Abstract
Free full text

Complete Genomic Structure of the Cultivated Rice Endophyte Azospirillum sp. B510
Abstract
We determined the nucleotide sequence of the entire genome of a diazotrophic endophyte, Azospirillum sp. B510. Strain B510 is an endophytic bacterium isolated from stems of rice plants (Oryza sativa cv. Nipponbare). The genome of B510 consisted of a single chromosome (3 311 395 bp) and six plasmids, designated as pAB510a (1 455 109 bp), pAB510b (723 779 bp), pAB510c (681 723 bp), pAB510d (628 837 bp), pAB510e (537 299 bp), and pAB510f (261 596 bp). The chromosome bears 2893 potential protein-encoding genes, two sets of rRNA gene clusters (rrns), and 45 tRNA genes representing 37 tRNA species. The genomes of the six plasmids contained a total of 3416 protein-encoding genes, seven sets of rrns, and 34 tRNAs representing 19 tRNA species. Eight genes for plasmid-specific tRNA species are located on either pAB510a or pAB510d. Two out of eight genomic islands are inserted in the plasmids, pAB510b and pAB510e, and one of the islands is inserted into trnfM-CAU in the rrn located on pAB510e. Genes other than the nif gene cluster that are involved in N2 fixation and are homologues of Bradyrhizobium japonicum USDA110 include fixABCX, fixNOQP, fixHIS, fixG, and fixLJK. Three putative plant hormone-related genes encoding tryptophan 2-monooxytenase (iaaM) and indole-3-acetaldehyde hydrolase (iaaH), which are involved in IAA biosynthesis, and ACC deaminase (acdS), which reduces ethylene levels, were identified. Multiple gene-clusters for tripartite ATP-independent periplasmic-transport systems and a diverse set of malic enzymes were identified, suggesting that B510 utilizes C4-dicarboxylate during its symbiotic relationship with the host plant.
1. Introduction
Endophytes are microorganisms that are able to colonize the intercellular, and sometimes also intracellular, spaces of plant tissues, without causing apparent damage to the host plant. Gram-positive and Gram-negative bacterial endophytes have been isolated from several tissues in numerous plant species.1,2 Many endophytes have beneficial effects on plant growth and health.3–5 N2-fixing bacterial endophytes, such as Herbaspirillum seropedicae, Gluconacetobacter diazotrophicus, and Azoarcus sp., have been found within the tissues of some crops and grasses, and partially contribute to the nitrogen requirement of the host plants.6 Azoarcus sp. strain BH72, isolated from the salt marsh plant kallar grass, is best studied in terms of the molecular mechanisms of establishment inside plants and endophyte functions.7
Krause et al.8 reported the first full genome sequence of an endophyte, strain BH72 of Azoarcus species (4.38 Mb), and this sequence provided valuable insights into the life of bacterial endophytes, including information about interactions with host plants. Fouts et al.9 also reported the whole genome sequence of a N2-fixing endophyte, Klebsiella pneumoniae 342. Comparative genomics of naturally occurring bacterial endophytes provides information that can be used to develop enhanced bacterial endophytes.10
The genus Azospirillum consists of spirillum-shaped, N2-fixing, Gram-negative alpha-proteobacteria that often live in the plant rhizosphere.11 Since Azospirillum inoculation promotes plant growth, agronomic applications of this genus have been developed.12 Azospirillum sp. B510 was isolated on 23 August 1999 from the surface-sterilized stems of rice plants (Oryza sativa cv. Nipponbare) that were cultivated in the Kashimadai experimental paddy field of Tohoku University (Miyagi, Japan).13 The B510 strain is closely related to A. oryzae COC8, which was reported as a paddy soil bacterium (with 97.7% identity in their 16S rRNA gene sequences),14 and B510 is classified in the same cluster of the phylogenetic tree as A. oryzae COC8 (Supplementary Fig. S1). In addition to being a diazotroph under free-living conditions, B510 was found to have positive motility, and to be capable of degrading plant cell walls.13 Inoculation with Azospirillum sp. B510 was shown to promote plant growth under both laboratory and field conditions (Isawa et al., unpublished results). Specifically, the field experiment in a field in Hokkaido, Japan, indicated that B510 inoculation increases stem number resulting in an increase in seed yield (Isawa et al., unpublished results). Moreover, B510 inoculation enhanced disease resistance to virulent rice blast fungus and the bacterial pathogen Xanthomonas oryzae.15 Thus, Azospirillum sp. B510 is likely a beneficial bacterium with agronomic applications.
In this study, we demonstrated the endophytic characteristics of Azospirillum sp. B510 and its ability to fix N2 in planta. Then, we determined the complete nucleotide sequence of the Azospirillum sp. B510 genome and deduced the gene repertoire in the genome. This is the first report of the genome structure of the genus Azospirillum.
2. Materials and methods
2.1. A bacterial strain, inoculation of rice plants, and estimation of N2 fixation ability and of the internal Azospirillum sp. B510 population
Azospirillum sp. B510 is a diazotrophic endophyte that was isolated from the stem of cultivated rice, O. sativa cv. Nipponbare.13 Bacteria were cultured in Nutrient Broth (Difco, Detroit, MI, USA), collected by centrifugation at 5000g for 3 min, and washed twice with sterile saline (0.85% w/v NaCl). The bacterial cell suspension was adjusted to 2 × 107 cells ml−1 in saline solution just before inoculation.16
The hulls of rice seeds were carefully removed using forceps. After the hulled seeds were shaken in 10% (w/v) Ca(OCl)2 for 30 min at 28°C, they were washed more than three times with sterile distilled water. A surface-sterilized seed was placed in a sterilized test tube (16.5 mm in diameter, 150 mm in height) containing 9 ml of 0.325% (w/v) semi-solid agar solution with the sterilized inorganic nutrients,13,17 and the tube was covered with an aluminium cap. Each seed was inoculated with a bacterial cell suspension of 1 × 107 cells. The rice plants were cultivated at 25°C under long-day conditions (16-h light and 8-h dark) for 10 days in a plant growth cabinet (LH300; NK Systems Co. Ltd, Osaka, Japan) that provided 65 mmol photons m−2 s−1 of photosynthetically active radiation.18
To estimate N2-fixing activity, acetylene was introduced into test tubes, each containing a 10-day-old rice seedling, and the tubes were enclosed with a sterilized rubber stopper. After a 24-h incubation period, the ethylene concentration was determined by gas chromatography as described previously.13 Internal populations of inoculated bacteria inside rice tissues were estimated as follows. The 10-day-old rice seedlings were sampled from the test tubes. After the seed parts of the seedlings were removed using forceps, the remaining parts of the seedlings were weighed. The parts of seedlings were dipped in 70% (v/v) ethanol and then immersed in 1% NaOCl solution for 30 s. They were then quickly washed three times with sterilized distilled water and then once with sterilized saline solution. After the surface-sterilized plants were aseptically macerated in 1 ml of saline solution using a mortar and pestle, the macerates were serially diluted with saline solution and plated on Nutrient Agar (Difco) plates. After incubation at 30°C for 7 days, colony forming units (CFUs) were determined based on the fresh weight of the rice plants. Simultaneously, uninoculated plants were grown and subjected to CFU determination, as a negative control.
2.2. Genome sequencing
Total cellular DNA was purified according to standard procedures, and three genomic libraries, based on two types of cloning vectors, were constructed for sequencing. The IB5100/1 library contained inserts of ~3.0 kb cloned into pUC118 (Takara Bio Inc., Japan), the IB5102/3 library contained inserts of ~4.5 kb cloned into pUC118, and the IB510b library contained inserts with an average size of 58 kb cloned into a BAC vector, pCC1BAC (Epicentre Bio., USA).
Genome sequencing was performed using the whole-genome shotgun method in combination with BAC end-sequencing. The nucleotide sequences of both ends of the clones from the IB5100/1, IB5102/3, and IB510b libraries were analysed using a Dye-terminator Cycle Sequencing Kit and the 3730XL Sequencer (Applied Biosystems, USA). The end-sequence data from the BAC clones facilitated the gap-closure process and provided the scaffolding for reconstruction of the sequence of the entire genome. We filled the remaining gaps in the sequence by primer walking, using the plasmids or the BAC clones as templates. The integrity of the reconstructed genome sequence was assessed by chromosome walking using the end sequences of the BAC clones.
2.3. Gene assignment, annotation, and information analyses
RNA- and protein-encoding regions were assigned by a combination of computer prediction and similarity searches, as described previously.19
Genes for structural RNAs were identified by similarity searches against an in-house structural RNA database that had been constructed based on data available in GenBank (rel.167). tmRNA-, tRNA-, and rRNA-encoding regions were predicted using the ARAGORN 1.2.20 program,20 the tRNA scan-SE 1.23 program,21 and the RNAmmer ver.1.2S program,22 respectively, in combination with similarity searches.
The prediction of protein-encoding regions was carried out with the Glimmer 2.13 prediction program.23 Prior to prediction, a matrix was generated for the B510 genome by training with a data set of 610 open-reading frames that showed a high degree of sequence similarity to a translated gene set registered as the genomic data for both Magnetospirillum magneticum AMB-1 (accession number AP007255) and Rhodospirillum rubrum ATCC 11170 (CP000230), which are bacteria closely related to Azospirillum species.24 All the predicted protein-encoding regions of 150 bp or more were translated into amino acid sequences, which were then subjected to similarity searches against the non-redundant (nr) protein database from NCBI (GenBank database rel. 167.0) using the BLASTP program.25 In parallel, all the predicted intergenic sequences were compared with sequences in the nr database using the BLASTX program to identify genes that were not detected by the prediction process. For predicted genes that did not show sequence similarity to known genes, only those equal to or longer than 150 bp were considered candidates.
To annotate the functions of the assigned genes, the KAAS system, which is based on bi-directional best-hit information from sequence similarity against the KEEG GENES database and on heuristics, was first applied to all predicted protein-encoding genes.26 Next, the group escaped from KAAS was deduced based on the sequence similarity of their translated protein products to those of genes of known function and to the protein motifs in the InterPro database (ver. 17.0).27 A BLAST score of 10−5 was considered significant. Assignment of Clusters of Orthologous Groups of proteins (COGs) of predicted gene products was carried out by BLASTP analysis against the COG reference data set (http://www.ncbi.nlm.nih.gov/COG/).28 A BLAST E-value of less than 10−10 was considered significant. After filtering, COG assignments of the putative gene products were generated according to COG identification, using the best-hit pair in the reference data set.
Comparison between two genomic nucleotide sequences was performed using GenomeMatcher V.1.270.29 The GC-skew analysis was performed as described by Lobry.30 Phage_Finder ver. 4.6 was used to detect the prophage region inserted into the B510 genome.31
The FtsK Orienting Polar Sequences (KOPS) motif is specifically oriented toward the replication terminus of the genomic sequences in alpha-proteobacteria.32 The cumulative distribution of the KOPS sequence patterns (GGGNAGGG) was calculated along each replicon of B510, and the distribution of these patterns in the genome was plotted. Multicopy DNA elements of longer than 500 bp that have the capacity to encode a putative transposase were identified as insertion sequences (ISs), using the BLAST2 program, and then classified using RECON1.0533 and the IS finder (www-is.biotoul.fr).
3. Results and discussion
3.1. Colonization and N2 fixation in rice plants
The internal population of B510 was evaluated using surface-sterilized rice seedlings and the plate counting method. We calculated that there were 1.5–5.7 × 104 CFU g−1 fresh weight of inoculated seedlings. In contrast, no colonies were detected in uninoculated rice plants. These data indicate that B510 cells colonized internal rice tissues, although the colonization level was lower than that reported for other endophytes, including Herbaspirillum sp. B501 (~106 CFU g−1 fresh weight).13,34 Indeed, Yasuda et al.15 also observed colonization of Azospirillum sp. B510 around the basal parts of shoots of cv. Nipponbare using gusA-tagged B510.
To evaluate the in planta N2-fixing activity of Azospirillum sp. B510, acetylene reduction activity was assayed using rice seedlings inoculated or not with the bacterium. In the presence of acetylene, the seedlings inoculated with Azospirillum sp. B510 showed marked acetylene reduction activity compared with the activity in uninoculated plants and in plants inoculated in air (control) (Supplementary Table S1). Acetylene reduction activity of Azospirillum sp. B510 in planta (43 nmol h−1 g−1 fresh weight) was similar to that of Herbaspirillum sp. B501 (67 nmol h−1 g−1 fresh weight), a well-characterized N2-fixing endophyte isolated from rice plants.13,17,34
3.2. Sequencing and structural features of the genome
The nucleotide sequence of the entire B510 genome was deduced initially by assembling a total of 66 554 sequence files, which corresponded to approximately six genome equivalents, according to the method described in the Materials and methods section. To ensure that the nucleotide sequence was sufficiently accurate for further analysis of gene structure and function, finishing was carried out by visually editing the draft sequences and by additional sequencing to close the gaps. The genome of B510 consists of a single chromosome and six circular plasmids designated as pAB510a, pAB510b, pAB510c, pAB510d, pAB510e, and pAB510f. The total size of the genome is 7 599 738 bp, and the average GC content is 67.6%. The size and the percentage of GC content of each replicon are summarized in Table 1. The integrity of 99.9% of the final genome sequence was assessed by comparing the insert length of anchored BAC clones with the computed distance between the end sequences of the clones. The integrity of the remaining region (334 299–341 799 nt on the chromosome) where no BAC clone was anchored was confirmed using the sequence and insert length information of the plasmid clones.
Table 1
Features of replicons in Azospirillum sp. B510 genome
| Chromosome | pAB510a | pAB510b | pAB510c | pAB510d | pAB510e | pAB510f | |
|---|---|---|---|---|---|---|---|
| Size (bp) | 3 311 395 | 1 455 109 | 723 779 | 681 723 | 628 837 | 537 299 | 261 596 |
| G + C content (%) | 67.8 | 67.6 | 67.5 | 67.4 | 68.0 | 67.5 | 65.9 |
| Prophage | 2 | ND | ND | ND | ND | ND | ND |
| Genomic island | 6 | ND | 1 | ND | ND | 1 | ND |
| tRNA genes | 45 | 14 | 2 | 3 | 6 | 9 | ND |
| rRNA genesa | 2 (rrn1,2) | 4 (rrn4,5,6,7) | 1 (rrn8) | 1 (rrn9) | ND | 1 (rrn3) | ND |
| Protein genes | 2893 | 1131 | 631 | 533 | 519 | 415 | 187 |
| COG assignmentb | 2020 | 896 | 525 | 441 | 389 | 309 | 138 |
| Not in COGs | 873 | 235 | 106 | 92 | 130 | 106 | 49 |
ND means ‘not identified’.
aThe parenthetic references show IDs for the rRNA gene cluster (Supplementary Fig. S2).
bThe numbers of genes classified into 19 COG categories except for ones in ‘function unknown’ are shown.
The nucleotide position was numbered from one nucleotide upstream of the predicted ATG start codon, based on the predicted translational initiation site of the homologue of hemE (AZL028930) in the chromosome. Nucleotide positions for the plasmids were assigned based on the predicted translational initiation site of AZLa11310 in pAB510a, AZLb06310 in pAB510b, AZLc05330 in pAB510c, AZLe04150 in pAB510e, AZLf01870 in pAB510f, and the termination of AZLd05190 in pAB510d, respectively.
Ten Azospirillum species have been examined for their genome composition, using pulsed-field gel electrophoresis.35 Multiple replicons were identified in 10 Azospirillum species as with B510. However, the chromosome size of each Azospirillum strain (<2.7 Mb) was smaller than one of B510 (3.3 Mb). Linear plasmids were detected in several Azospirillum strains, such as A. brasilense and A. lipoferum,35 but similar structural units were not found in the B510 genome.
3.3. Structural features of the genome
3.3.1. Putative replication origin
A GC-skew analysis was performed to locate the probable origin of DNA replication. We established that the shift of GC-skew values occurred in two regions of the chromosome, at coordinates 35 and 1710 kb, as shown in Fig. 1 (the innermost circle). The hemE locus, which is known to associate with the origins of replication in alpha-proteobacteria,36 was found to be adjacent to the shift point of the GC skew. A cluster of nine genes, rho–hypothetical–hemH–hemE–hypothetical–maf–aroE–coaE–dnaQ (AZL028900–AZL028930–AZL000010– AZL000050: these codes are hereinafter defined in the Protein-encoding genes section), occurring at ~0 kb on the B510 chromosome (Supplementary Fig. S2) was commonly found in the Magnetospirillum sp. AMB-1 genome.37 parA (AZL000140) and parB (AZL000150) were found downstream of dnaQ (AZL000050; Supplementary Fig. S2). These findings strongly suggest that the ori region of the chromosome is located between AZL028930 and AZL000010. This location of the ori is also supported by the KOPS motif distribution analysis (Supplementary Fig. S3).
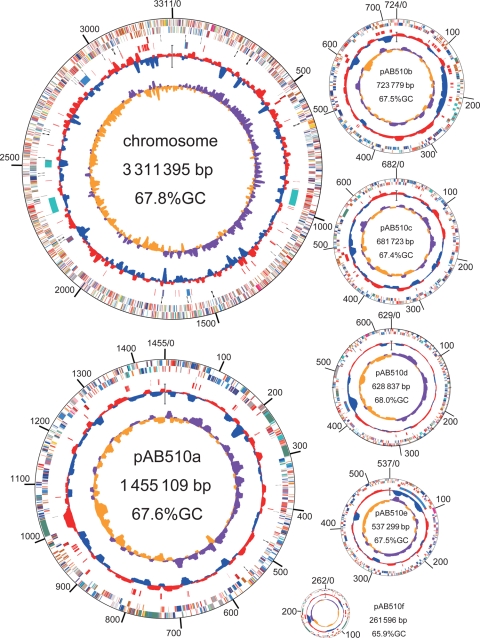
Schematic representation of seven circular replicons in the Azospirillum sp. B510 genome. The scale for all plasmids is the same, and the scale for the chromosome is one-half that of the plasmids. The scale indicates the location (in kb) outside the map. The bars in the two outer circles, the outermost circle and the second circle, show the positions of the putative protein-encoding genes in clockwise and counter-clockwise directions, respectively. The putative genes are represented by 25 colours, based on COG assignments (Supplementary Fig. S2). In the third circles from the outside, positions of structural RNA genes are indicated by black (tRNAs) and red (rRNAs) bars. In the fourth circles from the outside, the red bars indicate the positions of ISs, and the pale-green and blue areas show the insertion of prophages and genomic islands, respectively. The innermost and second circles from the centre show the GC-skew values (yellow and purple) and the average GC percent (blue and red), respectively, calculated using a window-size of 10 kb. The scales for GC percent are presented on the second circles. Top and bottom of each scale are shown, as follows: 73.9/58.9% in the chromosome, 74.6/57.0% in pAB510a, 72.5/60.7% in pAB510b, 72.0/60.2% in pAB510c, 74.9/48.3% in pAB510d, 71.9/59.0% in pAB510e, and 71.1/52.7% in pAB510f.
The shift of the GC skew was clearly observed for two of the six plasmids, pAB510a and pAB510d (Fig. 1). The predicted origins of both of these plasmids were located at around 0 kb. The KOPS motif distribution analysis also indicates that these regions are the origins of replication (Supplementary Fig. S3). These regions accommodate repA-parAB genes (AZLa00020, AZLa00030, AZLa00040) in pAB510a and repB (AZLd00010) in pAB510d. However, no repC candidate was found in the B510 genome, suggesting that the two B510 plasmids, which contain typical origin of replication regions, may have distinct, unknown initiators of replication.
3.3.2. Mobile DNA elements
ISs are small mobile DNA elements capable of transposition via a self-encoded transposase and can be classified into various families based on their structure.38 Two-hundred and eighty IS copies were assigned in all seven replication units of the B510 genome. On the basis of the type of transposase present, these ISs could be classified into 29 groups of 12 families (www-is.biotoul.fr; Supplementary Tables S2 and S3). As shown in Fig. 1, they are rather evenly distributed among the replicons, although there are several regions where a disproportionate number of ISs are located within the replicon. The remarkable frequency of ISs was occurred in pAB510f, in which a 33-kb region corresponding to 12.6% of the replicon is occupied by 30 ISs classified into 19 groups.
It is remarkable that 27 out of 280 ISs were located from 3164 to 3208 kb, between CRISPER-like sequences (described later) and one of the rRNA gene clusters on the chromosome (Fig. 1). Thirty-one ISs classified into 18 groups were also found between 80 and 157 kb on pAB510b (Fig. 1). The partial segments (AZL027610 and AZLb01330) of a gene similar to recombinase were identified at both ends of the above-mentioned high-density IS regions. This feature suggests that these regions may behave as a single large mobile genetic element.
3.3.3. ‘Phages’ and ‘a defense system’
The Phage_Finder program detected two independent putative prophages with their putative att sites at coordinates 915901–982641 (B510PP01) and 2490727–2522517 (B510PP02) on the Azospirillum sp. B510 chromosome (Fig. 1). The predicted sequences of att sites were assigned as 11 bases long (CAAGGCCGCCG) at both termini of B510PP01 and 12 bases (GCTGGGCGGCGGC) in B510PP02. The attL sites of these putative prophages (B510PP01 and B510PP02) were located near the potential genes encoding integrases AZL008670 and AZL022160, respectively. Moreover, a recombinase homologue (AZL008660) was found upstream of the AZL008670 integrase of B510PP01. Both B510PP01 and B510PP02 contained terminases (AZL008420 and AZL022110), capsid proteins (AZL008390, AZL008410, and AZL022100), and tail proteins (AZL008340 and AZL022050). These genetic traits found in two regions, B510PP01 and B510PP02, are characteristic of a horizontally transferred element.
A putative duplicated sequence of the B510PP01 prophage (at coordinates 915901–982641) was identified at coordinates 2296582–2356862 on the chromosome (Fig. 1). A detailed comparison between these two regions showed that this phage-like element lacked regions corresponding to both termini, namely a 4.8-kb region on the attL side that includes the integrase gene (AZL008670), and a 1.7-kb region on the attR side. The presence of this putative cryptic phage with deletions may indicate that the B510 cell was subjected to multiple infection events during the course of evolution.
The release of phage particles from B510 cells is induced by mitomycin C treatment.39 The DNA in the released phage was estimated to be ~10 kb in size. In the present study, however, we could not find a prophage-like region of this size in the entire B510 genome. A possible explanation for this is that diverse prophages in the genome were overlooked during the computer identification.
A search of the genome sequence by GenomeMatcher (ver.1.282) detected a large number of copies of short sequences at the coordinates 3158606–3163849 on the chromosome (Supplementary Fig. S4A and B). This region exhibited a structure typical of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) systems and is composed of 70 different spacers (36–42 bp), an identical repeat element (37 bp) containing an 8-bp palindrome (5′-CCTGGGCG), and cas genes encoding CRISPR-associated proteins (Supplementary Figs S4C and S5).40 When the whole genome sequence of B510 was subjected to CRISPRFinder (http://crispr.u-psud.fr/crispr), four additional CRISPR-like regions without complete cas genes were found exclusively at the coordinates 1016560–1018180, 1178059–1178606, 3035919–3038413, and 3199247–3200867 on the chromosome. Recently, it was reported that CRISPRs act as an antiphage defense system; bacteria integrate new spacers that are derived from phage genomic sequences during CRISPR-mediated phage resistance.40,41
3.3.4. Genomic islands
Genomic islands are regions that form syntenic groups consisting of multiple accessory genes, and they are inserted into the genome through horizontal transfer.42 They are often inserted into tRNA genes in various bacterial genomes.42 Duplicated portions of tRNA genes, which are a typical feature of genomic islands, were found at eight locations in the B510 genome (Table 1). These duplicated portions were separated from the corresponding genes by DNA segments ranging in size from 6.7 to 71 kb (B510GI01-06, d7, e8; Supplementary Table S4). This suggests that genomic islands were inserted into the tRNA genes of the B510 genome. ‘Supplementary Table S4’ summarizes the species of tRNA genes, their positions, the length of duplications, and the DNA regions that separate the perfect or near-perfect duplicated tRNA gene segments, which are 20–48 bp long. The presumptive genomic islands varied in size and contained putative genes for an integrase or a site-specific recombinase at one of their termini (Supplementary Table S4). The GC content of these elements (53–67%) was lower than the average GC content of the chromosome (67.8%). As this type of nucleotide bias is a general feature of genomic islands,42 the lower GC content of the presumptive genomic islands strongly suggests the horizontal origin of these elements.
3.4. Aspects of predicted genes and their organization
3.4.1. RNA-encoding genes
A total of 79 tRNA genes, representing 45 tRNA species, were identified (Fig. 1, Supplementary Fig. S6 and Table S5). These tRNA genes were dispersed throughout the genome, but did not occur in pAB510f (Table 1). Among them, 37 tRNA genes are likely to be transcribed as a single unit, whereas 20 (representing trnA-UGC, trnI-GAU, and trnfM-CAU) are likely to form a common transcriptional unit consisting of eight rRNA gene clusters, and the remaining 22 are tandemly arranged into nine clusters (Supplementary Table S6). Duplicated gene sets for trnK, trnT, trnP, and trnE were arrayed in pairs as complete copies. Thirty-four of the 79 tRNA genes are encoded by plasmids (Table 1). They were classified into 19 tRNA species, eight of which are specific to pAB510a or pAB510d (Supplementary Table S5), indicating that these plasmids are essential.
Nine rrns with a form of 16S-trnA-trnI-23S-5S were assigned in the genome, two of which are on the chromosome (rrn1 and rrn2) and seven on the plasmids (rrn3, rrn4, rrn5, rrn6, rrn7, rrn8, and rrn9) (Supplementary Fig. S2). The presence of nine rrns shows not to be typical copy number as alpha-proteobacteria genome, because alpha-proteobacteria generally possess five or less copies of rrn, whereas gamma-proteobacteria and firmicutes possess nine or more copies (http://ribosome.mmg.msu.edu/rrndb/).43
Of the five rrn-gene clusters, four (rrn2, rrn3, rrn6, and rrn9) were followed by trnfM (Supplementary Fig. S7). A genome island (B510GIe8) was inserted in trnfM in rrn3 located on pAB510e. One of the clusters lacking trnfM (rrn5) had a deletion of 522 nucleotides long, containing trnA and trnI, as well as a truncation of the 3′ region that encoded the 5S rRNA in addition to trnfM (Supplementary Fig. S7).
A gene-encoding transfer-messenger RNA (ssrA: AZLr028), which is known to be involved in the degradation of aberrantly synthesized proteins, was found in the B510 genome. Putative genes for two types of small RNAs, the B subunit of RNase P (rnpB: AZLr029), and the signal recognition particle (SRP) RNA (ffs: AZLr040) were also identified in the B510 genome (Supplementary Fig. S2).
3.4.2. Protein-encoding genes
The potential protein-encoding regions were assigned using a combination of computer prediction using the Glimmer program and a similarity search, as described in the Materials and methods section. By taking into account sequence similarity to known genes and relative position of predicted encoding region, to avoid overlaps, the total number of putative protein-encoding genes assigned to the genome was 6309 (Table 1). The average gene density was estimated as being one gene in every 1205 bp. The putative protein-encoding genes that start with ATG, GTG, TTG, or ATT codons were denoted by a serial number with the prefix ‘AZL’, representing the previous species name of this bacterium, Azospirillum lipoferum. It should be noted that the putative gene assignment used in this paper represents coding potential based on a defined set of assumptions.
We assigned functions to the 6309 potential protein-encoding genes by performing similarity searches, as described in the Materials and methods section. Seventy-five percent (4750) of the genes exhibited sequence similarity to genes of known function, and 25% (1559) showed sequence similarity to hypothetical genes.
We generated COG assignments of the translated gene products by conducting a BLASTP search against the COG reference data set. A total of 4718 putative gene products were classified into 23 COG categories, excluding gene products of unknown function, as shown in Table 1 and Supplementary Fig. S2.
3.5. Characteristic features of the protein-encoding genes
3.5.1. Genes involved in N2 fixation
B510 is a bacterium capable of performing N2 fixation in a modified Rennie semi-solid medium.13 Furthermore, we demonstrated its ability to fix N2 in planta in this study (see the Colonization and N2 fixation in rice plants section). The genes encoding the nitrogenase core and assembly proteins were separately clustered in three loci on the B510 chromosome. These three gene clusters were arranged in nifA [1 gene]–nifB [3 genes]–nifZ [1 gene]–nifST (AZL022440–AZL022530), nifWV [1 gene]–nifSU (AZL006520–AZL006560), and nifHDK [2 genes]–nifENX (AZL007710–AZL007640; Fig. 2A). nifQ (AZL010780) was located at a distance from above other nif gene clusters. The fixABCX (AZL006470–AZL006500) cluster, which encoded components participating in the transport of electrons to nitrogenase,44,45 was adjacent to the nifWVSU cluster and in an opposite orientation (Fig. 2A). Not all of the major components of N2 fixation were encoded by genes on the chromosome; AZLc04520, which is a homologue of nifJ encoding pyruvate–flavodoxin oxidoreductase, was solely found on the pAB510c plasmid. However, considering that a homologue of nifF, which encodes flavodoxin, an electron donor of NifJ, was not found in the B510 genome, AZLc04520 may not be involved in the major pathway for electron transfer to nitrogenase in B510.
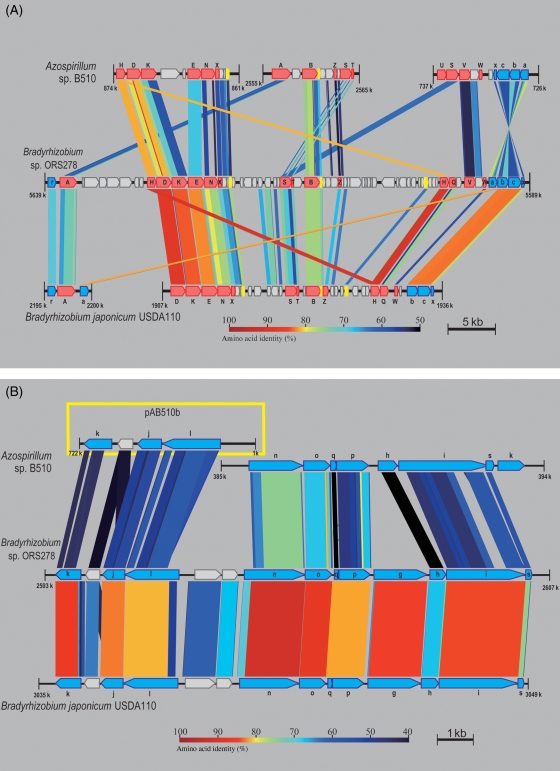
Schematic diagram of the N2-fixation gene clusters from Azospirillum sp. B510 and two Bradyrhizobium species. (A) nif gene cluster of Azospirillum sp. B510, Bradyrhizobium sp. ORS278, and Bradyrhizobium japonicum USDA110. The conservation of the gene arrangement of nif/fix is represented as boxes, with arrowheads indicating the direction of transcription. nif genes are surrounded by red boxes and are in capital letters. fix genes are in blue, lowercase font. Ferredoxin genes are shaded yellow. Black horizontal bars indicate the scale. The position on the chromosomal sequence is indicated in kb below both termini of the bars. The sequences were analysed using the tblastx function of GenomeMatcher V.1.270. The BLAST E-value of 10−10 was considered significant for the assignment of relationship. The bars connecting the maps represent close matches, with colour indicating the degree of amino acid identity determined in the BLAST alignment. The colour code is displayed at the bottom of the figure. (B) fix clusters of three bacteria. A box with a yellow border indicates that the gene cluster is located on the plasmid.
The structures of the three nif/fix gene clusters in B510 were mostly conserved with those of Bradyrhizobium sp. ORS278, which forms nodules on stems of aquatic legumes (Fig. 2A).46 However, B510 has the following distinctive features: the inversion of fixABCX, the insertion of two other predicted genes (AZL007670 and AZL007680) into a region between nifHDK and nifENX, and the transposition of nifQ from these clusters (Fig. 2A). These features may reflect the high frequency of genome rearrangements in B510.
Cytochrome c terminal oxidase is an enzyme essential for N2 fixation in rhizobial species.47 The oxidase components and the cation pump that is involved in oxidase activity are encoded in rhizobia by fixNOQP and fixGHIS, respectively. These gene clusters were located in tandem on the chromosome of B510 (AZL003350–AZL003410), although fixG was missing from the gene cluster (Fig. 2B). These gene clusters were located upstream of the fixK homologue (AZL003420: a probable transcriptional regulator). The fixG genes were present in triplicate and located at distinct positions on the chromosome (AZL016980), pAB510b (AZLb04870), and pAB510e (AZLe02910). Cytochrome c terminal oxidase is partially responsible for the N2-fixing capacity during microaerobic respiration in A. brasilense.48 It is possible that cytochrome c terminal oxidase plays a similar role in B510, although the N2-fixing capacity of B510 during microaerobic respiration has not yet been investigated.
A two-component regulatory system, fixLJ, is required to activate the expression of the nifA and fixK regulators of N2 fixation in rhizobia.49 We identified a gene cluster in pAB510b that included fixLJK (AZLb06310, AZLb06300, and AZLb06280; Fig. 2B). The domain organization of FixL (AZLb06310) in B510 was more similar to that of Bradyrhizobium japonicum (bll2760) than to that of Sinorhizobium meliloti (SMa1229).50 The genome structures of two diazotrophic endophytes, Azoarcus sp. BH72 and K. pneumoniae Kp342, have been reported,8,9 and the gene clusters encoding nitrogenase components in these genomes did not contain fixL. These findings suggest that the mechanism of N2 fixation in B510 is similar to that in rhizobia and differs from that in two diverse endophytes, Azoarcus sp. BH72 and K. pneumoniae Kp342.
3.5.2. Plant hormone-related genes
Plant-associated bacteria, such as endophytes and rhizosphere bacteria, often produce plant hormones or modulate their functions by producing inhibitors for phytohormone biosynthesis.51 In particular, the modulation of plant ethylene levels by bacterial 1-aminocyclopropane-1-carboxylate (ACC) deaminase interferes with the physiological functions of the host plant.51,52 B510 possesses the acdS gene (AZLb04170), which encodes ACC deaminase, and the acdR gene (AZLb04180: leucine-responsible regulatory protein; LRP), which regulates the expression of acdS via its putative LRP binding site, on the pAB510b plasmid (Supplementary Fig. S8).53,54 It is possible that B510 produces ACC deaminase in response to plants, which then reduces the level of ethylene in plants, and which might promote plant growth and alleviate signs of environmental stress.51,55 The acdS gene was not present in the previously reported genomes of Azoarcus sp. BH72 and K. pneumoniae Kp342.8,9
Biosynthesis of indole-3-actic acid (IAA) is widespread among plant-associated bacteria and has a beneficial impact on plant growth.56 In azospirilla and enterobacteria, IAA is generally synthesized from tryptophan via indole-3-pyruvic acid (IPyA). A key gene in this process is ipdC, which encodes indole-3-pyruvate decarboxylase (IPDC). No ipdC homologue was found in the B510 genome, irrespective of the presence of ipdC in the genome of the endophyte K. pneumoniae Kp342 (GKROPF_B2055)9 and an IAA-inducible ipdC gene in rhizosphere bacterium A. brasilense.57 We could not find an iaaC homologue in B510 either, even though the iaaC gene is co-transcribed with the ipdC gene in A. brasilense.57 Nevertheless, another pathway for IAA biosynthesis could exist in B510. The IAA biosynthetic pathway catalyses the decarboxylation of tryptophan into indole-3-actamide (IAM) and the hydrolysis of IAM to produce IAA. In Agrobacterium tumefaciens and Pseudomonas syringae, the iaaM and iaaH genes are known to be involved in these steps.56 In B510, AZLb03560 and AZLb03580 were assigned as candidates for these genes that participate in the IAM pathway.
3.5.3. Genes encoding transporters and iron-transport-related proteins
Transporters play indispensable roles in various cellular processes including the delivery of nutrients, elimination of waste products, and adaptation to environmental conditions. A variety of transporters are also thought to be involved in the symbiotic exchange of nutrients between host plant cells and intracellular rhizobia in symbiosis.
We identified a total of 354 possible transporters based on sequence similarity to known transporters and an InterPro search.58 These genes were subsequently classified into 62 families according to the transporter classification system used in Transport Classification Database (http://www.tcdb.org/).59 The gene composition of genes in these gene families in B510 is more similar to those in rhizobial species than in two other N2-fixing endophytes, Azoarcus sp. BH72 and K. pneumoniae Kp342 (Supplementary Table S7). Two of the families identified, the ATP-binding cassette (ABC) superfamily and the major facilitator superfamily (MFS), were composed of a large number of paralogs (141 and 57, respectively) that accounted for a significant portion of the transporter genes in the B510 genome. Although this feature is common among the rhizobia and the endophyte, there are significantly more ABC and MFS genes in B510 genome than in Azoarcus sp. BH72 (Supplementary Table S7). Moreover, members of the TC families, which include the Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) family and the Tricarboxylate Transporter (TTT) Family (Supplementary Table S7), were found in B510 and rhizobia, but not in the Azoarcus sp. BH72 genome.
TonB-dependent outer-membrane proteins, which are responsible for the specific uptake of ferric-siderophore complexes, are crucial for the perception of environmental signals and are associated with plant pathogens.60 Strain B510 possesses one set of tonB/exbB/exbD accompanied with anti-sigma and ECF sigma factor genes on pAB510d (Supplementary Table S8).60 In contrast, B510 carried nine genes encoding TonB-dependent outer-membrane proteins on three replicons, pAB510d, the chromosome, and pAB510a (Supplementary Table S8). The number of iron transport proteins in the B510 genome was not as high as that in other N2-fixing endophytes, such as Azoarcus sp. BH72 (22 genes), or in the rhizosphere bacterium, Pseudomonas fluorescens pf5 (45 genes). A homology search suggested that putative receptors for ferrioxamine, vitamin B12, hemin, and heme exist in B510. Because strain B510 produces siderophore (Kawaharada et al., unpublished results), the iron transport systems of B510 may be active in the bacterium in free-living and endophytic environments.
A C4-dicarboxylate, such as malate, is considered to be the major energy and carbon source of Rhizobium during symbiosis with host plants. The transport systems and metabolic enzymes of C4-dicarboxylates in Rhizobium are essential for their symbioses. With regard to the transport of carbon from the source to B510 during symbiosis, we identified five complete sets of gene cluster composed of three genes encoding the tripartite ATP-independent periplasmic-transport (TRAP-T) system (TC# 2.A.56) that transports C4-dicarboxylate (Supplementary Table S9). One of the gene cluster for TRAP-T system, AZLa08570, AZLa08580, and AZLa08600, is located upstream of a gene cluster containing citrate lyase subunits (AZLa08510, AZLa08500, AZLa08490, and AZLa08480) and a gene for malic enzyme (AZLa08520). Since citrate lyase works in citrate metabolic pathway under anoxic conditions in K. pneumoniae and Escherichia coli, it might allow B510 growing utilizing citrate as the energy source in planta. Malic enzymes are responsible for the respiratory metabolism in the N2 fixation as occurs during rhizobial symbiotic relationships with plants.61 The genome sequence of B510 revealed the presence of three genes encoding malic enzymes (AZL000200, AZLa05660, and AZLa08520). In a phylogeny, including genes from rhizobia and other endophytes, plasmid-encoding AZLa05660 and AZLa08520 are classified in a group that includes DME (NAD+-dependent malic enzyme), and AZL000200 is in a cluster harbouring TME (NADP+-dependent malic enzyme; Supplementary Fig. S9). Consequently, these findings suggest that a system for uptake and metabolism of C4 dicarboxylic acids exists in B510, as is the case in rhizobia, and this system may contribute a part of the carbon utilizations during the symbiosis.
3.5.4. Genes for the type IV secretion system
The B510 genome harboured a gene cluster with significant sequence similarity to that encoding the type IV secretion system (T4SS). Within this cluster was a gene encoding a putative coupling protein TraG/VirD4 (AZL011550), followed by the genes encoding components of secretion machinery TrbB/VirB11, TrbE/ViB4, TrbL/VirB6, TrbG/VirB9, and TrbI/VirB10 (AZL011560, AZL011580, AZL011600, AZL011620, and AZL011630, respectively). It is likely that this region is part of a genomic island, since an integrase gene (AZL011700) is coded in a region adjacent to this cluster; however, tRNA gene sequences were not detected in the border regions. The absence of genes encoding the other necessary components of T4SS might suggest that the genes in the cluster are not functional, but are remnants of genomic rearrangements. Alternatively, it is possible that there are functional homologues with less sequence similarity to prototypical T4SS proteins in the B510 genome.62
3.5.5. Genes involved in the photoresponse
Plant phytochromes are one of the photoreceptors that enable plants to adapt their growth and development to the light environment. The phytochromes are also widely distributed in diverse prokaryotes, including cyanobacteria and proteobacteria.63 The B510 genome has two genes, AZL019550 and AZLa05830, which encode distinct types of bacterial phytochromes, such as those found in the plant-tumour inducing bacterium A. tumefaciens and in the photosynthetic bradyrhizobium, Bradyrhizobium sp. ORS278.64,65 The translated amino acid sequence of the plasmid-encoded AZLa05830 protein, which shows significant similarity (47% amino acid identity) with AtBphP1 (Atu1990) in A. tumefaciens, is composed of 751 amino acid residues and contains domains that are typical of bacterial phytochromes (PAS-GAF-phytochrome-HisK). A gene encoding a response regulator (AZLa05820) in a CheY superfamily is located downstream of AZLa05830, suggesting that this gene is involved in signal transduction. In contrast, AZL019550 encodes a polypeptide of 853 amino acid residues that contains ‘PAS-GAF-phytochrome-HisK-RR’ domains. The translated gene product had 52% amino acid identity with that of BrBphP2 (BRADO2008) of Bradyrhizobium sp. ORS278. These two candidates for the light-sensing proteins may involve in the signalling pathway, although the actual function of these phytochromes remains to be analysed in B510.
3.5.6. Quorum-sensing
Quorum-sensing regulation in several strains of A. lipoferum modulates functions related to rhizosphere competence and adaptation, such as pectinase activity, siderophore synthesis, and IAA production.66 B510 produces acyl homoserine lactone (AHL) molecules in culture67 and carries genes related to AHL synthase (AZLa05890), AHL acylase (AZL013430), and AHL efflux protein (AZLd00800). A LuxR family transcriptional regulator was not found in the vicinity of AHL synthase gene (AZLa05890), although there were 22 luxR family transcriptional regulators on the genome.
3.5.7. Motility
Most endophytes are motile, because the endophytes systemically spread into the plant from infection sites.2 We found that B510 carried at least two sets of genes that encode proteins involved in flagella assembly (Supplementary Table S10). Thirty one flagella-related genes were clustered on the pAB510e plasmid (AZLe00750–AZLe01230), whereas 25 and 15 of flagella-related genes were present on the chromosome and pAB510a, respectively (Supplementary Table S10). Thirty redundant genes with roles in chemotaxis (e.g. cheAWR) or that encoded methyl-accepting chemotaxis proteins were found on the chromosome, pAB510a, pAB510c, pAB510d, and pAB510e.
3.5.8. Comparison with the pRhico plasmid
Azospirillum brasilense strain Sp7 has a pRhico plasmid that is responsible for the interaction with plant roots. pRhico has been incompletely sequenced with five gaps, and the presence of 32 genes encoding enzymes involved in surface polysaccharide biosynthesis has been reported.68 To examine the structural similarity between the pRhico plasmid and the B510 genome, a similarity search with the pRhico sequences as a query was conducted against the entire B510 genome sequence, using the BLAST program, with a threshold E-value of 10−10.
The search detected significant alignments between the B510 genome and 43 regions of pRhico, 23 of which were mapped on pAB510f. One of these sequences incompletely matched 881 nucleotides inside the putative replication protein gene (repA: AZLf00010) of pAB510f and one matched 138 nucleotides upstream of repA (Supplementary Fig. S10), and 17 mapped to regions on pAB510f that coincided with 11 genes that were presumably involved in cell envelope biosynthesis and outer membrane constitution (AZLf00040, AZLf00050, AZLf00060, AZLf00080, AZLf01050, AZLf01060, AZLf01640, AZLf01790, AZLf01050, and AZLf01810; Supplementary Fig. S10). The results suggest an evolutionary link and some functional relationship between pRhico and pAB510f. However, remaining portions of pRhico, such as those containing exoC and tRNA genes, were scattered throughout the B510 genome and most of the sequences were poorly conserved, indicating that drastic genome rearrangements had taken place since the two species diverged (Supplementary Fig. S10).
The sequence information is available in public DNA databases (DDBJ/GenBank/EMBL), under the following accession numbers: AP010946 for the B510 chromosome, AP010947 for pAB510a, AP010948 for pAB510b, AP010949 for pAB510c, AP010950 for pAB510d, AP010951 for pAB510e, and AP010952 for pAB510f. The nucleotide sequences of the genome and annotations describing the predicted genes are available online at RhizoBase (http://genome.kazusa.or.jp/rhizobase).
Funding
This work was supported by the Kazusa DNA Research Institute Foundation. This work was supported in part by Special Coordination Funds for Promoting Science and Technology and in part by a Grant-in-Aid for Scientific Research on Priority Areas ‘Comparative Genomics’ from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and in part by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, PMI-0002). Funding for open access charge: The Kazusa DNA Research Institute Foundation.
Acknowledgement
We are grateful to F. Wisniewski-Dyé (University Claude Bernard Lyon 1) for valuable comments concerning genes for quorum-sensing.
References
Articles from DNA Research: An International Journal for Rapid Publication of Reports on Genes and Genomes are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/dnares/dsp026
Subscription required at dnaresearch.oxfordjournals.org
http://dnaresearch.oxfordjournals.org/cgi/reprint/17/1/37.pdf
Free to read at dnaresearch.oxfordjournals.org
http://dnaresearch.oxfordjournals.org/cgi/content/abstract/17/1/37
Subscription required at dnaresearch.oxfordjournals.org
http://dnaresearch.oxfordjournals.org/cgi/content/full/17/1/37
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Efficacy of novel bacterial consortia in degrading fipronil and thiobencarb in paddy soil: a survey for community structure and metabolic pathways.
Front Microbiol, 15:1366951, 15 May 2024
Cited by: 0 articles | PMID: 38812693 | PMCID: PMC11133635
Uncovering microbiomes of the rice phyllosphere using long-read metagenomic sequencing.
Commun Biol, 7(1):357, 27 Mar 2024
Cited by: 1 article | PMID: 38538803 | PMCID: PMC10973392
Functional and morphological analysis of isolates of phylloplane and rhizoplane endophytic bacteria interacting in different cocoa production systems in the Amazon.
Curr Res Microb Sci, 2:100039, 25 May 2021
Cited by: 3 articles | PMID: 34841330 | PMCID: PMC8610332
Morphological and Metabolite Responses of Potatoes under Various Phosphorus Levels and Their Amelioration by Plant Growth-Promoting Rhizobacteria.
Int J Mol Sci, 22(10):5162, 13 May 2021
Cited by: 8 articles | PMID: 34068175 | PMCID: PMC8153024
Deciphering the Root Endosphere Microbiome of the Desert Plant Alhagi sparsifolia for Drought Resistance-Promoting Bacteria.
Appl Environ Microbiol, 86(11):e02863-19, 19 May 2020
Cited by: 19 articles | PMID: 32220847 | PMCID: PMC7237788
Go to all (56) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 9 of 9)
- (1 citation) ENA - AP010950
- (1 citation) ENA - AP010951
- (1 citation) ENA - AP010952
- (1 citation) ENA - CP000230
- (1 citation) ENA - AP010946
- (1 citation) ENA - AP010947
- (1 citation) ENA - AP010948
- (1 citation) ENA - AP010949
- (1 citation) ENA - AP007255
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Effects of Different Sources of Nitrogen on Endophytic Colonization of Rice Plants by Azospirillum sp. B510.
Microbes Environ, 33(3):301-308, 29 Aug 2018
Cited by: 8 articles | PMID: 30158365 | PMCID: PMC6167112
Azospirillum sp. strain B510 enhances rice growth and yield.
Microbes Environ, 25(1):58-61, 01 Jan 2010
Cited by: 26 articles | PMID: 21576855
Effects of colonization of a bacterial endophyte, Azospirillum sp. B510, on disease resistance in rice.
Biosci Biotechnol Biochem, 73(12):2595-2599, 07 Dec 2009
Cited by: 32 articles | PMID: 19966496
Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects.
FEMS Microbiol Rev, 24(4):487-506, 01 Oct 2000
Cited by: 243 articles | PMID: 10978548
Review




