with a green fluorescent wild type acceptor strain at a donor:acceptor ratio of 1:5. Over the course of infection, the number of bacteria per donor and acceptor cell was determined by visual inspection. The proportion of wild type infected donor cells decreased concomitantly with a sharp increase of infected acceptor cells at 21 hours post infection (hpi), and the number of bacteria per acceptor cell increased over time (Fig. 1B, bar shadings). This indicated successful transmission of bacteria and replication in acceptor cells. In contrast, the proportion of infected racH-donor cells remained relatively constant (above 50%). Strikingly, infection of acceptor cells from racH-donor cells was about 8-fold less than from wild type donor cells (Fig. 1C). The racH-cells were deficient in intercellular spreading of mycobacteria and it appears that a RacHdependent release mechanism is required for cell-to-cell transmission under these conditions.
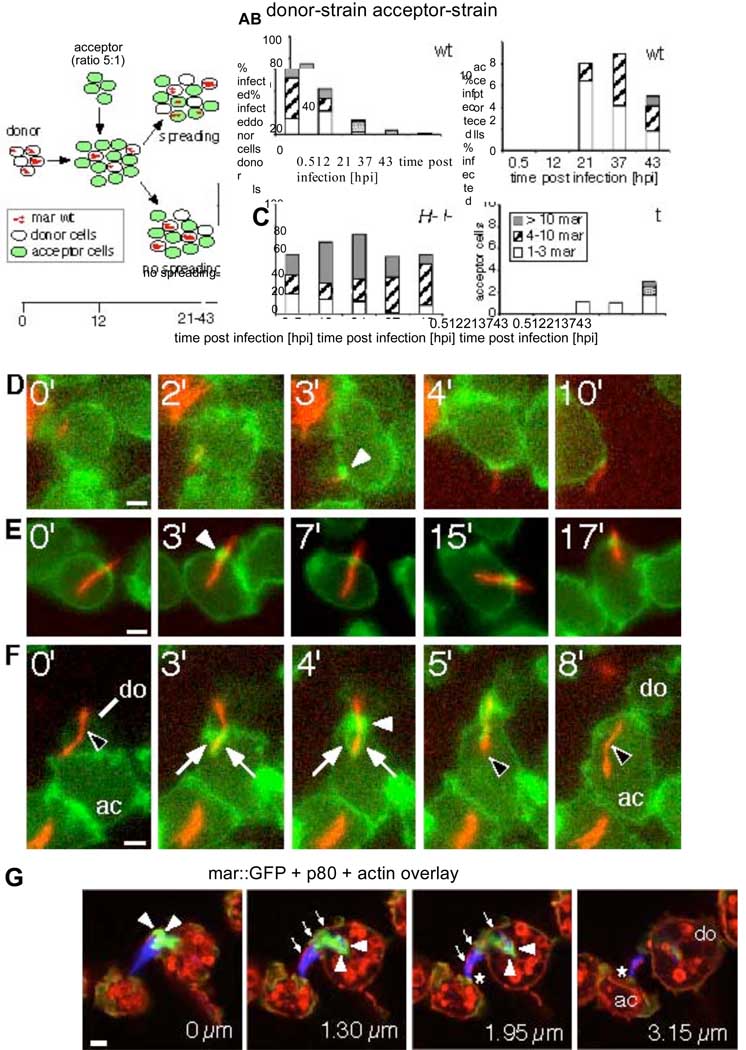
Direct cell-to-cell transmission of M. marinum occurs via ejectosomes and is RacH dependent. (A) Quantitative dissemination assay. A donor strain (wild type or racH-cells) was infected with DsRed-expressing M. marinum and, at 12 hpi, mixed with a green fluorescent acceptor strain. Presence of bacteria in donor and acceptor cells was scored. Dissemination efficiency for wild type (B) and racH-(C) donor cells. Number of bacteria per cell (classified into groups of 1–3, 4–10 and >10 bacteria/cell) are indicated. (D–F) Live GFP-ABD expressing Dictyostelium cells (green) infected with DsRed-expressing M. marinum (red) were imaged at 35 hpi for the indicated times (upper left corner). (D) Nonlytic ejection of mycobacteria occurs through actin-dense ejectosomes (white arrowhead). (E) Cells with mycobacteria spanning the plasma membrane retain normal motility. (F) Cell-to-cell transmission of a cytosolic bacterium (black arrowhead) from a donor cell (do) to an acceptor cell (ac) through an ejectosome (white arrowhead) into a phagocytic cup (white arrows). (G) A bundle of bacteria are ejected from a donor cell (do). The plasma membrane bulge (small arrows) is ruptured at the tip (asterisk), where it contacts an acceptor cell (ac). Actin tails stained by phalloidin (green, arrowheads), were polarized at the posterior of bacteria (blue). Vertical distance from the first section is indicated in µm. Scale bars 1 µm.
Cytosolic pathogens, such as Listeria and Shigella use actin-based tails and filopodia for intercellular spreading. During M. marinum infection of macrophages, unidentified mycobacterial proteins induce actin tails in a Wiscott Aldrich Syndrome protein (WASP)dependent manner (23), as well as structures reminiscent of Shigella-induced filopodia (9) that can be captured by neighboring cells ((9) and fig S4). Hence, we monitored F-actin dynamics in infected cells expressing a GFP-fusion with the actin binding domain (ABD) of filamin (24) by live microscopy (Movies S1–S7, Fig. 1D–F and fig. S5A,B). At late stages of infection, despite the presence of many cytosolic bacteria, cells exhibited apparently normal amoeboid motility (Movie S1) and cell division (Movie S2). In contrast to the observation in macrophages ((9) fig. S4), no persistent actin tails were visible on cytosolic bacteria, possibly due to the high rate of actin depolymerisation in Dictyostelium (25). However, transient actin flashes were produced when bacteria contacted the cell cortex (fig. S5A, Movie S3).
Bacteria were “ejected” through an F-actin-dense structure we called an “ejectosome” (Fig. 1D, Movie S4). Mycobacteria were also caught “spanning” the plasma membrane without inducing host cell lysis, a situation stable for the duration of the observation (e.g. 17 min, Fig. 1E, Movie S5). A significant proportion of cells harbored multiple similar structures (fig. S5B, Movie S6). Ejectosomes can apparently exert a contractile force, forming a tight septum around the bacteria as they are towed behind motile host cells (Fig. 1D, Movie S4). We also captured images of synchronized ejection from a donor cell and phagocytosis by an acceptor cell (Fig. 1F, Movie S7). Hence, an actin-based mechanism appeared to be responsible for nonlytic ejection of cytosolic mycobacteria, which, concomitantly with capture by neighboring cells, ensured efficient and antibiotic-insensitive intercellular spreading.
The live observations were confirmed by staining fixed infected cells with fluorescent phalloidin. During phagocytosis, the cell deforms towards the bacteria and extends lamellipodia that surround it along most of its length (Fig. 2A, ,3E).3E). By contrast, ejectosomes are short barrel-shaped structures usually found in flat membrane regions (Fig. 2B). Multiple ejectosomes were often observed to cluster (Fig. 2C, D). Cell fixation preserved the structure of ejectosomes “coupled” to phagocytic cups suggesting direct donor-to-acceptor transmission (Fig. 2E).
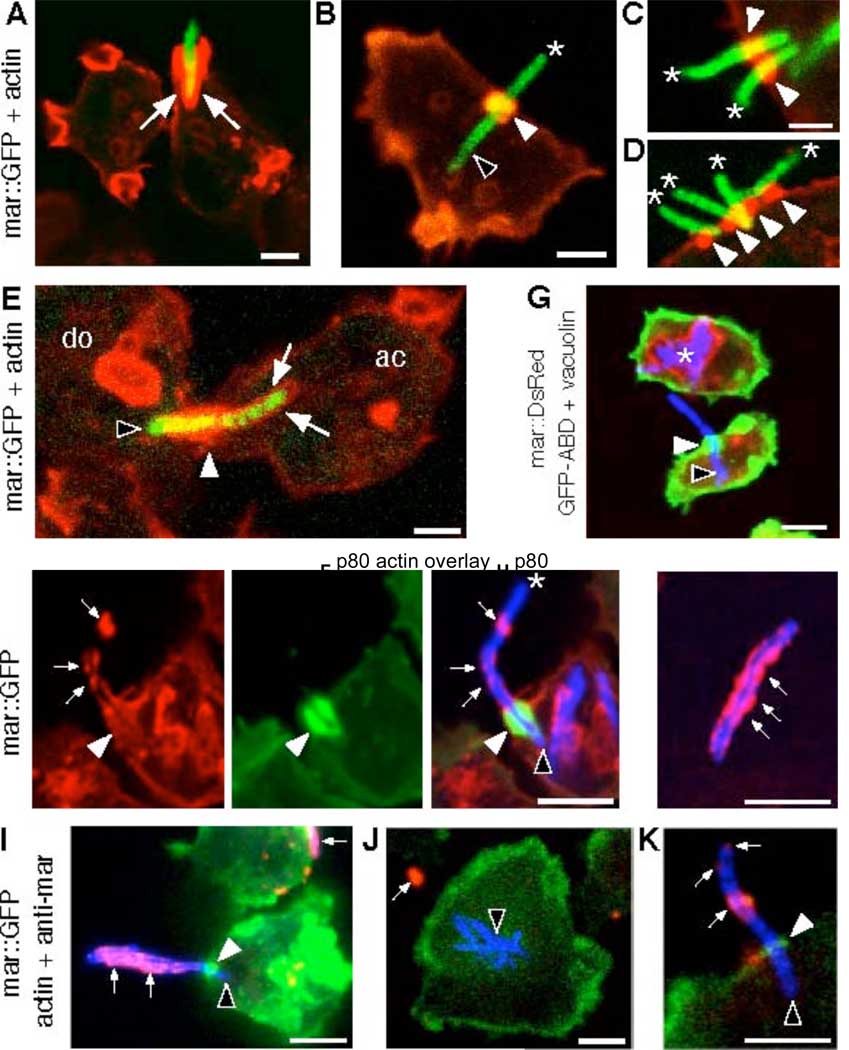
Biogenesis, structure and topology of ejectosomes. Paraformaldehyde-fixed cells infected with GFP-expressing M. marinum (green), stained for F-actin (red). (A) A phagocytic cup lined with F-actin (arrows). A single (B) or multiple bacteria (C, D) spanning the host cell plasma membrane (asterisks) through ejectosomes (white arrowheads) with the intracellular part of the bacterium devoid of actin-labeling (black arrowhead). (E) Cell-to-cell transmission of bacteria from a donor (do) to an acceptor (ac) cell through an ejectosome into a phagocytic cup (arrows). (F) Movement of cytosolic bacteria (blue) through ejectosomes (F-actin, green) induces a plasma membrane bulge (small arrows) that ruptures at the tip (F, asterisk). The intracellular part of the ejecting bacterium is devoid of p80, a plasma membrane marker (F), and vacuolin, a niche marker (asterisk, G). Some extracellular bacteria were positive for p80 (small arrows, H). (I–K) Live infected cells (M. marinum, blue, actin, green) were incubated in the presence of an anti-M. marinum serum. The extracellular parts of ejecting (small arrows, I, K) or outside (small arrow, J) bacteria were accessible to the antibody and labeled red, in contrast to intracellular bacteria (black arrowhead, J), confirming partial loss of plasma membrane integrity. Scale bars 1 µm.
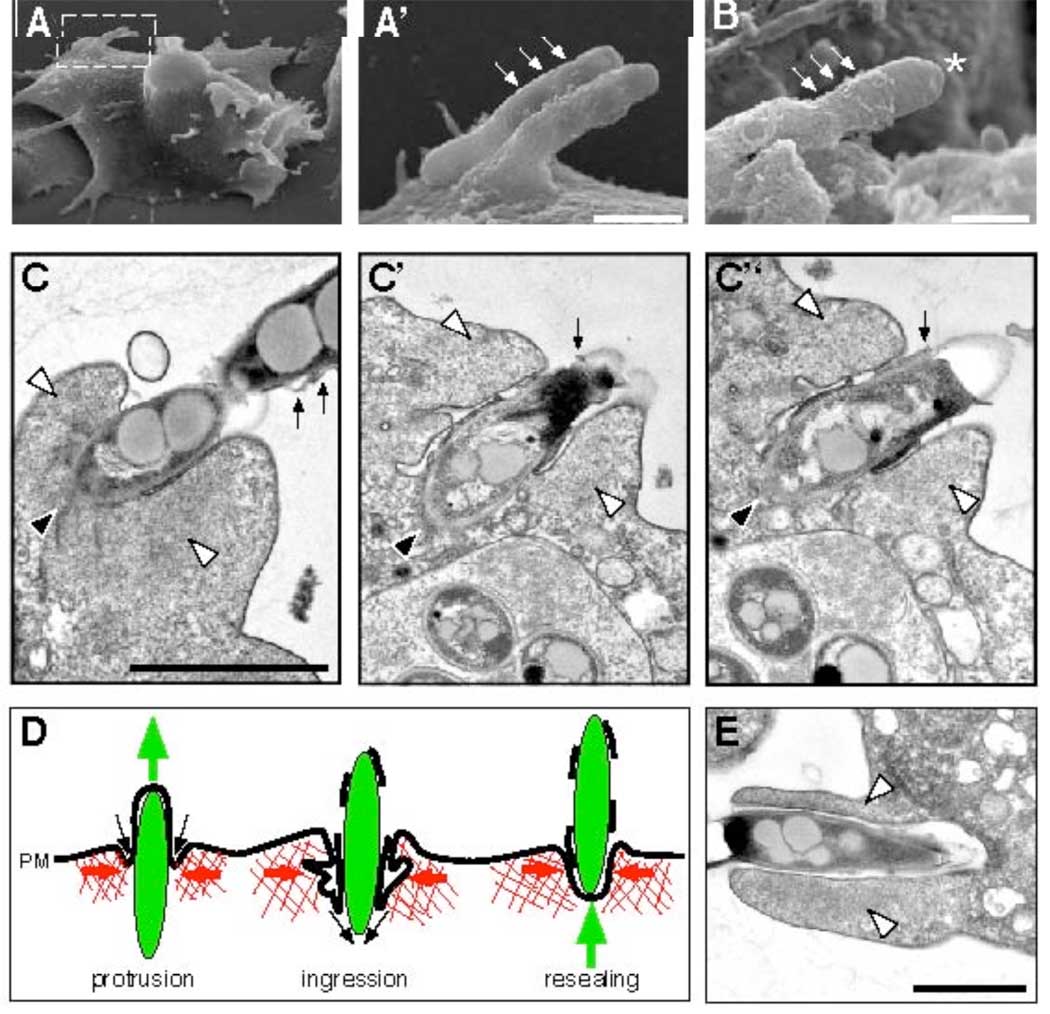
Electron microscopy of ejecting bacteria and schematic representation of an ejection event. Scanning electron microscopy (A–B, A’ is the magnified inset of A) showed bulges of the plasma membrane (small arrows), some ruptured at the tip of the ejecting bacterium (B, asterisk). Scale bars 0.5 µm. (C) Serial sections through an ejectosome revealed the organization of the Factin (white arrowheads) and the plasma membrane. The posterior of the ejecting bacterium (black arrowhead) was in the cytosol. The bacterium was separated from the F-actin by the invaginated plasma membrane, which was tightly apposed to its surface. Membrane fragments were scattered along the extracellular part of the bacterium (small arrows). (D) Schematic representation of an ejection. During outward movement of a bacterium (green), the F-actin barrel exerts contraction (red arrows), and the invaginating plasma membrane (PM) reseals at the posterior of the bacterium (black arrows), maintaining a tight septum despite partial membrane rupture. (E) Micrograph of a phagocytic cup. The actin-filled lamellipodia (white arrowheads) extend and engulf a bacterium. Scale bars 1µm.
What propels bacteria during ejection is unclear, because we rarely observed bacteria with typical F-actin-tails (Fig. 1G). Ejection may be powered by a mechanical process resulting from a combination of cortical tension, cytoplasmic pressure and a reaction of the actin-cortex to bacteria-induced deformation. Myosin IB and coronin were strongly enriched at the ejectosome (fig. S5G, H), whereas myosin II and the Arp2/3 complex were not or weakly enriched in the Factin barrel (fig. S5I, J). Thus, the ejectosome may either assemble de novo or result from a rearrangement of preexisting cortical structures.
In all cases of ejection, the part of the bacterium inside the cell was devoid of membrane markers from endosomes (Fig. 2F and fig. S5C) or the replication compartment (Fig. 2F, G), which excludes the phenomenon being an exocytic event. The protruding part of the bacterium induced a bulge in the plasma membrane (Fig. 2F, fig. S5C), demonstrating that the bacterium is on an outward journey through the cell surface. The membrane labeling often appeared patchy at the tip of the bulge (Fig. 2F, fig. S5C), suggesting local loss of integrity. Sometimes, extracellular mycobacteria were wrapped in plasma membrane remnants (Fig. 2H, fig. S5D). Outward plasma membrane deformation (Fig. 3A, C) and partial membrane rupture (Fig. 3B, C) were also observed by scanning and transmission electron microscopy.
Labelling of the protruding part of ejected M. marinum with an anti-M. marinum serum in non-permeabilised cells indicated accessibility of the bacterium surface, and hence local loss of plasma membrane integrity (Fig. 2I, K). However, infected cells do not lyse, as judged by live imaging (Fig. 1E, Movies S1–S7), and they are also not leaky, as demonstrated by the exclusion of a membrane-impermeant DNA-binding dye during a two hours-long incubation (fig. S5E). Serial sections through an ejectosome (Fig. 3C) showed a cytosolic bacterium protruding from a cell with the plasma membrane ruptured towards the tip of the bacterium, but at the same time invaginated between the bacterium and a zone of dense actin meshwork. We propose that, during ejection, the ingressing plasma membrane stays tightly apposed to the bacterium surface and finally reseals at the posterior of the bacterium, resulting in a dynamic but tight seal that prevents host cell lysis or leakage (schematically depicted in Fig. 3D).
During Dictyostelium infections, M. tuberculosis was first found in spacious vacuoles (fig. S7) that accumulated vacuolin, the Dictyostelium flotillin homologue (fig. S6A). Then, with an efficiency lower than that of M. marinum, the bacilli translocated into the cytosol (Fig. 4A, fig. S6A, S7) where they induced ejectosomes (Fig. 4A, fig. S6B, C). In Dictyostelium, M. avium also accumulated in spacious compartments decorated with vacuolin, but, in contrast to M. tuberculosis and M. marinum, no vacuole breakage was detected (Fig. 4A), and no ejectosome was observed. Vacuole escape and nonlytic ejection from the host cell may represent conserved strategies among tubercular mycobacteria that could play a prominent role in dissemination of infection, rather than intracellular survival per se.
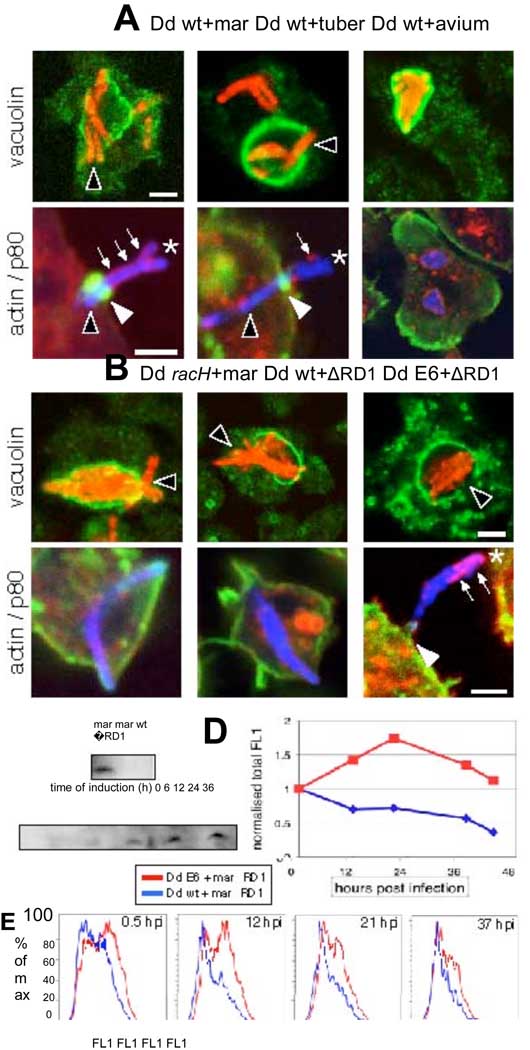
Ejection is a process involving both host and pathogen factors and is a strategy shared by tubercular mycobacteria. (A) In wild type cells (Dd wt) both, M. marinum (mar) and M. tuberculosis (tuber) were able to escape their vacuolin–coated (green) niche (upper row, black arrowheads) and form ejectosomes (lower row, green, white arrowheads). Plasma membrane fragments on ejecting bacteria were positive for p80 (red, small arrows), but the intracellular part of the bacteria was devoid of p80 labeling (black arrowhead). In contrast M. avium remained in a vacuolin-positive compartment (upper row) and did not form ejectosomes. (B) In racH-cells GFP-expressing M. marinum (red) translocated into the cytosol (upper row), but no ejectosome was detected (lower row; actin, green; p80, red). M. marinum ΔRD1 escaped its niche in wild type and ESAT-6 (E6) expressing cells; however, ejectosomes (white arrowhead) were only detected in ESAT-6 expressing cells. (C–E) Trans-complementation by expression of M. marinum ESAT-6 in the Dictyostelium cytosol. Removal of tetracycline-induced ESAT-6 expression (C). Wild type and ESAT-6 expressing cells were infected with GFP-expressing M. marinum ΔRD1 and the infection monitored by FACS. Quantification of total fluorescence of infected cells indicated increased replication of M. marinum ΔRD1 in ESAT-6 expressers (D, red curve, normalized to T=0.5 hpi). Infected ESAT-6 expressers with high fluorescence (FL1) persisted longer (E, red curve), compared to infected wild type Dictyostelium (D, E, blue curve).
We expected a direct correlation between the observed number of ejectosomes and the spreading of infection. Indeed, in wild type cells, there were 14–15 ejectosomes/100 cells at 37 hpi, whereas no ejectosome was ever observed in racH-cells (Fig. 4B, fig. S8B). This is not due to deficient vacuole escape, because at 37 hpi, over 90 % of M. marinum were cytosolic and free of vacuolin staining, compared to 75% in wild type cells (Fig. 4B, fig. S9). Complementation of racH-cells with GFP-RacH partially restored the capacity to form ejectosomes (fig. S8C). In accordance, membrane fragments carrying GFP-RacH polymerize actin around them upon addition of GTPγS in a cell-free assay (26).
In correlation with the capacity to induce ejectosomes, the RD1 locus is present in M. tuberculosis and M. marinum (13–15), but absent from M. avium (12). A M. marinum mutant deleted of its RD1 locus (ΔRD1) replicated inefficiently in Dictyostelium (Fig. 4D), and the population of infected cells steadily decreased with time (Fig 4E and fig. S8D). Vacuole escape was readily detectable (Fig. 4B and fig. S9), but was about 4-to 5-fold less efficient than for wild type M. marinum, as measured by colocalization with vacuolin or p80. This corroborates recent findings in macrophages using a collection of ESX-1 mutants (11). But the most striking observation was the complete absence of ejectosomes (Fig. 4B). If the ESX-1 locus secretes vacuole escape factors (11) it may also secrete factors that coordinate egress of the resulting cytosolic bacteria. It is possible that ESAT-6, one of the major secreted effectors, plays a role in both processes. We designed a trans-complementation strategy in which M. marinum ESAT-6 was conditionally expressed directly inside the cytosol of its host Dictyostelium (Fig. 4C). M. marinum ΔRD1 apparently replicated better in ESAT-6 expressing cells than in wild type cells (Fig. 4D) and showed a 1.5-to 2-fold increased frequency of niche escape. Most importantly, it induced ejectosome formation (Fig. 4B). Our data demonstrated that nonlytic ejection of tubercular mycobacteria from Dictyostelium is a concerted process that necessitates host and pathogen factor(s), and is crucial for the maintenance of an infection in a cell population.
Conceptually, the ejectosome bears intriguing analogies with the contracting actin-ring formed during purse-string closure of membrane wounds in Xenopus oocytes (27) and during the repair of toxin-induced macroapertures in endothelial cells (28). Because lytic release from a single-celled amoeba would be lethal, the ejectosome may have first evolved as a plasma membrane repair mechanism. This concerted and mutually benefitial strategy might have been conserved during evolution to ensure dissemination of mycobacteria between immune phagocytes of their metazoan host.
Dictyostelium can be thought of as a rudimentary innate immune phagocyte and this model has allowed us to identify a conserved strategy for egress and cell-to-cell spread that is shared by M. marinum and M. tuberculosis.
Footnotes
One-sentence summary: Mycobacteria exit host cells without lysing them via an ejectosome generated by the concerted action of host and pathogen factors.
References
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.1169381
Read article for free, from open access legal sources, via Unpaywall:
https://access.archive-ouverte.unige.ch/access/metadata/8ea59541-e61e-4db4-82a0-b70cb9d200a9/download
Subscription required at www.sciencemag.org
http://www.sciencemag.org/cgi/reprint/323/5922/1729.pdf
Free to read at www.sciencemag.org
http://www.sciencemag.org/cgi/content/abstract/323/5922/1729
Subscription required at www.sciencemag.org
http://www.sciencemag.org/cgi/content/full/323/5922/1729
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.1169381
Article citations
Exploring host-pathogen interactions in the Dictyostelium discoideum-Mycobacterium marinum infection model of tuberculosis.
Dis Model Mech, 17(7):dmm050698, 22 Jul 2024
Cited by: 1 article | PMID: 39037280 | PMCID: PMC11552500
Review Free full text in Europe PMC
Temporal genome-wide fitness analysis of Mycobacterium marinum during infection reveals the genetic requirement for virulence and survival in amoebae and microglial cells.
mSystems, 9(2):e0132623, 25 Jan 2024
Cited by: 1 article | PMID: 38270456 | PMCID: PMC10878075
A TRAF-like E3 ubiquitin ligase TrafE coordinates ESCRT and autophagy in endolysosomal damage response and cell-autonomous immunity to Mycobacterium marinum.
Elife, 12:e85727, 18 Apr 2023
Cited by: 11 articles | PMID: 37070811 | PMCID: PMC10181826
Physicochemical properties of intact fungal cell wall determine vesicles release and nanoparticles internalization.
Heliyon, 9(3):e13834, 17 Feb 2023
Cited by: 2 articles | PMID: 36873462 | PMCID: PMC9981904
Brucella Egresses from Host Cells Exploiting Multivesicular Bodies.
mBio, 14(1):e0333822, 09 Jan 2023
Cited by: 4 articles | PMID: 36622142 | PMCID: PMC9973279
Go to all (132) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cell biology. The art of making an exit.
Science, 323(5922):1678-1679, 01 Mar 2009
Cited by: 4 articles | PMID: 19325101
Modeling Tubercular ESX-1 Secretion Using Mycobacterium marinum.
Microbiol Mol Biol Rev, 84(4):e00082-19, 02 Sep 2020
Cited by: 14 articles | PMID: 32878966 | PMCID: PMC8570393
Review Free full text in Europe PMC
EspM Is a Conserved Transcription Factor That Regulates Gene Expression in Response to the ESX-1 System.
mBio, 11(1):e02807-19, 04 Feb 2020
Cited by: 22 articles | PMID: 32019792 | PMCID: PMC7002343
Distinct Mycobacterium marinum phosphatases determine pathogen vacuole phosphoinositide pattern, phagosome maturation, and escape to the cytosol.
Cell Microbiol, 21(6):e13008, 07 Feb 2019
Cited by: 19 articles | PMID: 30656819
Funding
Funders who supported this work.
NHLBI NIH HHS (3)
Grant ID: HL 055936
Grant ID: R01 HL055936
Grant ID: R01 HL055936-09
NIAID NIH HHS (3)
Grant ID: R01 AI067027
Grant ID: R01 AI067027-04
Grant ID: AI 067027





