Abstract
Free full text

Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion
Associated Data
Abstract
Plasmodium falciparum can invade erythrocytes by redundant receptors, some of which have variable expression. A P. falciparum clone Dd2 requiring erythrocyte sialic acid for invasion can be switched to a sialic acid-independent progeny clone Dd2NM by growing the Dd2 clone with neuraminidase-treated erythrocytes. The RH4 gene is transcriptionally up-regulated in Dd2NM compared to Dd2, despite the absence of DNA changes in and around the gene. We determined the epigenetic modifications around the transcription start site (TSS) at the time of expression of RH4 in Dd2NM (44 h) and at an earlier time when RH4 is not expressed (24 h). At 44 h, the occupancy of the +1 nucleosome site downstream of the TSS of the active RH4 gene in Dd2NM was markedly reduced compared to Dd2; no difference was observed at 24 h. At 44 h, histone modifications associated with up-regulation were positively correlated to the active RH4 gene of Dd2NM compared to Dd2; no differences were observed at 24 h. Histone H3K9 trimethylation (a marker for silencing) was higher in Dd2 than Dd2NM along the 5′-UTRs of the RH4 gene at both 44 and 24 h. Our data indicate that the failure of Dd2 to express the sialic acid-independent invasion receptor gene RH4 is associated with the epigenetic silencing mark H3K9 trimethylation present throughout the cycle.
The causative agent for the most severe human malaria, Plasmodium falciparum, invades erythrocytes by multiple redundant pathways (1, 2). Two P. falciparum receptor protein families, the Duffy Binding-Like (DBL) family and the Reticulocyte Homology (RH) family, play key roles in parasite invasion (1). A unique parasite clone Dd2 that initially invaded erythrocytes requiring sialic acid was able to be selected to a sialic acid-independent clone (Dd2NM) after maintaining the parental clone Dd2 in culture with neuraminidase-treated erythrocytes (3). Comparing the gene expression of Dd2 and Dd2NM, only RH4 and the DBL pseudogene PEBL, are transcriptionally up-regulated in Dd2NM (4, 5). The two genes are located contiguously on P. falciparum chromosome 4 and transcribed in opposite directions. The sequences of the exons and introns of the two genes, the 1.8 kb between the two ORFs, and 1.1 kb of genomic DNA 3′ to each gene’s coding region were identical in Dd2 and Dd2NM (4), suggesting that transcription of the two genes is epigenetically regulated.
Because a transcription-associated histone acetyltransferase from Tetrahymena was identified in 1996 (6), diverse histone modifications on different amino acid residues in the core histones have been discovered to play important roles in transcriptional regulation in various eukaryotic organisms (7). This led to a hypothesis of a program of epigenetic marks consisting of covalent histone tail modifications, chromatin remodeling, noncoding RNA, and DNA methylation (8). Thus far, no evidence for DNA methylation has been observed in P. falciparum. There are eight histone genes in P. falciparum that allow formation of a basic unit of chromatin, the nucleosome, where histone modifications and nucleosome remodeling occur (9). Histone modifications exist in P. falciparum. One example is a P. falciparum histone deacetylase, homologous to the yeast histone deacetylase silent information regulator 2 (PfSir2) (10, 11). Only a single gene of the approximately 60 var genes lacks Pfsir2, and it is exclusively expressed in the asexual blood stage of P. falciparum (10, 11). Furthermore, histone hypoacetylation linked to repression of other var genes was associated with var locus position on the nuclear periphery (10, 11). Acetylation of certain lysine residues in histone H3 and H4 is associated with activation of var and other genes (12–14). Recently, another type of histone modification, histone methylation, has also been identified in P. falciparum (12, 15, 16). Histone H3 lysine 4 (H3K4) di- and trimethylation have been shown as marks for var gene activation (12). In contrast, histone H3K9 trimethylation was linked to gene silencing of clonally variant gene families in P. falciparum (13, 17). These modifications in P. falciparum thus show the same correlations with gene activation or silencing seen in other organisms (7).
In this study, we investigated nucleosome occupancy in the intergenic region of the RH4 and PEBL genes for the low expressor (Dd2) and the high expressor (Dd2NM). Our data indicated that the +1 nucleosome occupancy downstream of the transcription start site (TSS) of the active RH4 gene was distinctly reduced in schizonts of Dd2NM. Furthermore, the histone modification distribution in the intergenic and the coding region of the RH4 and PEBL genes in Dd2 and Dd2NM was also studied by using ChIP. Histone H3K4 trimethylation and acetylation of histone H3 and H4 were highly enriched in the 5′-untranslated regions (5′-UTRs) of the two genes only in schizonts of Dd2NM at the time when the RH4 and PEBL genes were transcribed. Our data showed that the level of H3K9 trimethylation was higher in the schizont stage of Dd2 than in Dd2NM in the region between the TSSs and the coding regions of the RH4 and PEBL genes. H3K9 trimethylation of the 5′ region of the genes in Dd2 that was present throughout the cycle appears to be an epigenetic mark for silencing of RH4 and PEBL.
Results
The TSS Identified for RH4 and PEBL at 44 h.
P. falciparum genes RH4 (GenBank release no. AF420309) and PEBL (GenBank release no. AY032735) are contiguous genes that map to the subtelomeric region of chromosome 4 and transcribe in opposite directions (Fig. 1A). The region between the start codons of the two genes is 1,865 bp (GenBank release no. DQ100425). We amplified and sequenced the coding sequences of the RH4 and PEBL genes by reverse transcription-PCR. RH4 contains two exons, one encoding the signal sequence and a second containing the rest of the gene. PEBL is a pseudogene carrying a frame-shift in the 5′ end of the ORF. There is no intron in this region to allow read-through in the frame-shift.
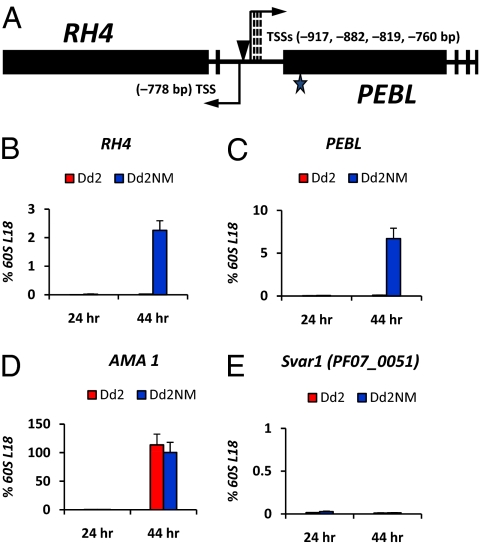
Transcription of the RH4 and PEBL genes at 24 h and 44 h of the 48-h asexual blood stage cycle. (A) Schematic structures of the RH4 and PEBL genes and identification of the transcription start sites (TSSs) of each gene. Exon-intron structures of the RH4 and PEBL genes are drawn based on gene sequences in GenBank (release no. AF420309 for RH4 and release no. AY032735 for PEBL) and from reverse transcription-PCR results. Black boxes represent the different exons in the two genes. A star indicates the frame-shift mutation in the 5′ end of the ORF of PEBL. Two arrows in opposite orientations indicate the transcription start sites and transcription orientations of the RH4 and PEBL genes. Three dashed lines indicate multiple TSS positions 3′ to the longest TSS in PEBL (−917 bp). A poly(dA)poly(dT) tract in the region between the transcription start sites of the two genes is indicated by an arrowhead. Total RNA samples from Dd2 and Dd2NM were collected at 24 or 44 h postinvasion for reverse transcription and real-time PCR analysis of transcription of the RH4 (B) and PEBL (C) genes. AMA1 (PlasmoDB ID: PF11_0344) (D), a gene expressed only at 44 h, and an asexual stage, silent var gene Svar1 (PlasmoDB ID: PF07_0051) (E) were studied at the same times. For each real-time PCR, the signal from each gene was normalized by a housekeeping gene 60S ribosomal protein L18 (60S L18, MAL13P1.209; Fig. S2) to generate the relative expression value for each gene. Experiments were independently performed at least three times. The SE is shown as an error bar.
In P. falciparum, histone modifications are mainly confined to the region surrounding the gene’s TSS (12). The studies described here were performed with parasites 44 h into a 48-h asexual blood stage cycle (the schizont stage) at a time when RH4 and PEBL are expressed. To accurately identify the TSSs of the RH4 and PEBL genes, 5′-Rapid Amplification of cDNA Ends (5′-RACE) analysis was adopted with three different gene specific primers for each gene as listed in Table S1. The RH4 gene has a 778-bp 5′-UTR upstream of the ATG start codon. Unlike RH4, there are multiple TSSs (760 bp, 819 bp, 882 bp, and 917 bp) upstream of the start codon of the PEBL gene (Fig. 1A). Between the TSS of the RH4 gene and the longest 5′-UTR of the PEBL gene, there are only 170 bp. In this region, there is a 30-bp poly(dA)poly(dT) tract (GenBank release no. DQ100425) 10 bp 5′ to the TSS of RH4 (Fig. 1A), suggesting that the structure of the nucleosome around this region may be dynamically unstable (18).
We further investigated the promoter activity of the 1,865-bp intergenic region between the start codons of the RH4 and PEBL genes. The reporter gene Luciferase replaced the RH4 coding region between the 1.8-kb intergenic region and an 800 bp sequence 3′ to the RH4 stop codon in the piggyback transposase system (Fig. S1A) (19). To avoid the effect of episomal Luciferase gene on its expression, the transfected parasite Dd2Luc that had lost the plasmid was cloned (Fig. S1 B and C). The Luciferase gene was stably integrated inside the coding region of a gene (PFF1065c), which is a gametocyte gene with unknown function and located around 900 kb and 500 kb away from the two telomeres of chromosome 6 (Fig. S1B). Under the selection of neuraminidase-treated erythrocytes, Dd2Luc was able to be switched to Dd2LucNM, which could invade the neuraminidase-treated erythrocytes. Interestingly, in Dd2Luc, the Luciferase gene was strongly transcribed at the schizont stage (Fig. S1D) to a level similar to Dd2LucNM, suggesting that the 1.8 kb intergenic region alone is not sufficient for gene silencing. Our data show that the 1.8-kb intergenic region presented strong promoter activity in late stage parasites.
Transcription profiles of the RH4 and PEBL genes at 24 and 44 h of the cycle were determined by real-time PCR analysis. At 44 h, the RH4 and PEBL genes were expressed in Dd2NM, but not in Dd2 (Fig. 1 B and C). Earlier, at 24 h, the genes were not expressed in either Dd2NM or in Dd2 (Fig. 1 B and C). AMA1 (PlasmoDB ID: PF11_0344), a gene required for parasite survival (20), was expressed at 44 h but not at 24 h in both Dd2 and Dd2NM (Fig. 1D). A silent var gene (PlasmoDB ID: PF07_0051, called Svar1 in this study) was silent in both Dd2 and Dd2NM at 24 and 44 h (Fig. 1E).
The TSS +1 Nucleosome of the RH4 Gene Is Depleted at 44 h in Dd2NM.
The nucleosome distribution over this region was determined by Solexa sequencing of DNA isolated using micrococcal nuclease digestion of cross-linked chromatin (21). The +1 nucleosome occupancy for the RH4 gene was greatly reduced in Dd2NM compared to Dd2 (Fig. 2A). No other nucleosome occupancy differences were observed, including at the +1 nucleosome site of the PEBL gene. To independently determine the nucleosome organization surrounding the TSSs of the RH4 and PEBL genes, a high-resolution nucleosome-scanning assay was performed based on real-time PCR (22). Analysis of the intergenic region between the start codons of RH4 and PEBL also revealed the presence of one nucleosome (+1) following the TSS of RH4 in Dd2 (Fig. 2B). Statistical analysis showed that the +1 nucleosome peak is significantly higher in Dd2 than in Dd2NM (P = 0.0093) (Fig. 2B). There were three more PCR positions showing differences between Dd2 and Dd2NM (Fig. 2B). Two of them were also in the +1 nucleosome region (P = 0.0377 and P = 0.0233, respectively) and another was in the intron region of RH4 (P = 0.0163). Thus, by both Solexa sequencing and real-time PCR scanning, the +1 TSS nucleosome occupancy of RH4 was reduced in Dd2NM, the expressed gene. This suggests that in the expressed gene, RH4, the reduced presence of the +1 nucleosome may be a factor in the increased expression.
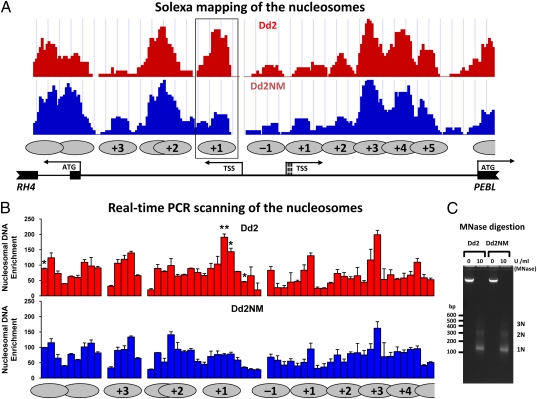
Nucleosome mapping in the intergenic region between the start codons for RH4 and PEBL for 44 h parasites. MNase-digested DNA samples from Dd2 and Dd2NM at 44 h were purified on 3.5% agarose gel and prepared for Solexa sequencing (A) and real-time PCR (B). The nucleosomes are shown by the gray ovals with marked numbers. The first nucleosome 3′ to the TSS of RH4 or PEBL was defined as +1. The nucleosome resides in the region between the two TSSs of RH4 and PEBL was defined as −1. The +1 nucleosome of RH4 is boxed and showed a lower number of sequence tags for Dd2NM than for Dd2. (B) Tiling-arranged PCR primer sets were used in real-time PCR to obtain the enrichment signals of nucleosomal DNA. Nucleosomal DNA enrichment values are presented as the percentage of a well-positioned nucleosome with the SE based on three biologically independent samples. Each PCR of Dd2 and Dd2NM were compared by the unpaired t test with Welch’s correction, **P < 0.01, *P < 0.05. (C) Enzymatic digestion of Dd2 and Dd2NM chromatin. The MNase digested (10 U/mL) or undigested (0 U/mL) DNA samples were analyzed on 2% agarose gel electrophoresis. DNA molecular weights were marked in bp on the left. Mononucleosome (1N), dinucleosome (2N), and trinucleosome (3N) released by MNase treatment were indicated on the right.
Histone H3 and H4 Acetylation and H3K4 Trimethylation at 44 h Are Associated with Expression of RH4 and PEBL in Dd2NM.
To determine the histone modification patterns along the RH4 and PEBL genes at 44 h, we designed 13 primer sets for real-time PCR following ChIP to cover the intergenic region and the coding regions of the RH4 and PEBL genes (Fig. 3A).
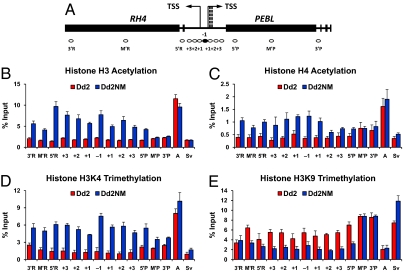
Histone modifications in the RH4 and PEBL genes for 44 h old parasites. Distribution of histone modifications determined by ChIP generated by real-time PCR (Fig. 2B) for the RH4 and PEBL genes from Dd2 and Dd2NM measured in the positions identified in A. After MNase digested chromatin samples were evenly divided, each aliquot was immunoprecipitated by various antibodies against acetylation at histone H3K9/K14 (B), acetylation at histone H4K5/K8/K12/K16 (C), histone H3K4 trimethylation (D), and histone H3K9 trimethylation (E). An active gene AMA1 (A) and a silent var gene Svar1 (Sv) in 44 h parasites were analyzed as controls. For each PCR product position, signals from the ChIP assay are presented as the percentage of input DNA. Experiments were independently performed at least three times to generate the average value of each histone modification signal at each position. The SE is shown as an error bar.
The antibody we used recognizes acetylation at lysines 9 and 14 in the histone H3 N-terminal tail. Histone H3 acetylation was enriched surrounding the TSSs and along the 5′-UTRs of the RH4 and PEBL genes in Dd2NM compared to Dd2 (Fig. 3B). This histone modification was greatly elevated in the 5′-UTR of AMA1, a late-stage-expressed gene, in both Dd2 and Dd2NM (Fig. 3B). In contrast, the silent var gene Svar1 had limited H3 acetylation in the 5′-UTR of the gene (Fig. 3B).
H4 acetylation at lysines 5, 8, 12, and 16 was highly enriched in the regions surrounding TSSs of the RH4 and PEBL in Dd2NM compared to Dd2 (Fig. 3C). In both Dd2 and Dd2NM, the 5′-UTR of the AMA1 gene, but not the silent Svar1 gene, contained this H4 histone modification (Fig. 3C).
Histone H3K4 trimethylation was increased along the 5′-UTRs and the coding regions of RH4 and PEBL in Dd2NM compared to Dd2 (Fig. 3D). The level of this modification was higher at the active gene AMA1, and lower for the silent gene Svar1 (Fig. 3D).
Taken together, our data are consistent with the previous finding that histone H3 and H4 acetylation and H3K4 trimethylation are highly enriched along the 5′-UTR of an active gene but not a silent gene in P. falciparum (12, 13).
At 44 h, H3K9 trimethylation was highly enriched along the 5′-UTR and the 5′ coding regions of the RH4 and PEBL in Dd2 compared to Dd2NM (Fig. 3E). At 44 h, H3K9 trimethylation in the middle and 3′ coding region of the gene was high in both Dd2 and Dd2NM. H3K9 trimethylation was also enriched in the 5′-UTR of Svar1, but not in that of AMA1, in both Dd2 and Dd2NM (Fig. 3E). Our data indicate that H3K9 trimethylation at the 5′-UTR and the 5′ coding region of the gene correlates with gene silencing of RH4 and PEBL in Dd2.
Epigenetic Modifications at 24 h When Genes Are Not Expressed.
The epigenetic modifications around the TSS of RH4 were further investigated at 24 h when RH4 was not expressed in either Dd2NM or Dd2. First, nucleosome occupancy was scanned by real-time PCR analysis. The density of the +1 TSS nucleosome of RH4 was similarly high at 24 h in Dd2 and Dd2NM (Fig. 4A). No significant difference of nucleosome occupancy between Dd2 and Dd2NM was observed by statistical analysis. Our data suggest that reduced nucleosome density at the +1 nucleosome position only occurs during gene expression.
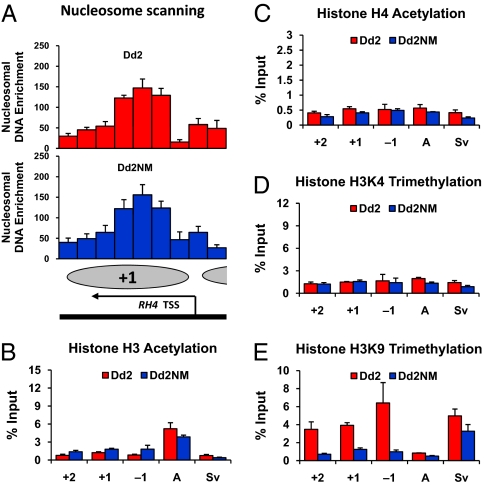
Histone H3K9 trimethylation is an epigenetic marker for silencing observed at 24 h for RH4 and PEBL. The chromatin samples from Dd2 and Dd2NM were prepared for nuclesome mapping (A) and ChIP assays for acetylation at histone H3K9/K14 (B), acetylation at histone H4K5/K8/K12/K16 (C), histone H3K4 trimethylation (D), and histone H3K9 trimethylation (E). Tiling-arranged PCR primer sets were used only for scanning occupancy of the +1 TSS nucleosome of the RH4. As described in the legend of Fig. 2, nucleosomal DNA enrichment values are presented as the percentage of a well-positioned nucleosome with the standard error based on three biologically independent samples. Enrichment of histone modifications at −1, +1, and +2 nucleosome positions surrounding the RH4 TSS was determined as described in the legend of Fig. 3. AMA1 and Svar1 genes were tested as control as described in the legend of Fig. 3.
Histone modifications around the TSS of RH4 were also investigated at 24 h. The active epigenetic marks, histone H3 acetylation (Fig. 4B), histone H4 acetylation (Fig. 4C), and histone H3K4 trimethylation (Fig. 4D), were similarly low around the TSS of RH4 at 24 h in Dd2 and Dd2NM. Histone H3 acetylation in the 5′-UTR of the AMA1 gene was higher in both Dd2 and Dd2NM than that in other genes at 24 h (Fig. 4B). H4 acetylation and H3K4 trimethylation were low at 24 h in the 5′-UTRs of the AMA1 and Svar1 genes in both Dd2 and Dd2NM (Fig. 4 B–D).
H3K9 trimethylation was present at high levels around the TSS of RH4 at 24 h in Dd2 (Fig. 4E). In contrast, this mark was low at 24 h around the TSS of RH4 in Dd2NM, at a time when RH4 was not yet expressed (Fig. 4E). This modification was also low in the 5′-UTR of AMA1 but was high in the silent var gene Svar1 at 24 h in Dd2NM (Fig. 4E). Our data indicate that H3K9 trimethylation plays a role in gene silencing at 44 h and as an epigenetic marker of gene silencing at 24 h in Dd2.
Discussion
Here we describe experiments to investigate the mechanism underlying the switch in invasion phenotype of two variants of one P. falciparum clone, Dd2 and Dd2NM. Our results reveal that a difference in one histone modification, H3K9 trimethylation, appears to distinguish the two variants. Histone H3K9 trimethylation is associated with RH4 silencing and persists around the TSS of RH4 throughout the asexual blood stage in Dd2 compared to Dd2NM.
Transcription in Plasmodium spp. is controlled, in part, by a family of Apicomplexan AP2 (ApiAP2) genes that derive from Apetala 2 (Ap2) of plants (23). The family of ApiAP2 is expressed differentially throughout the asexual blood stage and at other stages in the life cycle (23). One of the AP2 genes has been shown to control specific gene expression in the sexual stages of Plasmodium berghei (24). In addition, there is an epigenetic modification that is associated with transcription, demonstrated for P. falciparum var genes (12, 15), a family of approximately 60 genes, which are individually expressed in each individual parasite (25).
P. falciparum Dd2, a parasite clone that requires sialic acid on the erythrocyte surface for invasion of erythrocytes, was selected by growth on neuraminidase-treated erythrocytes (Dd2NM) that invades neuraminidase treated erythrocytes (3). Two contiguous genes, RH4 and the pseudogene PEBL, are markedly up-regulated in mRNA expression in Dd2NM compared to Dd2 (4, 5). In addition, the RH4 protein is only found in Dd2NM (4, 5). The two contiguous genes are expressed in opposite directions from a common intragenic region. This switch in expression occurs despite a completely unchanged DNA sequence in the exons and introns throughout the two genes, the 1.8 kb region between the start codons of the two genes, and 1.1 kb at the 3′ end of both genes (4). So far, no other gene that is associated with the switch from Dd2 to Dd2NM has been found to be up-regulated in the P. falciparum genome, as measured by gene expression microarrays (4, 5).
We describe the epigenetic modifications in the region during the late part of the asexual cycle when RH4 mRNA is expressed in Dd2NM but not in Dd2. The findings were as follows. First, the nucleosome immediately following the transcription start site (TSS) of RH4, the +1 TSS nucleosome, is greatly decreased in Dd2NM, when the gene is markedly up-regulated. One possible reason that the nucleosome is not completely absent is that the parasites are not perfectly synchronous and if the nucleosome change occurs late in the cycle, then there will be only a partial reduction of the nucleosome. The +1 nucleosome occupancy of PEBL is unchanged in Dd2NM as compared to Dd2.
Second, the acetylation of histones on H3 and H4 and the H3K4 trimethylation that are associated with up-regulation of other genes (12–14) were associated with the up-regulation of RH4 and PEBL in Dd2NM compared to Dd2. Third, H3K9 trimethylation, which is associated with gene silencing, had a stronger signal in the silent gene, Dd2, than in the up-regulated gene Dd2NM.
These histone modifications appear to constitute a specific pattern that characterizes the two up-regulated genes in the switch from Dd2 to Dd2NM and are associated with protein expression of RH4, the receptor for invasion of neuraminidase-treated erythrocytes by Dd2NM. The mechanisms that gave rise to these modifications are unclear. To explore this question, we asked when these characteristic marks first appeared. We studied 24 h old sychronized parasites, midway through the asexual cycle at the time when RH4 is not expressed. We found that at this earlier time H3K9 trimethylation was the only epigenetic mark that distinguished Dd2NM from Dd2. This mark was already high, but only in Dd2, as was found also at 44 h, late in the cycle in mature schizonts. The other histone modifications at RH4, which distinguish Dd2NM from Dd2 at 44 h, summarized above, were not seen at 24 h. These modifications presumably appear around the time of expression. Thus H3K9 trimethylation, the histone modification associated with silencing, marks RH4 for inactivation in Dd2 and is preserved throughout the asexual cycle. The mechanism by which H3K9 trimethylation persists through DNA replication is unknown in Plasmodium spp. In any case, the persistence of H3K9 trimethylation from one cycle to another indicates that the epigenetic state is present at 24 h and maintained through cell division.
After cloning Dd2 and growth in culture for various times, the frequency of parasites able to invade neuraminidase-treated erythrocytes (Dd2NM) is approximately 1 in 200 (26). If H3K9 trimethylation is the only basis of the silencing, then the rate of loss of H3K9 trimethylation from Dd2 is lower than the rate of H3K9 trimethylation in Dd2NM. We observed that we needed to grow Dd2NM in neuraminidase-treated erythrocytes because Dd2NM would slowly drift toward Dd2 as measured by reduced invasion rates into neuraminidase-treated erythrocytes, presumably moving toward an equilibrium of 1 in 200 cells containing Dd2NM (26).
The data for H3K9 trimethylation in silencing a P. falciparum var gene derives from the data by T. Chookajorn et al. (15), where the gene blasticidin-S-deaminase (BSD) for blasticidin resistance, is controlled by a var gene promoter. When the BSD gene is not selected by blasticidin and is silent, the H3K9 trimethylation is continuously present on the var promoter region and the BSD coding region. When the BSD gene is expressed through growth in the presence of blasticidin, the H3K9 trimethylation is no longer present on the gene.
Another data set on epigenetic modifications in P. falciparum is for var2CSA (12), a gene that encodes the erythrocyte membrane protein 1 (PfEMP1) that binds chondroitin sulfate A (CSA) in placenta (27). The P. falciparum that fails to express var2CSA is marked by H3K9 trimethylation early and late in the cycle around the TSS of var2CSA (12). H3K9 trimethylation has been known as an epigenetic marker of gene silencing in other organisms (7). The negative epigenetic regulator, H3K9 trimethylation around the RH4 TSS persists throughout the cycle in Dd2. The one modification that marks active expression of var2CSA throughout the cycle is H3K4 dimethylation (12, 28). Because var2CSA is expressed throughout the asexual blood stage cycle (29), it remains unknown if this histone modification is a positive marker for memory in P. falciparum. The positive marks in Dd2NM for RH4 expression at 44 h (Fig. 3) were not observed at 24 h when RH4 was not expressed (Fig. 4). In addition, the positive epigenetic modifications seen at 44 h in AMA1 were also not observed at 24 h when the gene is silent except possibly H3 acetylation (Fig. 4). This observation requires confirmation in other genes. In general, each gene at the time of expression is associated with nucleosome modifications. These positive marks of expression occur only when the gene is expressed, as in the case of a gene expressed late in the cycle, e.g., RH4 in our study, but not at the midterm of the asexual cycle when the gene is not expressed.
The turning off and on of genes involved in invasion (2, 30, 31) may be a survival strategy where redundant genes for invasion may be silenced by the mechanisms described in the present paper. This strategy may be similar to antigenic variation where the controlled expression of individual genes increases the parasite’s survival. Thus, as in the case of var genes and the RH4 gene, the genes for invasion that appear to be silenced in some cases may also be marked by H3K9 trimethylation as in RH4 in Dd2.
Materials and Methods
P. falciparum Parasites.
A single clone of P. falciparum parasite Dd2 used in this study was cloned in our lab previously (3). Parasite clone Dd2NM was derived from the Dd2 clone by incubating it with neuraminidase-treated erythrocytes (3). Parasites were cultured and synchronized as described previously (4). In particular, Dd2NM was maintained in neuraminidase-treated erythrocytes. At 24 h and 44 h postinvasion, synchronized Dd2 and Dd2NM were collected for subsequent analyses: gene expression, nucleosome mapping, and ChIP.
Solexa Analysis of Genome-Wide Nucleosome Mapping.
Nucleosomal DNA generated by Micrococcal nuclease (MNase) treatment was separated on 3.5% agarose gel and mononucleosome-size DNA was extracted and purified from the gel using the QiaGel Extraction kit (Qiagen). The mononucleosome-sized DNA fragments were blunt-ended using the End-It DNA End-Repair kit (Epicentre Biotechnologies). Poly-A was added to the blunt-ended DNA with Taq DNA polymerase (New England BioLabs) at 70 °C for 30 min. After purification using the QIAquick PCR purification kit (Qiagen), the modified DNA was ligated to paired-end adapters (Illumina) with T4 DNA Ligase (New England BioLabs) and purified again using the QIAquick PCR purification kit (Qiagen). PCR amplification was carried out by using Finnzymes High Fidelity DNA Polymerase Master Mix (New England BioLabs) and the PCR Primers PE 1.0 and 2.0 (Illumina). PCR products were purified again and sequenced using the Illumina 1G Genome Analyzer as described previously (21). Sequenced reads from the Solexa analysis were aligned to P. falciparum genome (3D7) on the Ares Lab Browser (http://areslab.ucsc.edu/).
Nucleosome Scanning by Real-Time PCR.
To determine nucleosome density by real-time PCR, gel-extracted mononucleosomal DNA was amplified by real-time PCR in three independent experiments as described previously (22). Each primer set was designed to generate a 100 ± 10-bp PCR fragment, which resides 30 ± 10 bp away from the next PCR fragment in the 1.8 kb intergenic region between the RH4 and PEBL genes; 1 ng of undigested genomic DNA was used as reference to normalize the PCR efficiency for each primer set. Each signal generated by all of the primer sets was normalized by that generated by a primer set (NR, Table S1) that amplifies a PCR product located in a well-positioned nucleosome that is located on chromosome 4, about 20 kb away from the RH4-PEBL loci and in the opposite direction to the telomere. Primers used for nucleosome scanning were listed in Table S1. Signals from Dd2 and Dd2NM at each primer set position were analyzed using the unpaired t test with Welch’s correction for three independent replicates in the software of GraphPad Prism version 5.0.
ChIP.
To measure the histone modification along the 5′UTRs and coding regions of the RH4 and PEBL genes, MNase digested chromatin samples were analyzed using the ChIP-IT Express Enzymatic kit (Active Motif) according to the product instruction manual; 40 μL of digested chromatin samples was incubated with 5 μg of antibodies against histone H3K9 trimethylation (17-625, Millipore), histone H3K4 trimethylation (07-473, Millipore), histone H3K9/K14acetylation (06-599, Millipore), and histone H4K5/K8/K12/K16 acetylation (06-866, Millipore), or without antibody (as input nucleosomal DNA control) at 4 °C with rotation overnight. DNA from ChIP was purified as described above and diluted with H2O to 400 μL. To measure the amount of immunoprecipitated DNA, 5 μL of purified nucleosomal DNA was amplified in each real-time PCR. The reaction was performed as described. Each of the primer sets used in ChIP was designed to amplify a 100 ± 10 bp PCR fragment that is located in a nucleosome position. The sequence of each primer is listed in Table S1.
Acknowledgments
We thank Dr. Susan Pierce, National Institute of Allergy and Infectious Diseases, for reviewing paper, Dr. Peter Crompton, National Institute of Allergy and Infectious Diseases, for statistic analysis of data, Dr. Thomas E. Wellems, National Institute of Allergy and Infectious Diseases, for plasmid pVLH, and Dr. John H. Adams, University of South Florida, for piggyback plasmids. This research was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913396107/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0913396107
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2836689?pdf=render
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/107/5/2224
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/107/5/2224.pdf
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/107/5/2224
Citations & impact
Impact metrics
Citations of article over time
Article citations
Heterochromatin dynamics during the initial stages of sexual development in Plasmodium falciparum.
Sci Rep, 14(1):23180, 05 Oct 2024
Cited by: 0 articles | PMID: 39369041 | PMCID: PMC11455859
PfSET2 Is Involved in Genome Organization of Var Gene Family in Plasmodium falciparum.
Microbiol Spectr, 11(1):e0389122, 05 Jan 2023
Cited by: 3 articles | PMID: 36602337 | PMCID: PMC9927267
Patterns of Heterochromatin Transitions Linked to Changes in the Expression of Plasmodium falciparum Clonally Variant Genes.
Microbiol Spectr, 11(1):e0304922, 14 Dec 2022
Cited by: 5 articles | PMID: 36515553 | PMCID: PMC9927496
Epigenetic and Epitranscriptomic Gene Regulation in Plasmodium falciparum and How We Can Use It against Malaria.
Genes (Basel), 13(10):1734, 27 Sep 2022
Cited by: 8 articles | PMID: 36292619 | PMCID: PMC9601349
Review Free full text in Europe PMC
Stochastic expression of invasion genes in Plasmodium falciparum schizonts.
Nat Commun, 13(1):3004, 30 May 2022
Cited by: 7 articles | PMID: 35637187 | PMCID: PMC9151791
Go to all (51) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (3)
- (2 citations) ENA - DQ100425
- (2 citations) ENA - AY032735
- (2 citations) ENA - AF420309
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Upregulation of expression of the reticulocyte homology gene 4 in the Plasmodium falciparum clone Dd2 is associated with a switch in the erythrocyte invasion pathway.
Mol Biochem Parasitol, 145(2):205-215, 25 Oct 2005
Cited by: 56 articles | PMID: 16289357
Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion.
Proc Natl Acad Sci U S A, 104(45):17789-17794, 30 Oct 2007
Cited by: 66 articles | PMID: 17971435 | PMCID: PMC2077073
Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes.
Science, 309(5739):1384-1387, 01 Aug 2005
Cited by: 191 articles | PMID: 16123303
Plasmodium falciparum: epigenetic control of var gene regulation and disease.
Subcell Biochem, 61:659-682, 01 Jan 2013
Cited by: 5 articles | PMID: 23150271
Review
Funding
Funders who supported this work.




