Abstract
Free full text

Identification and sequencing of cDNA clones encoding the granule-associated serine proteases granzymes D, E, and F of cytolytic T lymphocytes.
Abstract
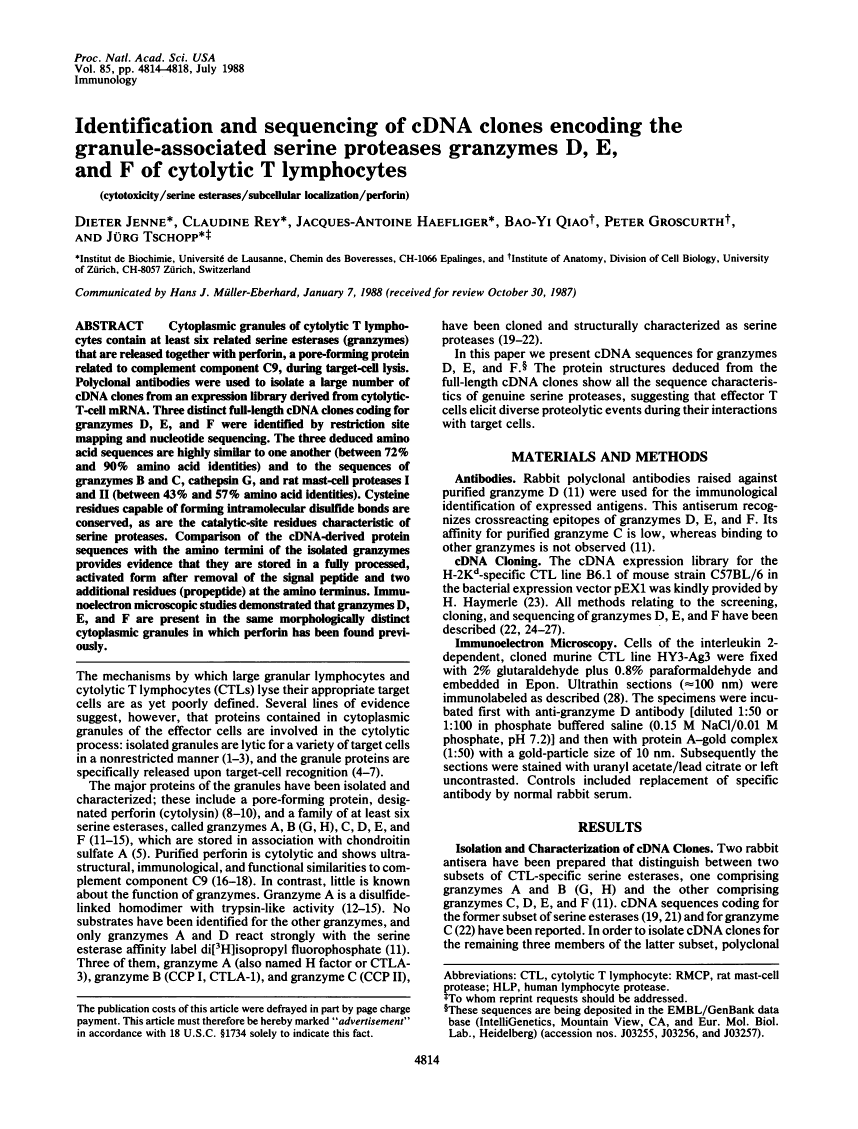
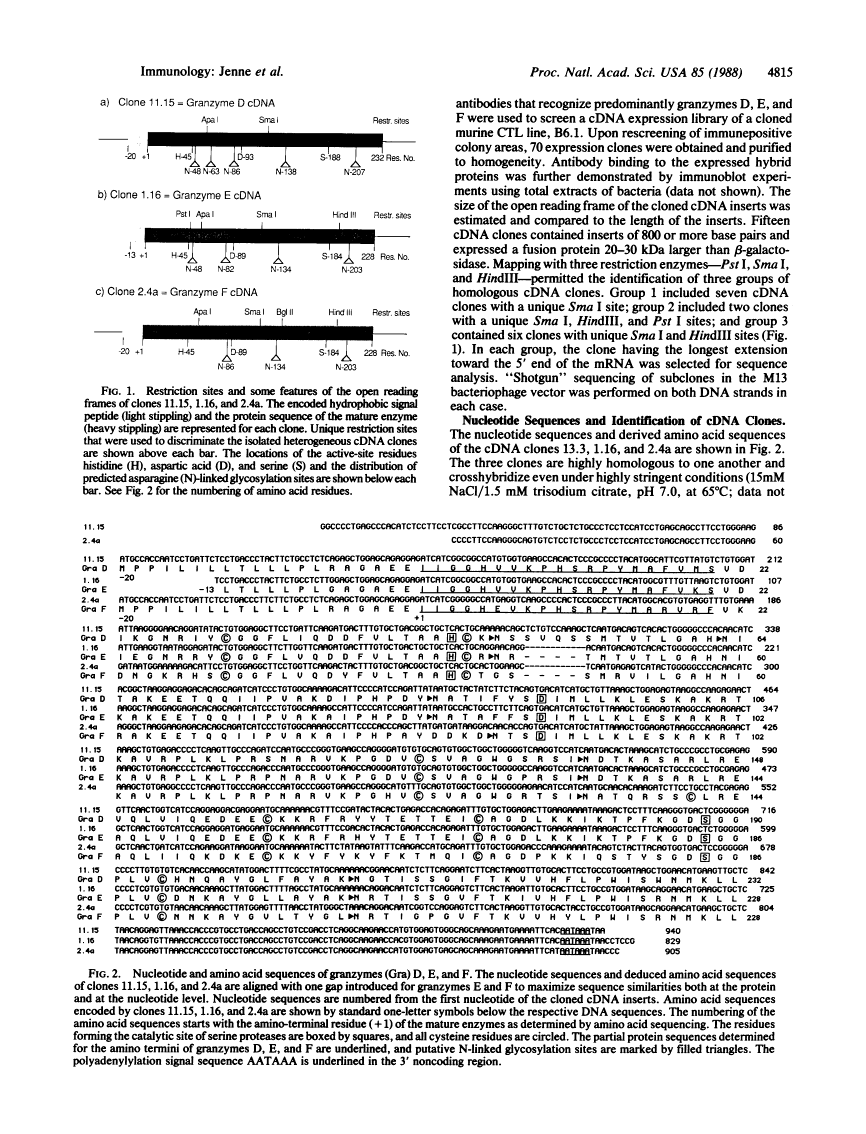
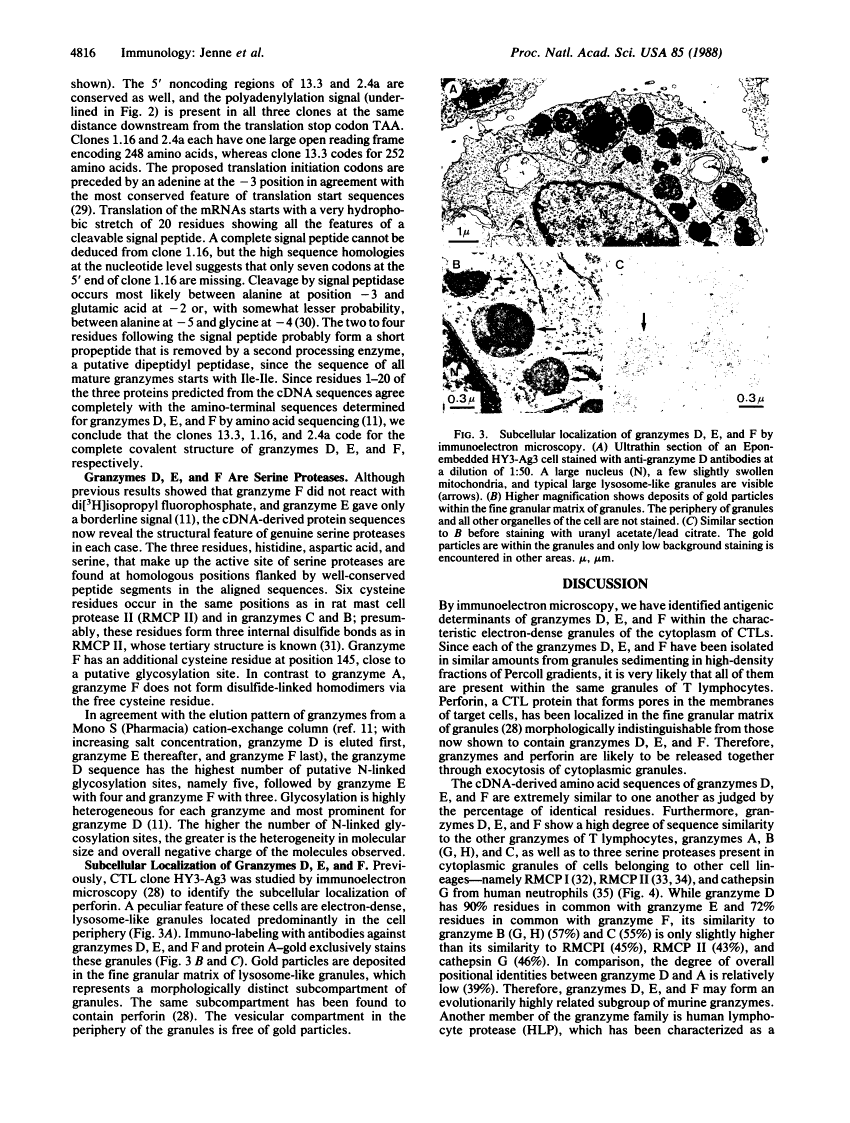
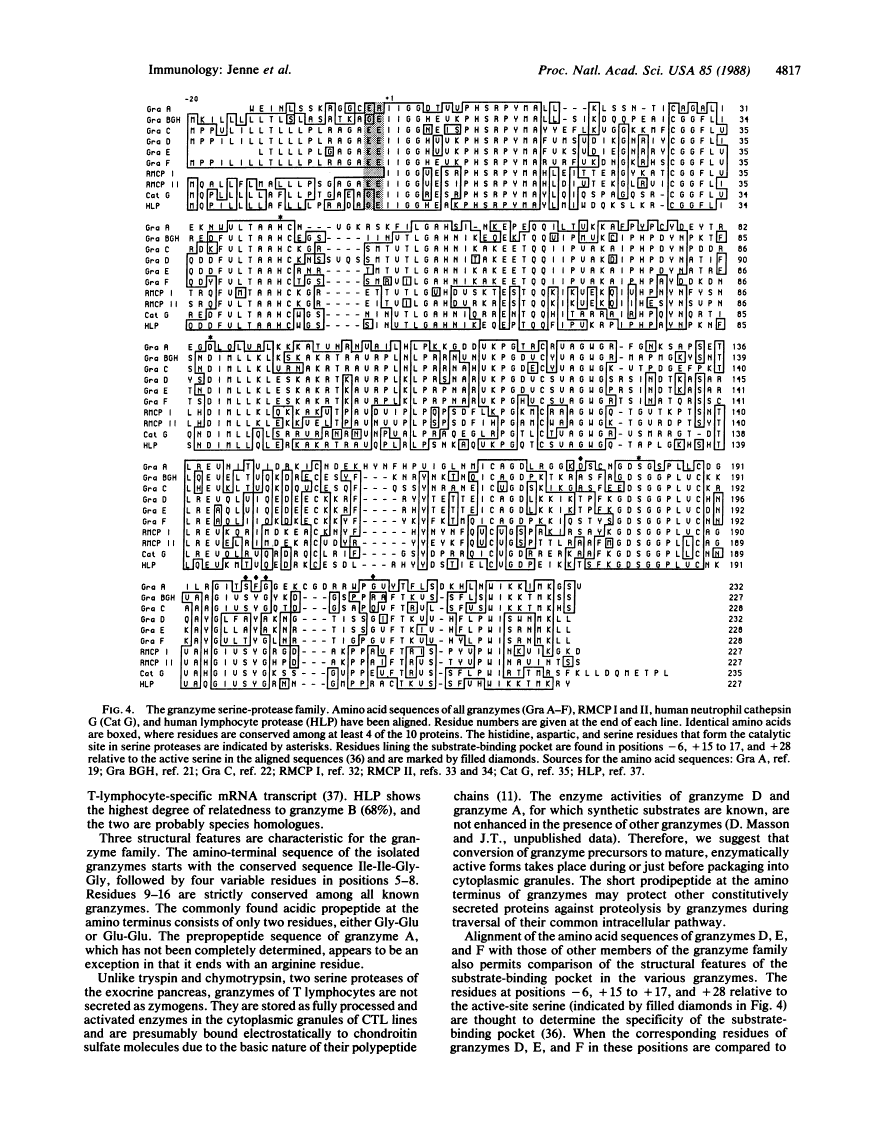
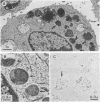
Cytoplasmic granules of cytolytic T lymphocytes contain at least six related serine esterases (granzymes) that are released together with perforin, a pore-forming protein related to complement component C9, during target-cell lysis. Polyclonal antibodies were used to isolate a large number of cDNA clones from an expression library derived from cytolytic-T-cell mRNA. Three distinct full-length cDNA clones coding for granzymes D, E, and F were identified by restriction site mapping and nucleotide sequencing. The three deduced amino acid sequences are highly similar to one another (between 72% and 90% amino acid identities) and to the sequences of granzymes B and C, cathepsin G, and rat mast-cell proteases I and II (between 43% and 57% amino acid identities). Cysteine residues capable of forming intramolecular disulfide bonds are conserved, as are the catalytic-site residues characteristic of serine proteases. Comparison of the cDNA-derived protein sequences with the amino termini of the isolated granzymes provides evidence that they are stored in a fully processed, activated form after removal of the signal peptide and two additional residues (propeptide) at the amino terminus. Immunoelectron microscopic studies demonstrated that granzymes D, E, and F are present in the same morphologically distinct cytoplasmic granules in which perforin has been found previously.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.4M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Henkart PA, Millard PJ, Reynolds CW, Henkart MP. Cytolytic activity of purified cytoplasmic granules from cytotoxic rat large granular lymphocyte tumors. J Exp Med. 1984 Jul 1;160(1):75–93. [Europe PMC free article] [Abstract] [Google Scholar]
- Podack ER, Konigsberg PJ. Cytolytic T cell granules. Isolation, structural, biochemical, and functional characterization. J Exp Med. 1984 Sep 1;160(3):695–710. [Europe PMC free article] [Abstract] [Google Scholar]
- Masson D, Corthésy P, Nabholz M, Tschopp J. Appearance of cytolytic granules upon induction of cytolytic activity in CTL-hybrids. EMBO J. 1985 Oct;4(10):2533–2538. [Europe PMC free article] [Abstract] [Google Scholar]
- Young JD, Leong LG, Liu CC, Damiano A, Cohn ZA. Extracellular release of lymphocyte cytolytic pore-forming protein (perforin) after ionophore stimulation. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5668–5672. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmidt RE, MacDermott RP, Bartley G, Bertovich M, Amato DA, Austen KF, Schlossman SF, Stevens RL, Ritz J. Specific release of proteoglycans from human natural killer cells during target lysis. Nature. 1985 Nov 21;318(6043):289–291. [Abstract] [Google Scholar]
- Pasternack MS, Eisen HN. A novel serine esterase expressed by cytotoxic T lymphocytes. Nature. 314(6013):743–745. [Abstract] [Google Scholar]
- Garcia-Sanz JA, Plaetinck G, Velotti F, Masson D, Tschopp J, MacDonald HR, Nabholz M. Perforin is present only in normal activated Lyt2+ T lymphocytes and not in L3T4+ cells, but the serine protease granzyme A is made by both subsets. EMBO J. 1987 Apr;6(4):933–938. [Europe PMC free article] [Abstract] [Google Scholar]
- Podack ER, Young JD, Cohn ZA. Isolation and biochemical and functional characterization of perforin 1 from cytolytic T-cell granules. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8629–8633. [Europe PMC free article] [Abstract] [Google Scholar]
- Masson D, Tschopp J. Isolation of a lytic, pore-forming protein (perforin) from cytolytic T-lymphocytes. J Biol Chem. 1985 Aug 5;260(16):9069–9072. [Abstract] [Google Scholar]
- Zalman LS, Brothers MA, Müller-Eberhard HJ. A C9 related channel forming protein in the cytoplasmic granules of human large granular lymphocytes. Biosci Rep. 1985 Dec;5(12):1093–1100. [Abstract] [Google Scholar]
- Masson D, Tschopp J. A family of serine esterases in lytic granules of cytolytic T lymphocytes. Cell. 1987 Jun 5;49(5):679–685. [Abstract] [Google Scholar]
- Simon MM, Hoschützky H, Fruth U, Simon HG, Kramer MD. Purification and characterization of a T cell specific serine proteinase (TSP-1) from cloned cytolytic T lymphocytes. EMBO J. 1986 Dec 1;5(12):3267–3274. [Europe PMC free article] [Abstract] [Google Scholar]
- Masson D, Nabholz M, Estrade C, Tschopp J. Granules of cytolytic T-lymphocytes contain two serine esterases. EMBO J. 1986 Jul;5(7):1595–1600. [Europe PMC free article] [Abstract] [Google Scholar]
- Young JD, Leong LG, Liu CC, Damiano A, Wall DA, Cohn ZA. Isolation and characterization of a serine esterase from cytolytic T cell granules. Cell. 1986 Oct 24;47(2):183–194. [Abstract] [Google Scholar]
- Pasternack MS, Verret CR, Liu MA, Eisen HN. Serine esterase in cytolytic T lymphocytes. Nature. 1986 Aug 21;322(6081):740–743. [Abstract] [Google Scholar]
- Zalman LS, Brothers MA, Chiu FJ, Müller-Eberhard HJ. Mechanism of cytotoxicity of human large granular lymphocytes: relationship of the cytotoxic lymphocyte protein to the ninth component (C9) of human complement. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5262–5266. [Europe PMC free article] [Abstract] [Google Scholar]
- Tschopp J, Masson D, Stanley KK. Structural/functional similarity between proteins involved in complement- and cytotoxic T-lymphocyte-mediated cytolysis. Nature. 322(6082):831–834. [Abstract] [Google Scholar]
- Young JD, Cohn ZA, Podack ER. The ninth component of complement and the pore-forming protein (perforin 1) from cytotoxic T cells: structural, immunological, and functional similarities. Science. 1986 Jul 11;233(4760):184–190. [Abstract] [Google Scholar]
- Gershenfeld HK, Weissman IL. Cloning of a cDNA for a T cell-specific serine protease from a cytotoxic T lymphocyte. Science. 1986 May 16;232(4752):854–858. [Abstract] [Google Scholar]
- Brunet JF, Dosseto M, Denizot F, Mattei MG, Clark WR, Haqqi TM, Ferrier P, Nabholz M, Schmitt-Verhulst AM, Luciani MF, et al. The inducible cytotoxic T-lymphocyte-associated gene transcript CTLA-1 sequence and gene localization to mouse chromosome 14. Nature. 1986 Jul 17;322(6076):268–271. [Abstract] [Google Scholar]
- Lobe CG, Finlay BB, Paranchych W, Paetkau VH, Bleackley RC. Novel serine proteases encoded by two cytotoxic T lymphocyte-specific genes. Science. 1986 May 16;232(4752):858–861. [Abstract] [Google Scholar]
- Jenne D, Rey C, Masson D, Stanley KK, Herz J, Plaetinck G, Tschopp J. cDNA cloning of granzyme C, a granule-associated serine protease of cytolytic T lymphocytes. J Immunol. 1988 Jan 1;140(1):318–323. [Abstract] [Google Scholar]
- Haymerle H, Herz J, Bressan GM, Frank R, Stanley KK. Efficient construction of cDNA libraries in plasmid expression vectors using an adaptor strategy. Nucleic Acids Res. 1986 Nov 11;14(21):8615–8624. [Europe PMC free article] [Abstract] [Google Scholar]
- Stanley KK. Solubilization and immune-detection of beta-galactosidase hybrid proteins carrying foreign antigenic determinants. Nucleic Acids Res. 1983 Jun 25;11(12):4077–4092. [Europe PMC free article] [Abstract] [Google Scholar]
- Biggin MD, Gibson TJ, Hong GF. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Groscurth P, Qiao BY, Podack ER, Hengartner H. Cellular localization of perforin 1 in murine cloned cytotoxic T lymphocytes. J Immunol. 1987 May 1;138(9):2749–2752. [Abstract] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. [Europe PMC free article] [Abstract] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. [Europe PMC free article] [Abstract] [Google Scholar]
- Le Trong H, Parmelee DC, Walsh KA, Neurath H, Woodbury RG. Amino acid sequence of rat mast cell protease I (chymase). Biochemistry. 1987 Nov 3;26(22):6988–6994. [Abstract] [Google Scholar]
- Woodbury RG, Katunuma N, Kobayashi K, Titani K, Neurath H, Anderson WF, Matthews BW. Covalent structure of a group-specific protease from rat small intestine. Appendix: crystallographic data for a group specific protease from rat intestine. Biochemistry. 1978 Mar 7;17(5):811–819. [Abstract] [Google Scholar]
- Benfey PN, Yin FH, Leder P. Cloning of the mast cell protease, RMCP II. Evidence for cell-specific expression and a multi-gene family. J Biol Chem. 1987 Apr 15;262(11):5377–5384. [Abstract] [Google Scholar]
- Salvesen G, Farley D, Shuman J, Przybyla A, Reilly C, Travis J. Molecular cloning of human cathepsin G: structural similarity to mast cell and cytotoxic T lymphocyte proteinases. Biochemistry. 1987 Apr 21;26(8):2289–2293. [Abstract] [Google Scholar]
- Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. [Abstract] [Google Scholar]
- Schmid J, Weissmann C. Induction of mRNA for a serine protease and a beta-thromboglobulin-like protein in mitogen-stimulated human leukocytes. J Immunol. 1987 Jul 1;139(1):250–256. [Abstract] [Google Scholar]
- Phillips M, Djian P, Green H. The nucleotide sequence of three genes participating in the adipose differentiation of 3T3 cells. J Biol Chem. 1986 Aug 15;261(23):10821–10827. [Abstract] [Google Scholar]
- Min HY, Spiegelman BM. Adipsin, the adipocyte serine protease: gene structure and control of expression by tumor necrosis factor. Nucleic Acids Res. 1986 Nov 25;14(22):8879–8892. [Europe PMC free article] [Abstract] [Google Scholar]
- Kelly RB. Pathways of protein secretion in eukaryotes. Science. 1985 Oct 4;230(4721):25–32. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.85.13.4814
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc280526?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
A transcriptomic study of myogenic differentiation under the overexpression of PPARγ by RNA-Seq.
Sci Rep, 7(1):15308, 10 Nov 2017
Cited by: 7 articles | PMID: 29127356 | PMCID: PMC5681552
Perforin and granzymes: function, dysfunction and human pathology.
Nat Rev Immunol, 15(6):388-400, 01 Jun 2015
Cited by: 582 articles | PMID: 25998963
Review
A quarter century of granzymes.
Cell Death Differ, 19(1):28-35, 04 Nov 2011
Cited by: 99 articles | PMID: 22052191 | PMCID: PMC3252830
Review Free full text in Europe PMC
Granzymes, cytotoxic granules and cell death: the early work of Dr. Jurg Tschopp.
Cell Death Differ, 19(1):21-27, 18 Nov 2011
Cited by: 15 articles | PMID: 22095283 | PMCID: PMC3252834
Review Free full text in Europe PMC
Perforin-mediated suppression of B-cell lymphoma.
Proc Natl Acad Sci U S A, 106(8):2723-2728, 05 Feb 2009
Cited by: 26 articles | PMID: 19196996 | PMCID: PMC2650333
Go to all (41) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Granzymes, a family of serine proteases released from granules of cytolytic T lymphocytes upon T cell receptor stimulation.
Immunol Rev, 103:53-71, 01 Mar 1988
Cited by: 149 articles | PMID: 3292396
Review
A family of serine esterases in lytic granules of cytolytic T lymphocytes.
Cell, 49(5):679-685, 01 Jun 1987
Cited by: 236 articles | PMID: 3555842
cDNA cloning of granzyme C, a granule-associated serine protease of cytolytic T lymphocytes.
J Immunol, 140(1):318-323, 01 Jan 1988
Cited by: 33 articles | PMID: 3257230
Isolation and sequence analysis of serine protease cDNAs from mouse cytolytic T lymphocytes.
J Exp Med, 168(5):1839-1854, 01 Nov 1988
Cited by: 23 articles | PMID: 3053963 | PMCID: PMC2189106