Abstract
Free full text

The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella
Abstract
Cilia and flagella are highly conserved microtubule (MT)-based organelles with motile and sensory functions, and ciliary defects have been linked to several human diseases. The 9 + 2 structure of motile axonemes contains nine MT doublets interconnected by nexin links, which surround a central pair of singlet MTs. Motility is generated by the orchestrated activity of thousands of dynein motors, which drive interdoublet sliding. A key regulator of motor activity is the dynein regulatory complex (DRC), but detailed structural information is lacking. Using cryoelectron tomography of wild-type and mutant axonemes from Chlamydomonas reinhardtii, we visualized the DRC in situ at molecular resolution. We present the three-dimensional structure of the DRC, including a model for its subunit organization and intermolecular connections that establish the DRC as a major regulatory node. We further demonstrate that the DRC is the nexin link, which is thought to be critical for the generation of axonemal bending.
Introduction
Cilia and flagella are important eukaryotic organelles that perform both motile and sensory functions. Most cell types in the human body assemble cilia, and they perform essential tasks throughout the life cycle, from early embryonic development to reproduction (Fliegauf et al., 2007). Cilia are also critical in the proper functioning of many organs, and thus, ciliary defects are associated with several diseases such as polycystic kidney disease, primary ciliary dyskinesia, Bardet-Biedl syndrome, infertility, and hydrocephalus (Fliegauf et al., 2007).
The core structure of most motile cilia and flagella, the 9 + 2 axoneme, is conserved from single-celled protists to mammals. It contains a central pair (CP) of singlet microtubules (MTs) surrounded by nine outer MT doublets (MTDs; Fig. 1 B). Each doublet contains a sequence of identical building blocks, which repeat longitudinally every 96 nm (Fig. 1 A). Neighboring doublets are interconnected circumferentially by nexin links and are attached to the CP of MTs through radial spokes (RSs; Fig. 1 B). The dynein motors responsible for motility are organized into two rows of outer and inner arms along the length of the doublets (Fig. 1, A and B). The axonemal dyneins are anchored to the A-tubule of each doublet in an ATP-insensitive manner and walk along the B-tubule of the neighboring doublet in an ATP-sensitive fashion, generating a local sliding motion between the doublets. Structures that restrict sliding such as the nexin links transform MT sliding into axonemal bending (Fig. 1, C and D; Satir, 1968; Summers and Gibbons, 1971).
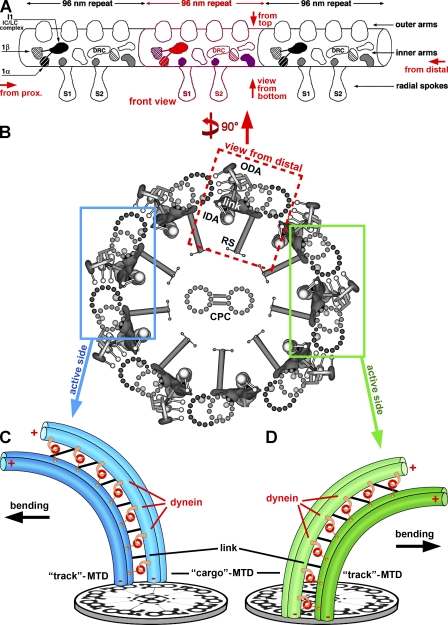
Schematic models showing the general organization of cilia and flagella and a model for the conversion of MTD sliding to bending. (A) Longitudinal view of an MTD with three 96-nm repeats shown in front view, i.e., looking from a neighboring B-tubule toward the A-tubule with its attached IDAs and ODAs. This is the classical view of the DRC density seen in previous EM studies (Mastronarde et al., 1992; Gardner et al., 1994). Red arrows indicate the orientation of additional views used in our figures: “view from bottom” looks at the 96-nm repeat from the center of the axoneme outwards, “from top” looks from the ODAs toward the axoneme center, “from prox.” is viewed from the flagellar base to its tip, and “from distal” is vice versa. (B) Simplified cross-sectional view of an axoneme with nine outer MTDs surrounding the CP complex (CPC); the viewing direction is from the flagellar tip (from distal). Three structures connect neighboring MTDs, the ODAs and IDAs and the nexin link (also called the circumferential link). One MTD is boxed in red and shown in longitudinal view in A. The blue and green boxes each highlight a pair of neighboring doublets from opposite sides of the axoneme. (C and D) Restricted interdoublet sliding model for ciliary and flagellar bend formation (Satir, 1968; Summers and Gibbons, 1971). Two pairs of MTDs from opposite sides of the axoneme, as highlighted in B, are shown. The dynein arms are anchored on the cargo MTD in an ATP-independent manner and walk toward the minus end (−) of the track MTD in an ATP-dependent manner, which causes sliding between neighboring MTDs. The links between MTDs are thought to be important for restricting and transforming MT sliding into bending. Note that the minus end–directed dynein motors (red) have to be active and produce MT sliding on alternate sides of the axoneme (switching between the blue and green side) to generate the effective and reverse bend (with opposite bending directions; black arrows). Panel A is adapted from Porter and Sale (2000). Panel B is adapted from Nicastro et al. (2006) with permission from Science.
Dyneins are minus end–directed motors (Sale and Satir, 1977), and their unidirectional movement along a B-tubule induces bending in one direction, whereas dyneins located on the opposite side of the axoneme induce bending in the opposite direction (Fig. 1, C and D; Satir, 1985). If all dyneins were active simultaneously, no effective beating and net propulsion would be produced. Therefore, to generate the ciliary and flagellar waveforms, dynein activity must be coordinated both along the length and around the circumference of the organelle (Satir, 1985; Smith and Sale, 1992). Structures important for the coordination of dynein activity include the CP complex, the RSs, the I1 inner arm dynein, and the dynein regulatory complex (DRC; Witman et al., 1978; Smith and Sale, 1992; Piperno et al., 1994; Smith, 2002; Bower et al., 2009). Signals are transmitted from the CP complex through the RSs to the inner dynein arms (IDAs). Both the intermediate chain–light chain (IC–LC) complex of the I1 dynein and the DRC are thought to distribute these signals to the IDAs and through the outer dynein–inner dynein (OID) linkers to the outer dynein arms (ODAs; Fig. 1; Nicastro et al., 2006).
Significant progress has been made on the identification of regulatory components within the CP (Mitchell, 2009) and RSs (Yang et al., 2006). Within the I1 dynein, the phosphorylation state of the intermediate chain IC138 is important for regulating MT sliding (Bower et al., 2009). Dynein activity also changes in response to changes in calcium concentration, perhaps a consequence of the calmodulin-containing complex associated with RSs (Dymek and Smith, 2007). Less is known about the structure and regulatory mechanism of the DRC. DRC4 is the PF2 gene product in Chlamydomonas reinhardtii and the only DRC component identified thus far (Rupp and Porter, 2003). Highly conserved orthologues have been identified in other species, in which they also play critical roles in ciliary and flagellar motility (Yeh et al., 2002; Rupp and Porter, 2003; Ralston and Hill, 2006; Colantonio et al., 2009).
The DRC is proposed to regulate dynein activity because mutations in DRC subunits (Table I) can suppress the paralysis caused by mutations in the RSs or CP (Brokaw et al., 1982; Huang et al., 1982; Piperno et al., 1994). The four drc mutants analyzed in this study exhibit distinct but overlapping defects in the assembly of at least seven axonemal polypeptides that are proposed to function as intermediates in the signaling pathway between the RSs and dynein arms (Table I). The structure of the DRC has previously been studied by EM of chemically fixed wild-type (WT) and drc mutant axonemes (Mastronarde et al., 1992; Gardner et al., 1994). When viewed in longitudinal section, the missing structure corresponded to a crescent-shaped density located between RS 2 and the ODAs (Fig. 1 A); however, specimen preparation limited image resolution (McIntosh et al., 2005). Recent advances in cryoelectron tomography (cryo-ET) now permit examination of the flagellum in its nearly native state (Nicastro et al., 2005, 2006; Bui et al., 2008). The 3D maps generated by cryo-ET have revealed new features such as the OID linkers and the first 3D views of the DRC in WT samples (Nicastro et al., 2006). However, to better understand how the DRC regulates dynein activity and flagellar beating, we need higher resolution images of the DRC and its interactions with other axonemal structures in both WT and mutant axonemes.
Table I.
Polypeptide deficiencies previously observed in drc mutants
| Polypeptides | Strain | ||||
| WT/pWT | sup-pf-4 | sup-pf-3 | pf2 | pf3 | |
| DRC component | |||||
| DRC1 (83) | + | + | + | + | − |
| DRC2 (70) | + | + | + | + | − |
| DRC3 (62) | + | + | +/− | − | +/− |
| DRC4 (55) | + | + | +/− | − | + |
| DRC5 (40) | + | − | − | − | − |
| DRC6 (29) | + | − | − | − | − |
| DRC7 (192)a | + | + | +/− | − | +/− |
| Other polypeptides | |||||
| Tektinb | +++ | +++ | +++ | +++ | + |
| Actinc | +++ | +++ | +++ | ++ | ++ |
| Centrinc | ++ | ++ | ++ | + | + |
| p28c | +++ | +++ | ++ | ++ | + |
| RSP13d | + | + | + | + | − |
| I2/I3 DHCsc | +++ | +++ | ++ | ++ | + |
| Dynein “e”e | + | + | +/− | +/− | − |
| Dynein “b”e | + | + | + | +/− | + |
DHC, dynein heavy chain. + indicates that the component is present; +/− indicates that the component is reduced; − indicates that the component is lacking. Unless noted otherwise, information is from Piperno et al. (1994). Molecular mass is indicated in parentheses (in kilodaltons).
The nexin link is important for the transformation of interdoublet sliding into axonemal bending (Summers and Gibbons, 1971; Warner, 1976). It was first discovered in EM images of axoneme cross sections as a structure linking neighboring MTDs (Gibbons, 1963). Later, freeze fracture experiments also visualized a candidate structure with bipartite ending (Bozkurt and Woolley, 1993). However, the identity of the nexin link has been a mystery for ~50 yr. No mutations or proteins have been unambiguously associated with the structure, and its mechanical properties are the subject of ongoing debate (Minoura et al., 1999; Lindemann et al., 2005). Although cryo-ET studies of the axoneme have provided new information, the precise location of the nexin link within the 96-nm repeat remains controversial (Nicastro et al., 2006; Bui et al., 2008).
Using cryo-ET, we studied the 3D structure of the DRC in WT and mutant axonemes of C. reinhardtii. Our study sheds light onto an old mystery: we demonstrate that the DRC is the nexin link. These results provide new insights into the protein composition of the nexin link and its potential functions. Moreover, we observed new connections between the DRC and other axonemal structures, indicating that the DRC is a central hub in the regulation of cilia and flagella.
Results
Based on their quality, 24 tomograms of intact, frozen-hydrated axonemes from six different strains were selected for tomographic averaging (Table II); selection criteria included good alignment and high signal to noise ratio as well as the absence of significant compression or distortion of the specimen (Fig. S1). For each sample, between 400 and 720 axonemal repeats from all MTDs were aligned and averaged in three dimensions, resulting in averaged volumes of the entire 96-nm repeat, including the MTs, RSs, IDAs, ODAs, and DRC (Fig. 2, A–I). The resolution of the final averages ranged from 3.2 to 4.0 nm, and, because we averaged multiple 96-nm repeats from different doublets around the axoneme, the resolution was relatively isotropic (Fig. S1 M and Table II).
Table II.
Characterization and image processing information for strains used in this study
| Name | Strain | Phenotype | Tomograms included | Number of particles averaged | Resolution |
| nm | |||||
| WT | CC-125, 137c mt+ | WT waveform, fast swimming | 4 | 610 | 4.0 |
| pWTa | pf2-4::PF2-GFP | WT waveform, fast swimming | 5 | 720 | 3.3 |
| sup-pf-4 | CC-2366, sup-pf-4 | WT waveform, slightly slower swimming | 3 | 500 | 3.8 |
| sup-pf-3 | CC-1399, sup-pf-3 | WT waveform, slow swimming | 3 | 500 | 3.7 |
| pf2b | pf2-4 | altered waveform, slow swimming | 6 | 400 | 3.9 |
| pf3 | CC-1026, pf3 | altered waveform, slow swimming | 3 | 500 | 3.8 |
Resolution is measured at the DRC. Unless otherwise noted, phenotype information is from Brokaw and Luck (1985), Brokaw and Kamiya (1987), and Gardner et al. (1994).

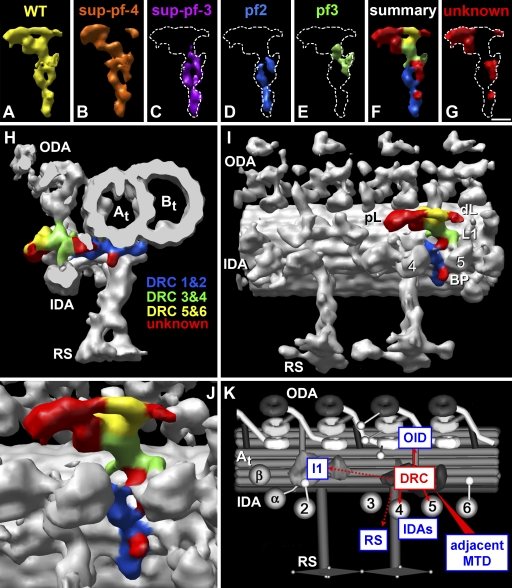
Views of the DRC in WT and a rescued drc mutant with WT phenotype (pWT). (A–F) Tomographic slices through averages of the 96-nm repeat obtained from WT (A–C) and a rescued pf2 mutant, pWT (D–F), showing the DRC in cross (A and D) and longitudinal sections (B, C, E, and F). The dashed red lines in A and D indicate the positions of the slices shown in the longitudinal views (B, C, E, and F); i.e., slices B and E are located close to the neighboring B-tubule (Bt; left), whereas slices C and F were taken closer to the A-tubule (At) of the doublet to which the DRC is attached. (G–K) 3D visualizations using isosurface rendering of averaged 96-nm repeats from WT (G and H) and pWT (I–K) in longitudinal front view (H and I), longitudinal bottom view (K), and cross-sectional view (G and J; for additional explanation of bottom and front views, see Fig. 1). (G–I) Overviews showing the general shape of the DRC in WT (G and H) and pWT (I) and its location within the 96-nm repeat in relation to other axonemal structures, including the two OID linkers (red arrowheads). Note that the IDAs just below the DRC are IA4 and -5. The light green–colored protrusion is an uncharacterized feature, which was previously proposed as a candidate for the nexin link (Bui et al., 2008). (J and K) Simplified surface-rendering visualization of the DRC (gold) represented by the pWT average, which has the highest resolution among our data. All structures except the DRC are either shown in gray transparency (J) or were removed (K). The DRC structures of WT and pWT are very similar (within the resolution limits) and share the same morphology, which consists of two parts, a base plate reaching from the B-tubule structure B11 to the A-tubule protofilament 4 and a linker that reaches from the A-tubule into the gap between neighboring MTDs. Please note that several different systems for numbering the MT protofilaments in the A- and B-tubules have been used (for review see Linck and Stephens, 2007). For consistency and clarity, we have chosen the system recently proposed by Linck and Stephens (2007). The linker ends in two lobes, the larger proximal lobe and the smaller distal lobe (dL). Four major protrusions can be distinguished: the base plate protrusion (BP), the OID linker (red arrowhead), and the two protrusions (L1 and L2) attached to the linker region. The only difference between WT and pWT is a small additional protrusion at the beginning of the base plate in pWT, which was not visible in WT or the other mutants; thus, it is not part of the common model of the DRC and was excluded from J and K. The inset explains the orientation of the DRC shown in K. Descriptions of the B11 structure have been controversial (Nicastro et al., 2006; Sui and Downing, 2006). Although we clearly observe density at this location, the structure appears thinner than a regular protofilament. The A-tubule is labeled as protofilaments 1–5; the B-tubule is labeled as protofilaments 8–10 and filament 11. Bar, 20 nm.
The architecture of the DRC in WT axonemes
Previous work showed that the DRC is located in the distal half of the 96-nm repeat (Mastronarde et al., 1992; Gardner et al., 1994). By comparing the two most severe drc mutants, pf2 and pf3, which together lack at least seven DRC components (Table I), we show in this study that the WT DRC is more complex and extended than previously reported (Fig. 2 and see Fig. 4), with a total length of ~50 nm (Fig. 2, A, D, G, and J; Fig. 3 C; and Video 1).

Connections within domains of the DRC and between the DRC and other axonemal structures. (A–G) A tomographic slice (A) and surface-rendering visualizations (B–G) of averaged 96-nm repeats of WT (C) and pWT (A, B, and D–G) are shown. A total of 10 connections are shown from different orientations. The connections are colored in magenta (isosurface and numbers 1–10). The DRC is colored yellow in WT (C) and gold in pWT (A, B, and D–G). (A–C) The DRC is the only structure in addition to the dynein arms that forms a continuous connection to the B-tubule (Bt) of the neighboring MTD (connections 1a and 1b), demonstrating that the DRC linker is the nexin link. (A and B) Similar bottom views of the DRC, but shown as tomographic slice (A) and surface-rendering representation (B). The position of the dynein stalks (arrowheads) and the IC–LC complex of the I1 dynein are indicated. (C) The cross-sectional view of WT shows that the DRC connects neighboring MTDs, whereas the uncharacterized protrusion (light green) distal of the DRC does not reach the neighboring B-tubule. (D–G) Surface renderings of the DRC and its connections to various axonemal structures from different viewpoints: distal (D), proximal (E), bottom (F), and top (G). Note that to allow better views of the DRC, different axonemal structures surrounding the DRC have been made transparent (D and E) or removed (F and G). Summary of connections: 1a and 1b, proximal and distal lobe (pL and dL) of the nexin link to neighboring B-tubule; 2, base plate to B11 structure; 3, OID link to ODA; 4, linker to tail of IA4; 5, L1 protrusion to the IA5 dynein head; 6 and 7, internal connections within the DRC linking the proximal and distal lobes (6) and the distal lobe and the L1 protrusion (7); 8, base plate to RS base; 9, DRC linker to an uncharacterized density that connects to the distal end of the IC–LC complex of I1 dynein; 10, DRC linker to an uncharacterized structure distal of the DRC. The light green protrusion shows an uncharacterized structure that was previously proposed as candidate for the nexin link (Bui et al., 2008). At, A-tubule; BP, base plate protrusion; L1 and L2, linker protrusions 1 and 2. Bar, 20 nm.

Comparison of the DRC structure between WT and drc mutants. (A–T) The WT DRC is displayed in yellow (A, F, and P; and pWT in K), and the mutant DRCs are shown in different colors and ordered from left to right by increasing size of structural changes: sup-pf-4 (B, G, L, and Q), sup-pf-3 (C, H, M, and R), pf2 (D, I, N, and S), and pf3 (E, J, O, and T). The averaged 96-nm repeats are visualized by isosurface rendering and are shown from four different viewing directions: longitudinal close-up view from the front (slightly rotated toward the bottom view; A–E), cross-sectional overview from distal end (F–J), and longitudinal close-up from the top (K–O) and bottom (P–T). Different parts of the DRC and associated structures are missing in the mutants. In sup-pf-4, the structural changes are small, with only the L2 protrusion (G and L) and most of the distal lobe (dL) missing (B and L). In all of the other mutants, both the proximal and distal lobes (pL and dL) and the L2 protrusion are missing; i.e., these strains have no DRC connection to the B-tubule (Bt) of the neighboring doublet (M–O). In addition, the DRC linker, including the OID linker (F and K, red arrowheads) to the ODAs, is reduced in length to varying degrees (compare H–J with M–O). In pf3, the entire DRC base plate (E, J, and T), including its protrusion (BP; J and T), connections 2 and 8 (see Fig. S2 J), and the hole (red arrow/dashed red circle) through the B11 structure (T), is missing. The base plate protrusion is also lacking in pf2 (I). The defects in the assembly of IDAs also increase from left to right; i.e., IA4 (A–E, left arrows) is reduced in sup-pf-3 (C) and pf2 (D) and missing in pf3 (E), whereas IA5 (right arrows) is reduced in pf2 (D) and missing in pf3 (E). A-tubule (At) includes protofilament A2; B-tubule includes filaments B10 and -11. The light green protrusion in A is an uncharacterized structure previously proposed as a candidate for the nexin link (Bui et al., 2008). L1, linker protrusion 1.
Transformation of the pf2 mutant with a GFP-tagged version of the WT PF2 gene rescues the motility defects (Bower, R., personal communication). Averages of WT and the rescued strain, which we denote pseudo-WT (pWT), are almost identical along the 96-nm repeat (Fig. 2, compare A–C, G, and H with D–F and I). However, some structural details such as the dynein stalks and tails are more easily visualized in the pWT averages (Figs. 2 D and 3 B and Fig. S3). Other minor variances between the WT and pWT structures such as differences in the thickness of some connections can be accounted for by differences in resolution. Therefore, we used averaged tomograms from both strains to identify the major domains within the structure of the WT DRC (Figs. 2 and and33 and Fig. S2).
The DRC contains two distinct domains, a base plate that is attached to the A-tubule and a linker that connects to the B-tubule of the neighboring doublet
The DRC can be subdivided into two distinct parts (Fig. 2 and Video 1). The first part is the base plate, which is attached to the underside of the A-tubule that faces the axoneme center (Fig. 2 J). The second part is the linker, which extends away from the A-tubule toward the B-tubule of the neighboring MTD (Fig. 2, G and J; Fig. 3, A–E; and Fig. S2, D–F). The base plate is located distal to RS 2 and extends from the junction of the A- and B-tubules to protofilament 4 of the A-tubule (Fig. 2 J). The base plate appears to be directly connected to the B11 density at the A-tubule–B-tubule junction (Fig. 2, G and J; and Fig. S2, I and J), and it also has one short appendage, the base plate protrusion (Fig. 2, J and K). Just proximal to the base plate connection, a 3–4-nm hole penetrates the MT wall at the B11 density (Fig. 4, P–S; and Fig. S2, G–J). From the volume measured in the tomographic averages, we estimated the molecular mass of the base plate to be ~300–350 kD (see Materials and methods for details).
The DRC linker begins at protofilament A4 and projects into the gap between neighboring doublets (Figs. 2 J and 3 C). The central region contains at least three different protrusions (L1, L2, and the distal OID linker; Fig. 2, H–J), and its total mass is estimated at ~1–1.2 MD. The L1 protrusion connects to inner arm dynein IA5 (Fig. 2 J and Fig. S3), and the OID linker connects the DRC to the ODAs (Nicastro et al., 2006). As the DRC linker approaches the neighboring doublet, it branches into two distinct lobes (Fig. 2 K). The proximal lobe is at least twice the size of the distal lobe, but both lobes connect to protofilament B8 of the neighboring B-tubule (Fig. 3, A–E; Fig. S2, D–F; and Fig. S3). Therefore, the DRC forms a continuous connection from the A-tubule to the B-tubule of the neighboring MTD (Fig. 3, C–E; Fig. S2, D–F; and Figs. S3 and S5 N). As the DRC is the only structure besides the dynein arms that connects adjacent outer doublets (Fig. 3, A–C; and Fig. S3 and S5, A–D), we conclude that the DRC is in fact the nexin link (Fig. S5, M–O).
Connections between the DRC and other structures in the 96-nm repeat
The DRC makes 10 connections to structures in the 96-nm repeat (Fig. 3, Fig. S2, and Video 2). Connections 1–3 are mentioned in the previous section (1a and 1b, proximal and distal lobes; 2, connection to the B11 structure; 3, OID linker) and are described in greater detail in Fig. 3 and Fig. S2. Connection 4 is the attachment of the dynein tail of IA4 to the base plate (Fig. 3 F and Fig. S2, C and E), and connection 5 is the connection between the L1 protrusion and IA5 dynein head domain (Figs. 2 J and 3, D and F; and Fig. S2, C and E).
Connections 6–10 are thin and most easily observed in the averages with the best resolution (Fig. 3, D–G; and Fig. S2, B–F). The proximal and distal lobes of the B-tubule linker appear to be interconnected by a thin bridge (6; Fig. 3, F and G; and Fig. S2, C–F), and the distal lobe is also connected to the L1 protrusion (7; Figs. 2 K and 3, D, F, and G; and Fig. S2, D–F). In addition, the base plate appears to connect to the base of RS 2 (8; Fig. S2 J). In pWT and sup-pf-4, we observed a fine connection (9) between the DRC linker and another structure just proximal of the DRC that connects to the IC–LC complex of the I1 inner dynein (Fig. 3, E and F; Fig. S2, A, B, E, and F; and Fig. S3). Finally, the DRC connects on its distal side to an uncharacterized structure (10; Fig. 3, A, B, D, F, and G; and Fig. S2, E and F).
In sup-pf-4, the distal lobe and its connection to the neighboring B-tubule are reduced
Cryo-ET of sup-pf-4 axonemes demonstrates the sensitivity of our approach for revealing structural details within the DRC. The sup-pf-4 strain was isolated as a suppressor of RS paralysis, and it is most similar to WT with respect to its motility phenotype (Table II). It only lacks two polypeptides (Table I; Huang et al., 1982), but a previous EM study of sup-pf-4 did not identify any clear differences in the DRC structure (Gardner et al., 1994). However, cryo-ET of sup-pf-4 axonemes has revealed that most of the distal lobe of the linker (Fig. 4, B and L; and Fig. S5, E and F) as well as the entire L2 projection (Fig. 4, G and L) are missing. This significantly reduces the size of distal lobe connection (1b) to the adjacent B-tubule (Fig. 4, B and L; Fig. 5 B; and Fig. S4, C–F). No differences have been observed in any other DRC domains. The estimated mass of missing density in sup-pf-4 is ~300 kD.
DRC subunit localization and model of the DRC interactions. (A–J) Averaged repeats from WT (A, F, and H–J) and drc mutants (B–E) are visualized by isosurface rendering. (A–E) Bottom views of the DRC structure in WT (A), sup-pf-4 (B), sup-pf-3 (C), pf2 (D), and pf3 (E; coloring as in Fig. 4). The dashed white line represents the shape of the WT DRC for better comparison (C–E and G). (F–J) Proposed locations of the DRC subunits, as suggested by our WT mutant comparisons. We correlated previously published data on missing subunits (Huang et al., 1982; Piperno et al. 1994) with structural data on the presence or absence of specific DRC features in the different drc mutants (see Discussion section “Location of DRC subunits within the NDRC”). The suggested locations of DRC subunits are summarized and colored (see color legend in H) within the WT DRC structure (F and H–J), which is shown from the bottom (F), in cross section (H), and as and overview (I) and close-up (J) in a longitudinal front-bottom view. The WT mutant comparison also revealed DRC structures that could not be correlated to the published biochemical DRC components, suggesting that these densities are unknown DRC subunits (colored red in F–J and highlighted in G). Note that subunit DRC7 was not included in the model because of uncertainty about its presence or absence in different mutants. (K) Simplified model that summarizes the structural connections between the WT DRC with other axonemal structures, indicating potential regulatory interactions. The IDA includes IA2–6 and I1 dynein with IC–LC complex and 1-α- and 1-β-dyneins. At, A-tubule; BP, base plate protrusion; Bt, B-tubule; dL, distal lobe; L1, linker protrusion 1; pL, proximal lobe. Bar, 10 nm.
sup-pf-3 and pf2 show significant changes in all parts of the DRC linker
Previous studies have shown that the sup-pf-3 mutation is associated with more significant polypeptide deficiencies and structural defects than sup-pf-4 (Table I; Huang et al., 1982; Gardner et al., 1994). Consistent with these observations, the tomographic average of sup-pf-3 showed large deficiencies in the DRC linker region. Its mass was reduced by ~1 MD, and missing structures included the L2 projection and both the proximal and distal lobes of the linker (Fig. 4, C, H, and M; and Fig. 5 C). Thus, the entire connection to the neighboring B-tubule was lost, and the total length of the linker was reduced to ~36 nm (Fig. 4, H and M; Fig. 5 C; Fig. S4, G–J; and Fig. S5, G and H). Relative to WT, both the OID linker and the L1 projection were reduced in density in the tomographic averages (Fig. 4, C, H, and M), as was the density of IA4 (see the smaller size of the IA4 dynein head in Fig. 4 C).
The tomographic average of pf2 axonemes (Fig. 4, D, I, N, and S; Fig. 5 D; and Fig. S4, K–N) displayed larger deficiencies in the DRC structure than those of either sup-pf-4 or sup-pf-3 (compare Fig. 5 [B–D] with Fig. S5 [E–J]), which is consistent with previous studies (Table I; Mastronarde et al., 1992; Gardner et al., 1994). In pf2, the DRC linker was almost completely reduced, and the missing structures included the proximal and distal lobe, all three linker projections (L1, L2, and the OID linker), and a major part of the central DRC linker (Fig. 4, D, I, and N). Only a small density of ~70 kD was observed at the transition between linker and base plate (Fig. 4, D and I). The pf2 averages also showed that the base plate protrusion was missing (Fig. 4, D, I, and S). We estimate the mass of the DRC remaining in pf2 to be ~240 kD, or only ~15% of the WT DRC structure. In addition, the two inner arm dyneins most closely associated with the DRC were mostly (IA4) and slightly (IA5) reduced (Fig. 4 D). These observations are consistent with previous studies of inner arm defects in pf2 (Gardner et al., 1994; Piperno et al., 1994; Bui et al., 2008).
Both the linker and base plate of the DRC are significantly reduced in pf3
The polypeptide defects observed in pf3 axonemes are the most severe reported thus far and also affected a different subset of DRC subunits (Table I). Tomographic averages of pf3 axonemes also showed the largest differences in the structure of the DRC. We estimate the missing density to be ~1.3 MD. The major difference between pf3 and other drc mutants was the extent of the defect in the base plate, which was completely missing (Fig. 4, E, J, and T; Fig. 5 E; Fig. S4, O–R; and Fig. S5, K and L), including the connection to the B11 density at the A-tubule–B-tubule junction (Fig. 4, J and T). Moreover, the pf3 average was the only one that did not have a hole through the B11 density (Fig. 4, compare T with P–S). The surface of the A-tubule normally occupied by the base plate appeared smooth and indistinguishable from neighboring regions (Fig. 4 T). Defects in the IDA of pf3 were more extensive than those of pf2: IA4 was completely missing, and IA5 was reduced (Fig. 4 E).
The tomograms of pf3 axonemes also showed significant differences in the linker region, although they did retain more of the linker (Fig. 4, E and J; and Fig. 5 E) than either sup-pf-3 or pf2. However, the pf3 average lacked both the proximal and distal lobes that connect to the neighboring B-tubule as well as the L2 projection (Fig. 4, E, J, and O). A small OID projection appeared to be present, but it was not connected to the ODAs (Fig. 4 J).
Discussion
Cryo-ET reveals that the WT DRC is a large and extended structure
Cryo-ET and volume averaging have been used to characterize the structure of the ODAs and IDAs (Nicastro et al., 2005; Ishikawa et al., 2007; Bui et al., 2008) and have revealed that the DRC is connected to other structures within the 96-nm repeat (Nicastro et al., 2006). However, the results shown in this study represent higher resolution structures (up to 3.2 nm) of intact axonemes (Fig. S1). Previous EM studies showed that the DRC is a large structure at the distal end of the repeat (Fig. 1 A), but the resolution was limited by the methods for structural preservation (Mastronarde et al., 1992; Gardner et al., 1994). Using rapid freezing, we preserved the structure in a close to native state and, with averaging, reconstructed the 96-nm unit with almost isotropic resolution from the RS heads to the ODAs (Fig. S1 M).
The increased resolution allowed us to see new domains within the complex structure of the DRC and to identify changes in the drc mutants not previously detected. The mutant structures were essential for determining the overall extension of the DRC structure. For instance, the presence of the DRC base plate and the extension of the DRC linker all the way to the neighboring B-tubule were both unexpected findings (Figs. 2 and and33 and Figs. S2 and S5). That the base plate is part of the DRC is supported by the observation that it is directly connected to the DRC linker (Fig. 2 and Fig. S3); however, the more crucial observation is that averages of the pf3 mutant lack the entire base plate and both lobes of the DRC linker (Fig. 4, E, J, O, and T). Therefore, the intact DRC stretches from the junction between the A- and B-tubules of one outer doublet across the interdoublet space to the B-tubule of the neighboring doublet, giving it an overall length of ~50 nm in cross section (Fig. 2 J and Fig. S5, N and O). Features of the DRC structure not seen previously include the hole through the B11 density at the B-tubule junction and the high connectivity to other axonemal structures (Fig. 3 and Fig. S2).
The DRC is the nexin link
The nexin link, originally seen by Gibbons (1963) in cross sections and later called the circumferential link (Fig. S5 M) is thought to be an elastic element involved in the conversion of sliding to bending (Satir, 1968, 1985), but the component proteins are unknown (Lindemann et al., 2005). Our images show that the DRC is the only structure, besides the IDAs and ODAs, that connects from the A-tubule of one doublet to the B-tubule of the neighboring doublet (Fig. 3, A–C; and Figs. S3 and S5) and therefore must be the nexin link. We suggest the term the nexin-DRC (NDRC).
The suggestion that the nexin link is in close proximity to or part of the DRC is not entirely new (Mastronarde et al., 1992; Gardner et al., 1994; Woolley, 1997; Nicastro et al., 2006). However, others have suggested that a density distal of the DRC is the nexin link (Bui et al., 2008). We also identify this distal density in our images (colored light green in Fig. 2, G–I; Fig. 3 C; and Fig. S2, D–F), but it is only ~17 nm in length, i.e., ~10 nm shy of connecting to the neighboring B-tubule (Fig. 3 C; Fig. S2, D–F; and Fig. S3). At our resolution, we see no evidence of connections between this distal density and the DRC or between this density and the neighboring B-tubule, which suggests that it is not part of the nexin link.
Location of DRC subunits within the NDRC
The drc mutants were previously characterized by SDS-PAGE, and to date, seven DRC subunits have been described as missing spots or bands in different mutants (Huang et al., 1982; Piperno et al., 1994; Piperno, 1995). By correlating published data with the distribution of missing densities in our images, we can identify the likely locations of the DRC subunits and other polypeptides whose assembly depends on their presence (Fig. 5, F–J; and Video 3). We also observed structures that cannot be easily correlated with any of the known subunits (Fig. 5 G and Table III). We estimate the total mass of the WT DRC to be ~1.4–1.5 MD. This is nearly three times larger than the sum of all seven DRC subunits (~550 kD; Piperno et al., 1994). Although there is no information about the stoichiometry of DRC1–7, it seems likely that the DRC contains other, unidentified proteins.
Table III.
Summary of structural components present or absent in WT and drc mutants
| Structural component | Strain | ||||
| WT/pWT | sup-pf-4 | sup-pf-3 | pf2 | pf3 | |
| DRC linker (1,000–1,200) | |||||
| Nexin link connection to neighboring B-tubule | |||||
    Proximal lobe (~350) Proximal lobe (~350) | + | + | − | − | − |
    Distal lobe (~120) Distal lobe (~120) | + | +/− | − | − | − |
| Linker projections | |||||
    L1 (50–80) L1 (50–80) | + | + | +/− | − | +/− |
    L2 L2 | + | − | − | − | − |
    OID linker OID linker | + | + | +/− | − | +/− |
| Central linker region | |||||
    Central linker Central linker | + | + | +/− | − | +/− |
    Docking subunit (~60) Docking subunit (~60) | + | + | + | + | + |
| DRC base plate (300–350) | |||||
    Base plate protrusion (~80) Base plate protrusion (~80) | + | + | + | − | − |
    B11-A3 section B11-A3 section | + | + | + | + | − |
    Connection to B11 Connection to B11 | + | + | + | + | − |
    B-tubule hole B-tubule hole | + | + | + | + | − |
| IDAs | |||||
    IA4 IA4 | + | + | +/− | +/− | − |
    IA5 IA5 | + | + | + | +/− | (−) |
+ indicates that the component is present; +/− indicates that the component reduced; (−) indicates that the component is almost completely reduced; − indicates that the component is lacking. Molecular mass is indicated in parentheses (in kilodaltons).
Averages of sup-pf-4 lack most of the distal lobe and portion of the B-tubule connection (1b) and the L2 projection of the DRC linker (Fig. 4, B, G, and L; and Fig. 5 B). These structures are missing in all drc mutants (Fig. 4, L–O; and Fig. 5, B–E). sup-pf-4 axonemes lack two subunits, DRC5 (40 kD) and -6 (29 kD); these are also missing in all other drc mutants (Table I; Huang et al., 1982; Piperno et al., 1994). Thus, DRC5 and -6 must be located within the distal lobe of the B-tubule connector and/or the L2 projection (Fig. 5, F and H–J, yellow). However, the estimated loss of mass in sup-pf-4 of ~200–300 kD is greater than the combined mass of DRC5 and -6 (70 kD). Thus, either one or both subunits must be present in multiple copies, or there are additional unidentified DRC subunits whose assembly depends on the presence of DRC5 or -6 (Table III). Perhaps because the proximal lobe of the linker and B-tubule connector is unchanged in sup-pf-4 (Fig. 4, B and L), only minor changes in swimming behavior have been observed (Brokaw and Luck, 1985; Gardner et al., 1994).
The structural changes in the other three drc mutants are larger. pf2 axonemes lack at least five DRC subunits (DRC3–7) but retain DRC1 and -2 (Huang et al., 1982; Piperno et al., 1994). These observations suggest that DRC3, -4, and -7 are located in the missing linker, whereas DRC1 and -2 are located in the base plate structure that remains in pf2 and that reaches from the B-tubule junction to protofilament A4 (Fig. 5, F and H–J, blue). Consistent with this hypothesis, the base plate structure is missing in pf3 axonemes, which lack DRC1 and -2 (Table I). However, two features of the base plate are probably not associated with either DRC1 or -2. The first is the ~80-kD base plate projection, which is missing in pf2 (Fig. 4 I) even though DRC1 and -2 are present (Tables I and III). The second is an ~60-kD structure located near protofilament A4 that is present in both pf2 and pf3 (Fig. 5, D and E). We refer to the latter density as the docking subunit (Table III) because it appears to be the last contact of the DRC linker with the A-tubule. If one subtracts this subunit from the total estimated mass of the DRC structure in pf2, the remaining ~190 kD corresponds well with the estimated size of the DRC1–DRC2 complex (Tables I and III). The identity of the docking subunit, which is present in all of the mutants, is unknown.
Comparison of pf2 and sup-pf-3 indicates that DRC4 plays a pivotal role in the assembly of the DRC linker. pf2 mutations are null alleles in the DRC4 gene, whereas sup-pf-3 is an unusual DRC4 mutation that results in the assembly of truncated DRC4 subunits (Rupp and Porter, 2003; Bower, R., personal communication). Loss of DRC4 blocks the assembly of DRC3–7 in pf2 (Piperno et al., 1994). sup-pf-3 also lacks DRC3–6, but studies vary regarding the extent of these defects (Huang et al., 1982; Piperno et al., 1994; Piperno, 1995). The truncated forms of DRC4 in sup-pf-3 appear to be sufficient for the assembly of a larger part of the DRC linker as compared with pf2 (Fig. 4, H vs. I). These results place DRC4 near the base of the linker, close to the unknown ~60-kD docking subunit (Fig. 5, F and H–J). Consistent with this hypothesis, transformation of pf2 with WT or epitope-tagged versions of DRC4 rescued the pf2 motility defects and restored the missing DRC structures (Fig. 2, D–F and I; Rupp and Porter, 2003)
The pf3 averages show the largest defects; the entire base plate and both linker lobes are missing, and only a small portion of the linker close to the A-tubule remains (Figs. 4 J and 5 E). We estimate the size of the pf3 linker to be ~130–200 kD, compared with 1.2 MD for the WT linker, which means a loss of >80% of the WT mass. However, the pf3 linker appears too small to contain the remaining DRC subunits (DRC3, -4, and -7 and the docking subunit). One possibility is that the loss of the base plate results in increased flexibility of the remaining DRC structures. Disorganization, structural flexibility, or polypeptides that are present only in a subset of the repeats could reduce the apparent size of the structures in the final average. In addition, the absence of the pf3 gene product appears to destabilize the assembly of several other axonemal polypeptides that may contribute to the assembly of the DRC linker (Table I).
To summarize, we predict that DRC1 and -2 are located within the base plate, DRC5 and -6 (and perhaps additional polypeptides) are located in the distal lobe of the B-tubule connector, and DRC3, -4, and -7 are located within the central region of the linker (Fig. 5, F and H–J). However, this means that the proximal lobe (estimated at ~350 kD) and a small portion of the distal branch (~50 kD) cannot be correlated with any known DRC subunits, even though they are missing in sup-pf-3, pf2, and pf3 (Fig. 5 G and Table III). We predict that at least 500 kD, i.e., approximately one third of the total mass of the DRC, is comprised of DRC components that were not previously detected as missing in earlier 1D or 2D PAGE analyses. Efforts to identify these other DRC subunits are underway.
The nexin link as a regulator of dynein activity
The conclusion that the DRC subunits are major components of the nexin link has important implications for nexin function. One surprising observation of our tomograms is that none of the drc mutants display defects in the arrangement of the nine outer MTDs, even though the nexin links no longer connect neighboring doublets (Fig. 4, M–O). These results indicate that other structures such as the dynein arms and/or RSs are sufficient to maintain axoneme integrity in the absence of the nexin links. More importantly, our findings agree well with the hypothesis that the primary functions of the nexin link are signal transduction and local restriction of interdoublet sliding (Lindemann et al., 2005). In the field, the DRC and nexin link were regarded as two distinct structures with different functions, dynein regulation versus limiting doublet sliding, we now realize that both functions are performed by one and the same complex, the NDRC. This conclusion is not only important for future work, but it might require reevaluation of previous studies, as e.g., protease treatment could have not only weakened the nexin linkage but also interfered with dynein regulation.
It is still unclear what happens to the NDRC links during interdoublet sliding. Do the nexin links stretch and extend (Warner, 1976; Olson and Linck, 1977), e.g., by protein unfolding (Lindemann et al., 2005), or do they release and reattach after sliding along the neighboring B-tubule (Bozkurt and Woolley, 1993; Minoura et al., 1999)? One model is that the nexin link has elastic properties that contribute to the passive (i.e., non ATP dependent) recovery of the axoneme after bending. However, Bozkurt and Woolley (1993) found no evidence of stretched nexin links in bull sperm and suggested instead a displacement mechanism. We did not find indications of stretching of the nexin link in bent regions of our axoneme samples. However, this is not conclusive, as our samples were not actively beating and any curvature present was presumably passively imposed during blotting and plunge freezing. Future studies using actively beating axonemes may resolve this important question.
The highly connected NDRC as the central hub of the regulatory system
The NDRC is highly connected with other axonemal structures (Figs. 3 and 5 K and Fig. S2), which is consistent with the proposed role of the DRC as a key intermediate in the signaling pathway between the CP–RS complex and the dynein arms (Brokaw et al., 1982; Huang et al., 1982; Mastronarde et al., 1992; Gardner et al., 1994; Piperno et al., 1994; Rupp and Porter, 2003; Nicastro et al., 2006). The highest resolution tomograms revealed delicate connections to RS 2 (Fig. S2 J) and inner arms IA4 and -5 (Fig. S2, C and E) as well as the previously described OID linker (Fig. 3, D and E; and Fig. S2, B and C; Nicastro et al., 2006). The CP–RS complex is thought to selectively activate dyneins on a subset of outer doublets during axonemal bending (Fig. 1, B–D; for review see Smith and Yang, 2004). In the absence of the CP or RSs, dynein activity and MT sliding is inhibited, resulting in flagellar paralysis (Witman et al., 1978; Huang et al., 1982; for review see Smith and Yang, 2004). Mutations in the DRC can partially suppress the paralysis observed in CP–RS mutants (Huang et al., 1982). These genetic interactions led to a model in which the WT function of the DRC is to inhibit dynein activity in the absence of signals from the RSs (Brokaw et al., 1982). Our experiments agree with this model and suggest that the DRC inhibits dynein activity and MT sliding in the distal half of the 96-nm repeat through the constraints imposed by the nexin link. How signals from the RSs alter the conformation or activities of the NDRC remains a fascinating question, but the NDRC is clearly one of two central hubs for signal transduction and integration that regulate dynein activity and flagellar motility (Fig. 5 K).
Our highest resolution images also provide evidence for a thin connection (9) between the DRC and its counterpart in the proximal half of the 96-nm repeat, the I1 inner arm complex (Fig. S2, A, B, and F). Recent work has demonstrated that the IC–LC complex of the I1 dynein is located above the base of RS 1 and also serves to regulate MT sliding in response to signals from the RSs (Bower et al., 2009; Wirschell et al., 2009a,b). A connection between the DRC and I1 dynein suggests the possibility of feedback or cross talk between these two regulatory modules within the 96-nm repeat.
The realization that the DRC is the nexin link also suggests the possibility of a circumferential signal pathway that could regulate dynein activity around the axoneme. One of our more surprising findings was the connection of the DRC base plate to the B11 density and the associated hole through the B-tubule wall at one end of the DRC (Fig. 4 P and Fig. S2, G–J). This hole suggests the presence of a discontinuity in the B11 protofilament that could potentially contribute to the formation of the 96-nm repeat. The other end of the DRC makes contact with the B8 protofilament of the neighboring outer doublet (Fig. 3, A–E; Fig. S2, D–F; and Figs. S3 and S5 N). One interesting possibility is that the NDRC communicates information about strain or tension around the circumference of the axoneme structure from one doublet to the next during axonemal bending. There are species with motile cilia and flagella such as eel sperm that don't have CP MTs or RSs but retain IDAs and nexin links and beat with a helical waveform (Gibbons et al., 1985; Woolley, 1997). How is dynein activity regulated between the opposite sides of the axoneme in these flagella? Biophysical studies have indicated that sequential activation of dynein arms occurs along the length of the outer doublets, but the complexity of the observed waveforms suggest that additional factors such as the nexin links contribute to the regulation of motility in these and other systems (Woolley, 1998a,b; Woolley and Vernon, 2001; Lindemann et al., 2005). As more information becomes available about the identity of DRC subunits, it will be interesting to determine their distribution in other types of cilia and flagella.
Understanding how proteins interact and are spatially arranged within functional modules in vivo to form a functioning organelle or organism is a great challenge in modern structural biology. The technical approach used in this study allowed us to integrate genetic, biochemical, and structural information to not only present a high resolution 3D structure and subunit map of the NDRC but also provide new insights into the inner workings of the regulatory system in cilia and flagella. More such studies should lead to a comprehensive, molecular blueprint of the axoneme, with a better understanding of protein interactions and regulatory mechanisms, and ultimately reveal the mechanism of this remarkable nanomachine and how it fails in ciliary diseases.
Materials and methods
Axoneme preparation
Strains used in this study are listed in Table II. The pWT strain (pf2-4::PF2-GFP) was provided by R. Bower (University of Minnesota, Minneapolis, MN). Preparation of axonemes was performed as previously described (Witman et al., 1972; Rupp et al., 1996; Harris et al., 2009). In brief, strains of C. reinhardtii were grown on solid Tris-acetate-phosphate medium for 5–7 d (Harris et al., 2009). Cells were resuspended in minimal medium for at least 1 h to allow regeneration of flagella and then collected by centrifugation, washed two times to remove the cell wall and other debris, and resuspended in 10 mM Hepes buffer, pH 7.4 (1 mM SrCl2, 4% sucrose, and 1 mM DTT). Flagella were detached from the cells using the pH-shock method (Witman et al., 1972) followed by the addition of 5 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA, and 1 µg/ml aprotinin, leupeptin, and pepstatin. After centrifugation (1,000 g at 4°C for 5–10 min), the flagella-containing supernatant was purified away from the cell bodies and debris by two additional centrifugation steps over a 20% sucrose cushion. Flagella were demembranated with 0.1% IGEPAL CA-630 (Sigma-Aldrich), and axonemes were collected by centrifugation at 35,000 g for 60 min. The pellet was washed and resuspended in HMEEN buffer (30 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA, 25 mM NaCl, 1 mM DTT, and 0.1 µg/ml aprotinin, leupeptin, and pepstatin). Axonemes were stored at 4°C, shipped overnight on wet ice, and usually plunge frozen within 24 h.
Preparation of cryo samples
Cryo samples were prepared using Quantifoil grids (Quantifoil Micro Tools GmbH) with holey carbon support film (copper; 200 mesh; R2/2) and a homemade guillotine device (plunge freezer). After glow discharging grids for 30 s at −40 mA, 10-nm colloidal gold (Sigma-Aldrich) was applied and dried onto the grid for later use as fiducial markers in the tilt series alignment process. 3 µl of axoneme sample and 1 µl of a 10-fold–concentrated 10-nm colloidal gold solution were applied to the grid, briefly mixed, and blotted from the front side for 1.5–2.5 s with filter paper. Immediately after this, the grid was plunge frozen into liquid ethane that was cooled by liquid nitrogen to achieve vitrification of the axoneme sample. All samples were stored in liquid nitrogen until examination by cryo-EM.
EM
All electron micrographs were digitally recorded using a sensitive 2,000 × 2,000 charge-coupled device camera (Gatan, Inc.) on a transmission electron microscope (Tecnai F30; FEI, Inc.) with field-emission gun, high tilt stage, and postcolumn energy filter (Gatan, Inc.). Cryo samples were loaded into the microscope using a cryo-transfer holder (Gatan, Inc.) and kept below devitrification temperature at all times. Tilt series were recorded of intact, noncompressed axonemes located within holes in the carbon film. For a single-axis tilt series, typically 60–100 images were recorded at a magnification of 13,500 while tilting the sample from about −65 to 65° with 1.5–2.5° tilting increments. The defocus values ranged from −6 to −8 µm, and the specimen exposure was limited to a total electron dose of ~100 e/Å2 to minimize radiation damage of the sample. The microscope was operated at 300 keV, and imaging was performed under low dose mode conditions using the energy filter in the zero-loss mode with a slit width of 20 eV to enhance contrast. Acquisition of overview maps and tilt series was accomplished using the microscope control program SerialEM (Mastronarde, 2005).
Image processing
The tilt series images were aligned, reconstructed into tomograms, and analyzed using the IMOD software package (Kremer et al., 1996). Repetitive structures, i.e., the 96-nm repeat unit of the axoneme, were aligned and averaged with missing wedge compensation using the software program PEET (particle estimation for ET; Nicastro et al., 2006). Details about number of tomograms and particles included in the 3D averaged structures as well as the resulting image resolutions using the 0.5 Fourier shell correlation criterion (Böttcher et al., 1997) are summarized in Table II. Contrast transfer function and modulation transfer function corrections were calculated using the IMOD software (Xiong et al., 2009). The UCSF Chimera package (Pettersen et al., 2004) was used for 3D visualization by isosurface rendering, analysis, mass estimations, and the calculation of difference maps (Fig. S4). Mass estimations of the DRC complex and subvolumes were calculated using the average density of 1.43 g/cm3 for proteins (Quillin and Matthews, 2000) and after normalizing the isosurface-rendering threshold to the mass of MTs.
Online supplemental material
Fig. S1 shows the quality of the tomographic reconstructions of frozen-hydrated axonemes in this study, and it displays details on the resolution measurements at different locations within the averaged 96-nm repeat of pWT. Fig. S2 provides additional details to Fig. 3 about the connections of the DRC. Fig. S3 shows a gallery of sequential tomographic slices through the same averaged repeat from this mutant. Fig. S4 shows that difference maps between the WT and mutant averages highlight both the general consistency of the axonemal averages and the changes in the DRC structure. Fig. S5 shows that the presence or absence of the NDRC in WT and drc mutant strains can be seen in close-up EM and ET images. Videos 1–3 show isosurface rendering of averaged axonemal repeats from WT (Videos 1 and 3) and pWT (Video 2). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908067/DC1.
Acknowledgments
We wish to thank Raqual Bower for providing the pf2 rescue strain and discussions of unpublished data and Claudia G. Vasquez and Matthew R. Rebesco for their help with tomogram reconstruction and image processing. We are grateful to Dr. Chen Xu for his excellent management of the EM facility at Brandeis University and to Dr. David DeRosier for proofreading the manuscript.
This work was supported by grants from the National Institutes of Health (GM083122 to D. Nicastro and GM55667 to M.E. Porter) and the National Science Foundation (DMR-MRSEC-0820492) and by a Pew Scholars Award (to D. Nicastro).
Footnotes
Abbreviations used in this paper:
- CP
- central pair
- DRC
- dynein regulatory complex
- ET
- electron tomography
- IC–LC
- intermediate chain–light chain
- IDA
- inner dynein arm
- MT
- microtubule
- MTD
- MT doublet
- NDRC
- nexin-DRC
- ODA
- outer dynein arm
- OID
- outer dynein–inner dynein
- pWT
- pseudo-WT
- RS
- radial spoke
- WT
- wild type
References
- Böttcher B., Wynne S.A., Crowther R.A. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 386:88–91 10.1038/386088a0 [Abstract] [CrossRef] [Google Scholar]
- Bower R., VanderWaal K., O'Toole E., Fox L., Perrone C., Mueller J., Wirschell M., Kamiya R., Sale W.S., Porter M.E. 2009. IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol. Biol. Cell. 20:3055–3063 10.1091/mbc.E09-04-0277 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bozkurt H.H., Woolley D.M. 1993. Morphology of nexin links in relation to interdoublet sliding in the sperm flagellum. Cell Motil. Cytoskeleton. 24:109–118 10.1002/cm.970240204 [Abstract] [CrossRef] [Google Scholar]
- Brokaw C.J., Kamiya R. 1987. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton. 8:68–75 10.1002/cm.970080110 [Abstract] [CrossRef] [Google Scholar]
- Brokaw C.J., Luck D.J.L. 1985. Bending patterns of chlamydomonas flagella: III. A radial spoke head deficient mutant and a central pair deficient mutant. Cell Motil. 5:195–208 10.1002/cm.970050303 [Abstract] [CrossRef] [Google Scholar]
- Brokaw C.J., Luck D.J., Huang B. 1982. Analysis of the movement of Chlamydomonas flagella: the function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella. J. Cell Biol. 92:722–732 10.1083/jcb.92.3.722 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol. 183:923–932 10.1083/jcb.200808050 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Colantonio J.R., Vermot J., Wu D., Langenbacher A.D., Fraser S., Chen J.N., Hill K.L. 2009. The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature. 457:205–209 10.1038/nature07520 [Abstract] [CrossRef] [Google Scholar]
- Dymek E.E., Smith E.F. 2007. A conserved CaM- and radial spoke–associated complex mediates regulation of flagellar dynein activity. J. Cell Biol. 179:515–526 10.1083/jcb.200703107 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 10.1038/nrm2278 [Abstract] [CrossRef] [Google Scholar]
- Gardner L.C., O'Toole E., Perrone C.A., Giddings T., Porter M.E. 1994. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J. Cell Biol. 127:1311–1325 10.1083/jcb.127.5.1311 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gibbons B.H., Baccetti B., Gibbons I.R. 1985. Live and reactivated motility in the 9+0 flagellum of Anguilla sperm. Cell Motil. 5:333–350 10.1002/cm.970050406 [Abstract] [CrossRef] [Google Scholar]
- Gibbons I.R. 1963. Studies on the protein components of cilia from Tetrahymena pyriformis. Proc. Natl. Acad. Sci. USA. 50:1002–1010 10.1073/pnas .50.5.1002 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Harris E.H., Stern D.B., Witman G.B. 2009. The Chlamydomonas Sourcebook. Second edition Academic Press, Oxford, UK: 2000 pp [Google Scholar]
- Huang B., Ramanis Z., Luck D.J. 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for Flagellar function. Cell. 28:115–124 10.1016/0092-8674(82)90381-6 [Abstract] [CrossRef] [Google Scholar]
- Ishikawa T., Sakakibara H., Oiwa K. 2007. The architecture of outer dynein arms in situ. J. Mol. Biol. 368:1249–1258 10.1016/j.jmb.2007.02.072 [Abstract] [CrossRef] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [Abstract] [CrossRef] [Google Scholar]
- Linck R.W., Stephens R.E. 2007. Functional protofilament numbering of ciliary, flagellar, and centriolar microtubules. Cell Motil. Cytoskeleton. 64:489–495 10.1002/cm.20202 [Abstract] [CrossRef] [Google Scholar]
- Lindemann C.B., Macauley L.J., Lesich K.A. 2005. The counterbend phenomenon in dynein-disabled rat sperm flagella and what it reveals about the interdoublet elasticity. Biophys. J. 89:1165–1174 10.1529/biophysj.105.060681 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 10.1016/j.jsb.2005.07.007 [Abstract] [CrossRef] [Google Scholar]
- Mastronarde D.N., O'Toole E.T., McDonald K.L., McIntosh J.R., Porter M.E. 1992. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J. Cell Biol. 118:1145–1162 10.1083/jcb.118.5.1145 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- McIntosh R., Nicastro D., Mastronarde D. 2005. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 15:43–51 10.1016/j.tcb.2004.11.009 [Abstract] [CrossRef] [Google Scholar]
- Minoura I., Yagi T., Kamiya R. 1999. Direct measurement of inter-doublet elasticity in flagellar axonemes. Cell Struct. Funct. 24:27–33 10.1247/csf.24.27 [Abstract] [CrossRef] [Google Scholar]
- Mitchell D.R. 2009. The flagellar central pair apparatus. In The Chlamydomonas Sourcebook: Cell Motility and Behavior, vol. 3. Second edition. E.H. Harris, D.B. Stern, and G.B. Witman, editors Academic Press, Oxford, UK: 235–252 [Google Scholar]
- Nicastro D., McIntosh J.R., Baumeister W. 2005. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. USA. 102:15889–15894 10.1073/pnas.0508274102 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., McIntosh J.R. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 313:944–948 10.1126/science.1128618 [Abstract] [CrossRef] [Google Scholar]
- Olson G.E., Linck R.W. 1977. Observations of the structural components of flagellar axonemes and central pair microtubules from rat sperm. J. Ultrastruct. Res. 61:21–43 10.1016/S0022-5320(77)90004-1 [Abstract] [CrossRef] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [Abstract] [CrossRef] [Google Scholar]
- Piperno G. 1995. Regulation of dynein activity within Chlamydomonas flagella. Cell Motil. Cytoskeleton. 32:103–105 10.1002/cm.970320206 [Abstract] [CrossRef] [Google Scholar]
- Piperno G., Mead K., Shestak W. 1992. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 118:1455–1463 10.1083/jcb.118.6.1455 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Piperno G., Mead K., LeDizet M., Moscatelli A. 1994. Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell Biol. 125:1109–1117 10.1083/jcb.125.5.1109 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Porter M.E., Sale W.S. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:37–42 10.1083/jcb.151.5.37 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Quillin M.L., Matthews B.W. 2000. Accurate calculation of the density of proteins. Acta Crystallogr. D Biol. Crystallogr. 56:791–794 10.1107/S090744490000679X [Abstract] [CrossRef] [Google Scholar]
- Ralston K.S., Hill K.L. 2006. Trypanin, a component of the flagellar Dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathog. 2:e101 10.1371/journal.ppat.0020101 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rupp G., Porter M.E. 2003. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest–specific gene product. J. Cell Biol. 162:47–57 10.1083/jcb.200303019 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rupp G., O'Toole E., Gardner L.C., Mitchell B.F., Porter M.E. 1996. The sup-pf-2 mutations of Chlamydomonas alter the activity of the outer dynein arms by modification of the γ-dynein heavy chain. J. Cell Biol. 135:1853–1865 10.1083/jcb.135.6.1853 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sale W.S., Satir P. 1977. Direction of active sliding of microtubules in Tetrahymena cilia. Proc. Natl. Acad. Sci. USA. 74:2045–2049 10.1073/pnas.74.5.2045 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Satir P. 1968. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 39:77–94 10.1083/jcb.39.1.77 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Satir P. 1985. Switching mechanisms in the control of ciliary motility. Modern Cell Biology. Satir P., editor Alan R. Liss, Inc., New York: 1–46 [Google Scholar]
- Smith E.F. 2002. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskeleton. 52:33–42 10.1002/cm.10031 [Abstract] [CrossRef] [Google Scholar]
- Smith E.F., Sale W.S. 1992. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 257:1557–1559 10.1126/science.1387971 [Abstract] [CrossRef] [Google Scholar]
- Smith E.F., Yang P. 2004. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 57:8–17 10.1002/cm.10155 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sui H., Downing K.H. 2006. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 442:475–478 10.1038/nature04816 [Abstract] [CrossRef] [Google Scholar]
- Summers K.E., Gibbons I.R. 1971. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA. 68:3092–3096 10.1073/pnas.68.12.3092 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Warner F.D. 1976. Ciliary inter-microtubule bridges. J. Cell Sci. 20:101–114 [Abstract] [Google Scholar]
- Wirschell M., Nicastro D., Porter M.E., Sale W.S. 2009a. The regulation of axonemal bending. The Chlamydomonas Sourcebook: Cell Motility and Behavior, vol. 3. Second edition Harris E.H., Stern D.B., Witman G.B., Academic Press, Oxford, UK: 253–282 [Google Scholar]
- Wirschell M., Yang C., Yang P., Fox L., Yanagisawa H.-A., Kamiya R., Witman G.B., Porter M.E., Sale W.S. 2009b. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol. Biol. Cell. 20:3044–3054 10.1091/mbc.E09-04-0276 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Witman G.B., Carlson K., Berliner J., Rosenbaum J.L. 1972. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54:507–539 10.1083/jcb.54.3.507 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Witman G.B., Plummer J., Sander G. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747 10.1083/jcb.76.3.729 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Woolley D.M. 1997. Studies on the eel sperm flagellum. I. The structure of the inner dynein arm complex. J. Cell Sci. 110:85–94 [Abstract] [Google Scholar]
- Woolley D.M. 1998a. Studies on the eel sperm flagellum. 2. The kinematics of normal motility. Cell Motil. Cytoskeleton. 39:233–245 10.1002/(SICI)1097-0169(1998)39:3<233::AID-CM6>3.0.CO;2-5 [Abstract] [CrossRef] [Google Scholar]
- Woolley D.M. 1998b. Studies on the eel sperm flagellum. 3. Vibratile motility and rotatory bending. Cell Motil. Cytoskeleton. 39:246–255 10.1002/(SICI)1097-0169(1998)39:3<246::AID-CM7>3.0.CO;2-2 [Abstract] [CrossRef] [Google Scholar]
- Woolley D.M., Vernon G.G. 2001. A study of helical and planar waves on sea urchin sperm flagella, with a theory of how they are generated. J. Exp. Biol. 204:1333–1345 [Abstract] [Google Scholar]
- Xiong Q., Morphew M.K., Schwartz C.L., Hoenger A.H., Mastronarde D.N. 2009. CTF determination and correction for low dose tomographic tilt series. J. Struct. Biol. 168:378–387 10.1016/j.jsb.2009.08.016 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yanagisawa H.A., Kamiya R. 2004. A tektin homologue is decreased in Chlamydomonas mutants lacking an axonemal inner-arm dynein. Mol. Biol. Cell. 15:2105–2115 10.1091/mbc.E03-11-0854 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M., Agrin N.S., King S.M., Sale W.S., Kamiya R., et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119:1165–1174 10.1242/jcs.02811 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yeh S.D., Chen Y.J., Chang A.C., Ray R., She B.R., Lee W.S., Chiang H.S., Cohen S.N., Lin-Chao S. 2002. Isolation and properties of Gas8, a growth arrest-specific gene regulated during male gametogenesis to produce a protein associated with the sperm motility apparatus. J. Biol. Chem. 277:6311–6317 10.1074/jbc.M106941200 [Abstract] [CrossRef] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.200908067
Read article for free, from open access legal sources, via Unpaywall:
http://jcb.rupress.org/content/jcb/187/6/921.full.pdf
Free after 6 months at intl.jcb.org
http://intl.jcb.org/cgi/reprint/187/6/921.pdf
Free to read at intl.jcb.org
http://intl.jcb.org/cgi/content/abstract/187/6/921
Free after 6 months at intl.jcb.org
http://intl.jcb.org/cgi/content/full/187/6/921
Citations & impact
Impact metrics
Article citations
Protofilament-specific nanopatterns of tubulin post-translational modifications regulate the mechanics of ciliary beating.
Curr Biol, 34(19):4464-4475.e9, 12 Sep 2024
Cited by: 1 article | PMID: 39270640 | PMCID: PMC11466076
Primary Ciliary Dyskinesia: A Clinical Review.
Cells, 13(11):974, 04 Jun 2024
Cited by: 0 articles | PMID: 38891105 | PMCID: PMC11171568
Review Free full text in Europe PMC
Chlamydomonas as a model system to study cilia and flagella using genetics, biochemistry, and microscopy.
Front Cell Dev Biol, 12:1412641, 30 May 2024
Cited by: 0 articles | PMID: 38872931
Review
Homozygous variant in DRC3 (LRRC48) gene causes asthenozoospermia and male infertility.
J Hum Genet, 69(8):401-409, 20 May 2024
Cited by: 0 articles | PMID: 38769386
Effects of Tcte1 knockout on energy chain transportation and spermatogenesis: implications for male infertility.
Hum Reprod Open, 2024(2):hoae020, 04 Apr 2024
Cited by: 0 articles | PMID: 38650655 | PMCID: PMC11035007
Go to all (200) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Detailed structural and biochemical characterization of the nexin-dynein regulatory complex.
Mol Biol Cell, 26(2):294-304, 19 Nov 2014
Cited by: 43 articles | PMID: 25411337 | PMCID: PMC4294676
Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella.
J Biol Chem, 286(33):29175-29191, 23 Jun 2011
Cited by: 68 articles | PMID: 21700706 | PMCID: PMC3190725
In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography.
J Biol Chem, 290(9):5341-5353, 06 Jan 2015
Cited by: 32 articles | PMID: 25564608 | PMCID: PMC4342452
Ciliary Proteins: Filling the Gaps. Recent Advances in Deciphering the Protein Composition of Motile Ciliary Complexes.
Cells, 8(7):E730, 17 Jul 2019
Cited by: 23 articles | PMID: 31319499 | PMCID: PMC6678824
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (5)
Grant ID: GM083122
Grant ID: R37 GM055667
Grant ID: R01 GM055667
Grant ID: R01 GM083122
Grant ID: GM55667

 1,2
1,2



