Abstract
Free full text
Detecting gene-gene interactions that underlie human diseases
Associated Data
Abstract
Following the identification of several disease-associated polymorphisms by whole genome association analysis, interest is now focussing on the detection of effects that, due to their interaction with other genetic (or environmental) factors, may not be identified by using standard single-locus tests. In addition to increasing power to detect association, there is also a hope detecting interactions between loci will allow us to elucidate the biological and biochemical pathways underpinning disease. Here I provide a critical survey of the current methodological approaches (and related software packages) used to detect interactions between genetic loci that contribute to human genetic disease. I also discuss the difficulties in determining the biologcal relevance of statistical interactions.
The search for genetic factors that influence common, complex traits, and the characterisation of the effects of those factors is both a goal and a challenge for modern geneticists. In the last couple of years, the field has been revolutionised by the success of genome-wide association (GWA) studies 1 2 3 4 5. Most such studies have used a single-locus analysis strategy, whereby each variant is tested individually for association with some phenotype. However, an oft-cited reason for the lack of success in genetic studies of complex disease 6 7 is the existence of interactions between loci. If a genetic factor operates primarily through a complex mechanism involving multiple other genes, and possibly environmental factors, the fear is that the effect will be missed if one examines it in isolation, without allowing for its potential interactions with these other (unknown) factors. For this reason, several methods and software packages 8 9 10 11 12 13 14 15 have been developed to consider statistical interactions between loci, when analysing data from genetic association studies. Although, in some cases, the motivation for such analyses is to increase the power to detect effects 16, in other cases the motivation has been to detect statistical interactions between loci that are informtive about the biological and biochemical pathways underpinning the disease 7. We return to this complex issue of biological interpretation of statistical interaction later in the article.
The purpose of this Review article is to provide a survey of the current methodological approaches and related software packages that are currently used to detect interactions between genetic loci that contribute to human genetic disease. Although the focus is on human genetics, many of the concepts and approaches are strongly related to methods used in animal and plant genetics. I begin by describing what is meant by (statistical) interaction, and setting up definitions and notation for following sections. I then explain how one may test for interaction between two (or more) known genetic factors, and how one may address the slightly different question of testing for association with a single factor, while at the same time allowing for interaction with other factors. In practice, one rarely wishes to test for interaction purely between known factors, unless perhaps to replicate a previous finding or to test a specific biological hypothesis. More common is the desire to search for interactions, or for loci that may interact, given genotype data at a potentially very large number of sites (e.g. from a GWA analysis or from a more focussed candidate gene study). I therefore continue the article by outlining different methods (and software packages) that search for such interactions, ranging from simple exhaustive search to various DATA-MINING/MACHINE LEARNING approaches to BAYESIAN MODEL SELECTION approaches. Throughout these sections I take as a recurring example the analysis of a publically available genome-wide data set on Crohn's disease from the Wellcome Trust Case Control Consortium (WTCCC) 1. I conclude the article with a section discussing the biological interpretation of results found from such statistical interaction analysis.
The investigation of interactions has had a long history in genetics, ranging from classical quantitative genetic studies of inbred plant and animal populations 17 18 19 to evolutionary genetic studies 20 and, finally, to linkage and association studies in outbred human populations. In this article I focus primarily on human genetic association studies: readers are referred to references 20 21 22 23 24 25 for a discussion of interactions in the context of evolutionary genetics or in human genetic linkage analysis.
Definition of statistical interaction
Interaction as departure from a linear model
The most common statistical definition of interaction relies on the concept of a linear model describing the relationship between some outcome variable and some predictor variable(s). We propose a particular model for how we believe the predictors might relate to the outcome, and we use data (i.e. measurements of the relevant variables on a number of individuals) to determine how well the model fits our observed data, and to compare the fit of different models. Arguably the most well-known form of this type of analysis is simple linear or least squares regression 26, where we relate an observed quantitative outcome y (e.g. weight) to a predictor variable x (e.g. height) via a ‘best fit’ line or regression equation y = mx+c. More generally, we may use multiple regression 26 to include several different predictor variables (e.g. x1, x2, x3, representing height, age and gender).
From a statistical point of view, interaction signifies departure from a linear model describing how two (or more) predictors, B and C say, predict a phenotype outcome A (Box 1). For a disease outcome and case-control data, rather than modelling a quantitative trait y, the usual approach is to model the (expected) log-odds of disease as a linear function of the relevant predictor variables 26 27. Given genotype data, we may evaluate the likelihood of the data under this model and use MAXIMUM LIKELIHOOD (or other) methods to estimate the regression coefficients and test hypotheses, such as the hypothesis that the interaction term (i in the mathematical formulation of Box 1) equals 0.
Supplementary Text S1 describes some specific models that follow this general formulation, including the SATURATED ‘genotype’ model. Although this model provides the best possible fit to the data, it includes many parameters. We may make parameter restrictions to generate fewer degrees of freedom (df) and thus increase power. Although written in terms of nine or fewer regression parameters, the models of Supplementary Text S1 actually represent an infinity of different models, depending on the values taken by those parameters. There has been some interest in categorizing these models 28 29 30 in such a way as to aid either mathematical or biological interpretation. As discussed later, biological interpretation is usually easiest when the PENETRANCE values all equal either 0 or 1, leading to a clear relationship between genotype and phenotype. This situation, however, is unlikely for complex genetic diseases.
Marginal effects
An important issue in genetic studies is whether there are factors that display interaction effects, without displaying so-called MARGINAL EFFECTS 6 31. The problem with factors that display interaction effects, without displaying marginal effects, is that these factors will be missed in a single-locus analysis, as they do not lead to any marginal correlation between genotype and phenotype when each locus is considered individually. It is not clear in practice how often this might occur, as many models that include an interaction term even in the absence of main effects (α and β in the mathematical formulation of Box 1) do, in fact, lead to significant marginal effects i.e. they show correlations between genotype and phenotype that are detectable in a single-locus analysis. Thus, although one may derive mathematical models (sets of specific values for the regression coefficients) that lead to single-locus models displaying no marginal effects 6, it remains to be seen whether such models represent common underlying scenarios – and thus a potentially serious problem – in complex genetic diseases.
For simplicity, I have concentrated here on defining interaction in relation to two genetic factors (two-locus interactions). In practice, however, for complex diseases we might expect three-locus, four-locus and even higher-level interactions to operate as well. Mathematically, such higher-level interactions are simple extensions to the two-locus models described earlier. The problem with these models is that they contain a large number of parameters, which would require extremely large data sets to estimate accurately. Interpreting the resulting parameter estimates is also complicated, except perhaps in some simple cases – for example, when risk alleles at all loci are required to alter disease risk (i.e. when only the full multilocus interaction term differs from zero).
Testing for interaction between known factors
Regression models
Given two or more known (or hypothesised) genetic factors influencing disease risk, arguably the most natural way to test for statistical interaction (on the log-odds scale) is simply to fit a LOGISTIC REGRESSION MODEL that includes main effects and relevant interaction term(s) and then to test whether the interaction term(s) equal zero or not. A similar approach can be used for quantitative phenotypes, in which case linear rather than logistic regression is used. These analyses can be performed in virtually any statistical analysis package after construction of the required genotype variables. Alternatively, the --epistasis option in the whole-genome analysis package PLINK 12 provides a logistic regression test for interaction that assumes an allelic model both for main effects and interactions.
A more powerful approach in case-control studies is to use a ‘case-only’ analysis 32 33 34. Case-only analysis exploits the fact that, under certain conditions, an interaction term in the logistic regression equation corresponds to dependency or correlation between the relevant predictor variables within the population of cases. A case-only test of interaction can therefore be performed by testing the null hypothesis that there is no correlation between alleles or genotypes at the two loci, in a sample restricted to cases alone. This test can easily be performed via a simple χ2 test of independence between genotypes (a 4 degree of freedom (df) test) or alleles (a 1df test), or via logistic or MULTINOMIAL REGRESSION, in any statistical analysis package.
The main problem with the case-only test is its requirement that the genotype variables be uncorrelated in the general population – indeed it is this assumption, rather than the design per se, that provides the increased power compared to case-control analysis. The case-only test is therefore unsuitable for loci that are either closely linked or show correlation for some other reason (e.g. if certain genotype combinations are related to viability). Unlike epidemiological studies of environmental factors, where correlation and CONFOUNDING between variables is common, in genetic studies the assumption of independence between unlinked genetic factors would seem fairly reasonable. One could use a two-stage procedure to test first for correlation between the loci in the general population, and then use the outcome to determine whether to perform a case-only or case-control interaction test. However, this procedure has potential bias 35.
A preferable approach is to incorporate the case-only and case-control estimators into a single test. In this vein, Zhao et al. 36 proposed a test based on the difference in inter-locus allelic association between cases and controls, an idea originally suggested by Hoh and Ott 37. The --fast-epistasis option in PLINK 12 performs a similar test. Zhao et al. 36 found their test had greater power than a 4df logistic regression test of gene-gene interaction; however, this power increase may be largely due to the lower df in their allelic (rather than genotypic) test. Mukherjee and Chatterjee 38 35 proposed an EMPIRICAL BAYES PROCEDURE that uses essentially a weighted average of the case-control and case-only estimators of the interaction. This approach exploits the gene-gene independence assumption (and thus the power) of case-only analysis, while additionally incorporating controls, allowing the estimation of main effects. Routines that implement this procedure are available in Excel and/or Matlab.
Other approaches
Although regression-based tests of interaction would seem most natural (given the definition of interaction as departure from some linear regression model), alternative approaches have been proposed. Yang et al. 39 proposed a method based on partitioning of χ2 values that, similar to 36, contrasts inter-locus association between cases and controls. Their method showed higher power than logistic regression when the loci had no marginal effects. Recently there has been interest in INFORMATION-THEORETIC or ENTROPY-BASED approaches for modelling genetic interactions 40 41 42 43. It is unclear whether this framework offers any advantage over more standard statistical modelling of the same predictor variables, as in most cases the conditional probability statements implied by the two approaches are entirely equivalent 44.
Family-based studies
Here I have focussed on testing for interaction in the context of case-control or population-based studies. Several related methods have been proposed to test for interaction in the context of family-based association studies 45 46 47 48 49. The case-pseudocontrol 46 approach offers a regression-based framework that allows interaction tests very similar to those described here. Given the large sample sizes that are required when testing for interaction as opposed to main effects, 50 51, it is unclear whether investigators will have family-based cohorts of sufficient size to provide high power for detection of interactions. However, such cohorts may provide a useful resource for replication and characterisation of interaction effects that have been found using alterative means.
Testing for association while allowing for interaction
Rather than testing for interaction per se, many researchers are interested in allowing for interaction (with other genetic or environmental factors) when testing for association at a given genetic locus. The rationale is that if the test locus influences disease or phenotype outcome via interaction with another factor, then allowing for this interaction should increase the power to detect the effect at the test locus. From a mathematical point of view, a test for association at a given locus C, while allowing for interaction with another locus B (a ‘joint’ 16 test), corresponds to comparing the fit (to the observed data) of a linear model in which main effects of B, C and their interactions are included, to a model in which all terms (main or interaction) involving locus C are removed (Box 1).
Theoretically, if no interaction effects exist, these joint tests will be less powerful than marginal single-locus association tests. However, if interaction effects do exist, then the power of joint tests can be higher than that of single-locus approaches 52. Kraft et al. 16 showed that the joint test of a genetic effect, while allowing for interaction with a known environmental factor, performed nearly optimally over a wide range of plausible underlying models. This test uses case-control data to test the combination of a main effect at locus C and an interaction effect; since case-only analysis provides a more powerful test for the interaction effect 32 33 34, Chapman and Clayton 53 proposed using a version of the joint test that combines a case-control main effect component with a case-only interaction component.
The joint test of association, while allowing for interaction, assumes that one has some known (or hypothesised) measured factor with which the test locus may interact. In the absence of a specific factor of this type, a natural approach is to average over all other (potentially interacting) genetic factors when performing a test at a given locus. A Bayesian approach for doing this, in the context of GWA studies, is in development 14 and a beta version of the associated BIA software is available in limited release from its authors on request. Rather than averaging over all possible interacting loci, Chapman and Clayton 53 proposed using the maximum value of the joint test, evaluated over a pre-defined set of potentially modifying (interacting) loci, with significance assessed using a PERMUTATION argument.
Here I have concentrated on the issue of testing (either for interaction, or for association while allowing for interaction) at one or two specific genetic variants of interest. Rather than testing a single variant, it is now quite common to have genotype data at a large number of variants that may or may not have any prior evidence for involvement with disease. Given such data, various model selection approaches have been proposed that allow one to essentially step through a sequence of regression models searching for significant effects, both main effects and interactions 37 8 9 10 13 54 55 56. These approaches will be described in more detail in subsequent sections. First, I describe an approach that is feasible provided the number of main and interaction effects to be examined is not too large, namely, simple exhaustive search.
Exhaustive search
Two-locus interactions
Given genotype data at a number of different loci, arguably the simplest way to search for interactions between these loci is by exhaustive search. For example, to test all two-locus interactions, one could consider all possible pairs of loci and perform the desired interaction test for each pair. Similarly if testing for association while allowing for interaction, one could perform the relevant 3df or 8df 52 test (Box 1, Supplementary Text S1). Clearly an exhaustive search of this type raises a MULTIPLE TESTING issue somewhat analogous to the multiple testing issue encountered in single-locus analysis of GWA studies 1. If all tests are independent, a BONFERRONI CORRECTION is appropriate 52; however, LD between loci will induce correlation between many of the tests. If testing for association while allowing for interaction, additional correlation occurs due to the fact that the main effect of a locus will be a component of all tests involving that locus. Theoretically, one can use permutation 53 to assess significance while allowing for the multiplicity of (and correlation between) the tests performed, but, for large numbers of loci, this may be computationally prohibitive.
A pragmatic approach to the multiple testing issue in single-locus analysis of GWA studies is to use a relatively stringent significance threshold (e.g. p = 5 × 10−7) coupled with replication in an independent data set, to avoid generating large numbers of false positives. Stringent significance thresholds can also be motivated by Bayesian arguments concerning the low prior probability of any given variant being associated with disease 1. In practice, the Q-Q PLOT 1 has emerged as the tool of choice for visualising the results from an entire genome-scan.
Exhaustive search of all two-locus interactions from a genome-scan is time-consuming but computationally feasible. Marchini et al. 52 quote a time of 33 hours on a 10 node cluster to perform all pairwise tests of association (allowing for interaction) at 300,000 loci in 1000 cases and 1000 controls. The PLINK 12 website quotes 24 hours to test (using the --fast-epistasis option) all pairwise interactions at 100,000 loci typed in 500 individuals. Given that genome-wide studies now routinely generate between 500,000 and 1 million markers in 5000 or more individuals, these times will need to be scaled upwards by several weeks or even months, but exhaustive search of all two-locus interactions still remains feasible. In addition, the fact that each test can be computed independently of all other tests means that the entire search can be split up into several separate jobs to make use of parallel processing facilities, if available.
Higher-order interactions
The problem with exhaustive search is that it does not scale up to consideration of higher-order interactions. Since the number of tests (and therefore the time taken to perform the analysis) increases exponentially with the order of interaction considered, exhaustive search of all three-way, four-way or higher-level interactions would seem impractical in a genome-wide setting. For this reason, two-stage procedures have been proposed 57 52 58, whereby a subset of loci that pass some single-locus significance threshold are chosen, and exhaustive search of all two-locus interactions (or higher order if required, perhaps conditional on significant lower order effects 58) is carried out on this ‘filtered’ subset. The obvious drawback with this approach is that loci will only make it into the second (or subsequent) stages of the testing procedure if they show some marginal association with phenotype. Therefore this procedure would not be expected to be useful for detecting interactions that genuinely occur in the absence of marginal effects.
Use of a single-locus significance threshold is not the only way to reduce the number of markers for testing. Several of the machine learning approaches described in the next section (in particular ReliefF and Random Forests) could be used, as they do not require a locus to have a significant marginal effect. Biological plausibility offers an alternative strategy. Bochanovits et al. 59 used evidence of co-adaptation between loci in the mammalian genome to inform their selection of genes to undergo interaction testing in a human study. Emily et al. 60 used experimental knowledge on biological networks to reduce the number of interaction tests from 125 billion to 71,000, when analysing genotype data from the WTCCC 1. In their analysis of seven disease cohorts they found four significant interaction effects, including one (p = 1 × 10−9) between rs6496669 on chromosome 15 and rs434157 on chromosome 5 in Crohn's disease. An example of applying semi-exhaustive testing to this same data set, using the --fast-epistasis and --case-only options in PLINK 12, is shown in Figure 1.
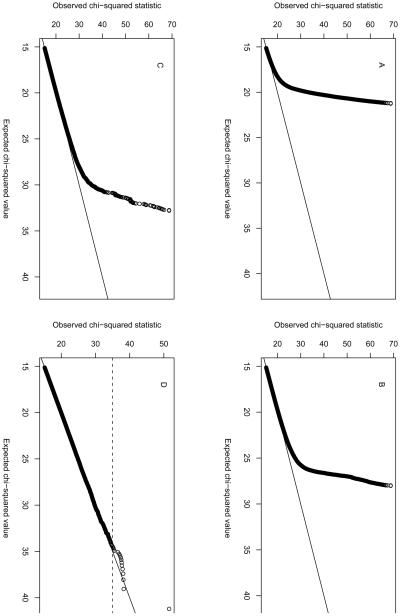

I used the --fast-epistasis and --case-only options in PLINK to analyse the WTCCC Crohn's disease and control samples. I used the same quality control procedures as the WTCCC to remove poor-quality SNPs and samples prior to analysis. I additionally discarded 561 SNPs that had been analysed by WTCCC but were subsequently discarded based on visual inspection of the SNP intensity cluster plots (Jeff Barrett, personal communication). To reduce the number of interaction tests to be performed I selected a set of 89294 SNPs that passed a single-locus p value threshold of 0.2. Analysis of the 89294 SNPs on a single node of a computer cluster took 14 days. Unfortunately, neither SNP in the interaction detected by Emily et al. 60 had the opportunity to appear in my analysis, as neither had a single-locus p value <=0.2.
(A) Results from --case-only analysis, in which SNP pairs were discarded if they were < 1Mb apart (Panel a), <5Mb apart (Panel b), and <50Mb apart (Panel c). The default in PLINK is to exclude tests of pairs of SNPs that are less than 1Mb apart. Even when extreme separations of 5Mb or 50Mb are enforced (Panels b and c), we find an excessive number of apparently significant results. Closer inspection revealed that in many cases these significant results result from correlation (within the sample of cases) between alleles at loci on different chromosomes. Given the general departure from the expected distribution, it seems likely that these significant --case-only results are artifacts rather than genuine interaction effects. Panel d: Q-Q plot of all results from the --fast-epistasis with p value < 0.0001. These results lie much closer to the expected line: indeed only one result appears to show strong departure from expected significance. The top ranking results (those with χ2 > 35, as indicated by the dashed line on Panel d) are shown in Supplementary Table 1. Interestingly, most of the SNPs involved in the putative interactions show little single-locus significance, apart from rs4471699 on chromosome 16. This SNP was not reported as significantly associated by WTCCC 1.
(B) Single-locus association results across chromosome 16. rs4471699 at position 30227808 shows the highest significance, but is far removed from the bulk of the significant results which are situated close to the NOD2/CARD15 gene (around position 49297083) Further investigation revealed that this SNP had been excluded from the WTCCC analysis owing to poor genotype clustering (Jeff Barrett, personal communication), even though it passed the stated WTCCC exclusion criteria and had not appeared in the original list of additional exclusions I was given. It therefore seems highly likely that both the single-locus and interaction results at rs447169 represent false positives.
Data-mining/machine learning and related approaches
Traditional regression-based methods are often criticised 8 61 31 for their inability to deal with non-linear models and with HIGH-DIMENSIONAL DATA (containing many potentially interacting predictor variables, leading to sparse contingency tables with many empty cells). For this reason, machine learning or data-mining methods, developed in the field of computer science, are sometimes preferred. The selection of predictor variables, and interactions between them, that predict an outcome variable is a well-known problem in these fields. Data-mining approaches do not fit a single pre-specified model, nor do they attempt exhaustive search, but rather they attempt to step through the space of possible models (including potentially large numbers of main effects and multi-way interactions) in some computationally efficient way. Many data-mining approaches are, in fact, equivalent to stepping through a particular sequence of regression models and attempting to find the model that best fits the data; the distinction that is often made between data mining and regression models is therefore, to some extent, a false one. Non-linearity is not an issue when fitting a SATURATED MODEL (although it may be an issue for more restricted models). One common theme in data-mining is the use of CROSS-VALIDATION 62 to avoid problems of OVERFITTING.
Data-mining methods typically have problems dealing with incomplete and/or unbalanced data sets (e.g. when the number of cases and controls are unequal 63). They also do not always deal particularly well with correlated predictors showing colinearity. This has been addressed in the mainstream statistics literature by the introduction of penalized regression approaches 64 65 that allow large numbers of predictor variables to be included in a regression model, but with many estimated regression coefficients ‘shrunk’ towards zero. In genetics, use of such techniques is just starting to emerge, including penalized logistic regression 66 67 and least angle regression 68 for identifying gene-gene interactions 69 70 in binary traits.
A good overview of several machine learning approaches for detecting gene-gene interactions is given by McKinney et al. 31. For the remainder of this section, I will focus on several methods that have become particularly popular and/or appear to show particular promise for detection of gene-gene interactions, or, more precisely, for detection of genes that may interact.
Recursive partitioning approaches
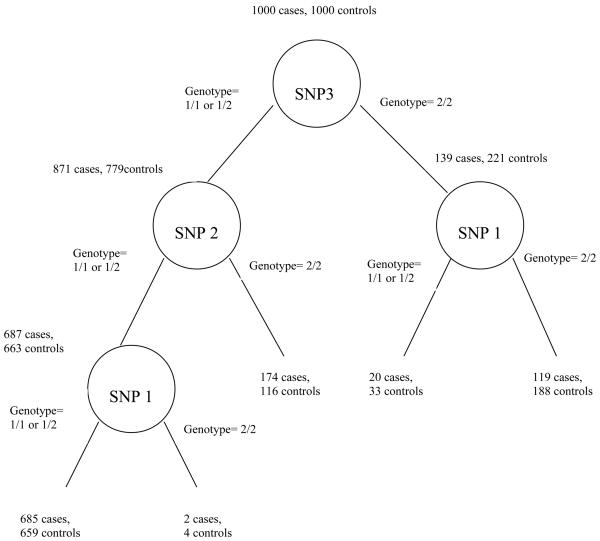
Recursive partitioning approaches (Box 2) have been used as an alternative to traditional regression methods for detecting genetic loci (and their interactions) that influence a phenotypic outcome 71 72 73. These approaches produce a graphical structure (resembling an upside-down tree) that maps possible values of certain predictor variables (e.g. SNP genotypes) to a final expected outcome (e.g. disease status). Each vertex or node of the tree represents a predictor variable, and from each node there are arcs or edges leading down to so-called ‘child’ nodes, where each edge corresponds to a different possible value that could be taken by the variable in the ‘parent’ node. A path through the tree represents a particular combination of values taken by the predictor variables appearing within that path.
Recursive partitioning approaches do not include interaction variables per se in the model. Rather, the nature of the trees constructed allows for interaction in the sense that each path through a tree corresponds to a particular combination of values taken by certain predictor variables, thus including potential interactions between them. The aim of tree-based approaches therefore corresponds most closely to testing for association while allowing for interaction rather than testing for interaction per se. One limitation of recursive partitioning is that, since it conditions on main effects of variables at the first stage (and on main effects conditional on previously selected variables at subsequent stages), pure interactions in the absence of main effects can be missed 74.
Rather than using a single tree, significant improvements in classification accuracy can result from growing an ensemble of trees. A popular ensemble tree approach is random forests 75 (Box 2). This approach has been used in several genetic studies 76 77. Apart from the classification of future observations (not our focus of interest), the main result of a random forests analysis is a list of variable importance measures. These measure the impact of each predictor variable both individually and via multi-way interactions with other predictor variables, and therefore have an advantage over a list of significance values from single-locus association testing.
Random forests provide a parallelizable and relatively fast algorithm for measuring variable importance, partly because at each split only a small random subset of predictors is used. To allow each predictor the opportunity to enter the model and to produce accurate prediction, one must choose carefully a number of key parameters such as the number of trees in the forest, the number of randomly-chosen SNPs considered at each node and the number of permutations used to assess variable importance. In an ideal world one might repeat the analysis several times to assess sensitivity to choice of these parameters. An example of applying random forests to the WTCCC Crohn's disease and control data, using the software package Random Jungle 78, is shown in Figure 2.

I used the software package Random Jungle 78 to perform a random forests analysis of the 89294 SNPs passing a single-locus p value threshold of 0.2 in the WTCCC Crohn's and control data. Since Random Jungle, in common with many other machine-learning approaches, prefers not to have missing (incomplete) genotype data, missing genotypes were imputed as the single most likely value on the basis of the genotype frequencies in the case-control data set. Analysis of the 89294 SNP set took approximately 5 hours, using 6000 trees in the forest and
Multifactor Dimensionality Reduction method
A variety of other data-mining approaches have been used for detection of interactions or potentially interacting variables in genetic association studies, including logic regression, 79 80 genetic programming 81, neural networks 54 55 and pattern-mining 82 83. One particularly popular method is Multifactor Dimensionality Reduction (MDR) 8 9 10. MDR has been used to identify putatively interacting loci in several phenotypes including breast cancer 8, type 2 diabetes 84, rheumatoid arthritis 85 and coronary artery disease 86, although, to date, it is unclear whether any of these identified interactions have been replicated in larger samples.
The MDR algorithm is described in Box 3 and in detail elsewhere 8 9 10 11 49. Rather than testing for interaction per se, MDR seeks to identify combinations of loci that influence a disease outcome, possibly via interactions rather than (or in addition to) via main effects. MDR achieves dimension reduction by converting a high-dimensional (multi-locus) model to a one-dimensional model, thus avoiding the issues of sparse data cells and over-parameterised models that can cause problems for traditional regression-based methods. MDR classifies genotypic classes as either ’high risk’ or ’low risk’ according to the ratio of cases and controls that are represented in each class. This could be considered overly simplistic: improvements that embed a more traditional regression-based approach into the cell classification step, allowing application to continuous as well as binary traits and adjustment for covariates, have been proposed 87 88.
The main problem with MDR (in common with other exhaustive search techniques) is that it does not scale up to consideration of large numbers of predictor variables (e.g. large numbers of loci from a genome-wide association study) 8 9. By performing exhaustive search for the best n-locus combination (within each of ten cross-validation replicates), anything more than a two-locus screen on more than a few hundred variables will be computationally prohibitive. An additional problem with early versions of the widely-used Java implementation of the MDR software (although note that other software implementations do exist 11 88) is that it was not designed with genome-wide data sets in mind, and thus could fail due to memory and disc-usage issues; these problems, however, appear to have been addressed in the most recent version of the software.
For investigation of higher-order interactions, MDR is therefore perhaps best suited for use with small numbers of loci (up to a few hundred), perhaps from a candidate gene study or selected from a larger set of potential predictors via a prior pre-processing or filtering step 40. This step could be as simple as using a single-locus significance threshold, but that would seem counter-intuitive if the goal is to detect interactions in the absence of marginal effects. Perhaps more appealing would be to use a measure of variable importance that allows for possible interactions, such as the variable importance measure from a random forests analysis or from one of the alternative filtering methods described below.
ReliefF, Tuned ReliefF and Evaporative Cooling
One promising filtering algorithm that has been proposed 40 is ReliefF 89, or its modified version, Tuned Relief (TuRF) 90. This approach uses a measure of proximity between observations (individuals) – calculated, for example, on the basis of the genome-wide genetic similarity between individuals – to determine each individual's nearest neighbours from within his or her own phenotype class, and from within the opposite phenotype class. For each predictor variable, its difference in value between pairs of neighbouring individuals, weighted negatively or positively according to whether the individuals come from the same or different phenotype classes, can be used to construct an importance measure for that variable 90. The algorithm is relatively simple and scalable and so should be applicable to large numbers of predictor variables and observations; an in-house C++ implementation was able to analyse 1 million loci in 200 individuals in approximately four minutes 90.
ReliefF and TuRF have both been implemented in the Java version of the MDR software. One problem with ReliefF is that it can be sensitive to large backgrounds of variants that are irrelevant to phenotype 74. This has motivated development of an alternative approach, Evaporative Cooling 91 74, that can be used to combine the strengths of ReliefF with those of random forests 74.
An example of analysis using the Java implementation of TuRF and MDR, applied to the WTCCC Crohn's disease data, is shown in Figure 3.
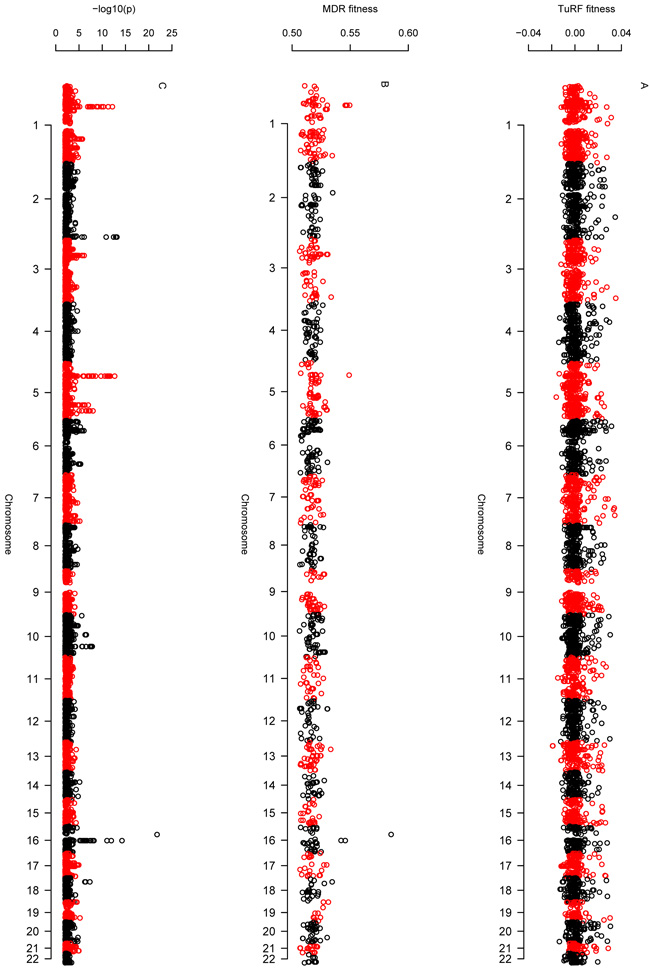
I used the Java implementation of MDR to analyse 6113 SNPs passing a single-locus p value threshold of 0.01 in the WTCCC Crohn's and control data, with missing (incomplete) genotypes imputed as described in the legend to Figure 2. Examination of all pairwise combinations in the entire 6113 SNP set proved computationally prohibitive but analysis via use of a prior filtering step with ReliefF or TuRF, which reduced the data set for MDR analysis to 1000 SNPs, was achievable. The best single-locus model identified was rs4471699, providing testing accuracy of 0.5852 and cross validation consistency of 10/10. The best two-locus model identified was rs4471699 and rs2076756, providing testing accuracy of 0.5879 and cross validation consistency of 4/10. MDR, in common with the other methods investigated, has clearly been dominated by the false positive result at rs4471699. Interestingly, however, this SNP is not selected by TuRF when filtering down the set of SNPs for MDR analysis to include only 100 SNPs. With the 100 SNP set, the best single-locus model identified was rs931058, providing testing accuracy of 0.5114 and cross validation consistency of 5/10. The best two-locus model identified was rs931058 and rs10824773, providing testing accuracy of 0.5205 but cross validation consistency of only 2/10. With the 100 SNP set it was computationally feasible to fit 3-locus and 4-locus models, however the resulting best models had similarly low cross validation consistencies. I also found extreme sensitivity (in both TuRF and MDR) to the choice of random number seed (data not shown), suggesting that, overall, these results should be interpreted with caution. A problem with MDR is that it outputs only the ‘best’ model rather than a measure of significance for all models or variables considered. Some idea of the ‘importance’ of variables can be determined by examining the ‘fitness landscape’ output from the program, shown here. Panel A: Fitness landscape scores from TuRF analysis of all 6113 SNPs Panel B: Fitness landscape scores from MDR analysis using top 1000 out of 6113 SNPs (filtered using TuRF) Panel C: Results from single-locus association analysis of all 6113 SNPs using the trend test implemented in PLINK. It is unclear whether the fitness landscape results from TuRF (Panel A) or MDR (Panel B) offer any great advantage over standard single-locus analysis (Panel C) with respect to determining the importance of variables.
Bayesian model selection approaches
Bayesian model selection techniques 92 offer an alternative approach for selecting predictor variables, and interactions between them, that best predict phenotype. The key difference between Bayesian model selection and simple comparisons of nested regression models via FREQUENTIST (non-Bayesian) procedures, lies in the specification of prior distributions for the unknown regression parameters as well as for a dimension parameter, specifying how many non-zero predictors are to be included in the regression equation. A posterior distribution for these parameters, given the observed data, can then be calculated through use of Markov chain Monte Carlo (MCMC) 93 simulation techniques, in which one traverses the space of possible models (sets of parameter values), sampling realisations at intervals. Although MCMC is an extremely flexible approach, it can require some care with respect to the choice of prior distributions, proposal schemes (determining how one moves between models) and the number of iterations required to achieve convergence.
Lunn et al. 56 propose essentially a Bayesian version of stepwise regression, implemented in the software WinBUGS. This method focuses on main effects of loci rather than interactions, but inclusion of interaction effects represents a relatively straightforward extension. The main problem with this method is that it can deal with at most only a few hundred variables 56 and does not scale to the large numbers of predictor variables that might be encountered in a genome-wide study. However, related approaches that can deal with data sets of higher dimensionality have been proposed 94.
Bayesian Epistasis Association Mapping
A recently-proposed MCMC approach specifically designed for the detection of interacting (as well as non-interacting) loci is Bayesian Epistasis Association Mapping 13, implemented in the software package BEAM. In BEAM, predictors in the form of genetic marker loci are divided into three groups: group 0 contains markers that are unassociated with disease, group 1 contains markers that contribute to disease risk via main effects only, and group 2 contains markers that jointly influence (i.e. interact) to cause disease via a saturated model. Given prior distributions concerning the membership of each marker in each of the three groups, and prior distributions for values of the relevant regression coefficients given group membership, a posterior distribution for all relevant parameters can be generated using MCMC simulation. As well as making inferences in a fully Bayesian inferential framework, one may use the results from BEAM in a frequentist hypothesis testing framework via calculation of a so-called ‘B-statistic’ 13 that tests each marker or set of markers for significant association with disease phenotype.
BEAM can handle relatively large numbers of markers (e.g. 100,000 SNPs typed in 500 cases and 500 controls 13) although, in practice, some modification to the default parameters (namely the BURN-IN PERIOD, number of starting points and number of MCMC iterations) may be required in order to apply the method in reasonable time. BEAM does not currently handle the 500,000 - 1 million markers that are now routinely being genotyped in genome scans of perhaps 5000 or more individuals. In theory, BEAM can account for LD between adjacent markers 13. However, it is unclear whether LD between non-adjacent markers is fully accounted for, suggesting that some ‘thinning’ of the marker set may be required, not only for computational reasons, but also to ensure that the markers are in low LD. An example of applying BEAM to the WTCCC Crohn's data is shown in Figure 4.

I used BEAM to analyse a set of 47724 SNPs passing a single-locus p value threshold of 0.1 in the WTCCC Crohn's and control samples. Analaysis of the 47724 SNPs took 8 days (with some modification to the default settings, most notably imposing a maximum of 5 × 107 MCMC iterations 13 as opposed to the default value of n2, where n is the number of loci). I estimated that analysis of the 89294 SNP set (passing a single-locus p value threshold of 0.2) with a similar number of MCMC iterations would have taken more than five weeks. Panel A: ‘B-statistic’ p values for the 1321 single-locus associations detected by BEAM. Panel B: Results from single-locus association analysis of all 47727 SNPs using the trend test implemented in PLINK. BEAM detects essentially the same loci as are detected by single-locus analysis. BEAM additionally detects (with quoted p = 0.000000) four two-locus interactions, each involving an interaction of rs2532292 on chromosome 17 with a nearby SNP (either rs12150547, rs17689882, rs17650381 or rs17574824) within the same cluster. None of these SNPs shows particularly strong single-locus association and so this putative interaction is intriguing. However, none of these pairs of SNPs showed significant (defined as p value < 0.0001) interaction in the PLINK --fast-epistasis analysis. Closer inspection of these SNPs in the control sample indicated that they are in strong LD (D′ > 0.99) with one another, suggesting that the detected interactions may in fact correspond to marker dependencies due to LD, rather than to genuine interaction effects.
Biological interpretation
The extent to which statistical interaction implies biological or functional interaction has been extensively debated in both the genetics 95 21 96 97 19 98 99 and epidemiological 100 101 102 literature. One problem has been the inherently different nature of definitions of interaction, and use of a common term, ‘epistasis’, to encapsulate these definitions 95 21 (Supplementary Text S2). In a recent review, Phillips 20 defines three different forms of epistasis – COMPOSITIONAL, STATISTICAL and FUNCTIONAL – that capture rather different concepts often lumped together under this single term. A unified framework, the natural and orthogonal interactions (NOIA) model, was proposed by Alvarez-Castro and Carlborg for modelling both statistical and functional epistasis. However, Alvarez-Castro and Carlborg's definition of ‘functional’ seems rather far removed from that of Phillips. The NOIA model is actually a mathematical model that is essentially a reparameterization of classical quantitative genetics models 19 (Supplementary Text S2) that allows main effects to be defined with respect to a different reference point, and interaction effects to be defined with respect to different definitions of ‘independence’ of main effects, in order to allow mapping of models between different experimental populations. Since, in a sense, the whole issue in interaction modelling is how one defines the ‘effect’ of a variable, and therefore how one measures ‘departure’ from ‘independence’ of effects (Supplementary Text S2), this reparameterization does not seem especially biologically enlightening.
Although it seems reasonable to assume that functional epistasis in the form of biomolecular or protein-protein interaction is a ubiquitous component of the underlying biological pathways determining disease progression 103 7, it does not follow that it will be detected as a mathematical or statistical interaction 102 104 - particularly if the variables being examined are, as in many cases, simply surrogates for the true underlying causal variants, correlated with these variants because of LD. The historical lack of success in genetic studies of complex disease can largely be attributed, not to ignored biological interactions 6 61 7, but rather to under-powered studies that surveyed only a fraction of genetic variation; the recent success of GWA studies 1 2 3 4 5 has demonstrated that single-locus association analysis in sufficiently large sample collections can detect modest genetic effects reliably and with robust replication 105 106.
Although the extent to which biological interaction can be inferred from statistical interaction may be limited 102, some interesting recent work 107 108 109 has focussed on whether, given a strong prior biological model (or set of models), one can use genetic and/or genomic data from outbred populations or inbred strains, to assess model fit and compare the fit of competing models. This is, in a sense, a more modest goal in that it relies on some prior understanding (or at least a strong biological hypothesis) concerning the action of the relevant predictors.
Conclusions
As we have seen, there are numerous methods, and an even larger number of software implementations, that allow investigators to examine or test for interaction between loci, given data of the type currently generated from large-scale genotyping projects. Although precise details of the methodologies differ, in many cases there are close conceptual links between the different approaches, an understanding of which can perhaps best be obtained through understanding the difference between testing for interaction versus testing for association while allowing for interaction.
From a practical point of view, probably the main difference between the methods I have described is the computational time required to implement the analysis. As data sets become ever larger, development of efficient and parallelizable computational algorithms will become increasingly more important. On this note, the use of ‘filtering’ approaches, that allow one to pre-select a subset of potentially interesting loci for input to a more computer-intensive exhaustive or stochastic search algorithm, may hold promise. In my application of various methods to the WTCCC Crohn's disease data, I found semi-exhaustive search of two-locus interactions (implemented in PLINK 12) and a random forests analysis (implemented in Random Jungle 78) to be the most computationally feasible of the methods examined. Bayesian Epistasis Association Mapping (implemented in BEAM 13) was feasible only for a filtered data set and with some modification to the default (recommended) input parameter settings: it is unclear what effect (if any) this will have had on the reliability of the results. MDR was feasible for examining two-locus interactions in a drastically filtered data set, or for examining higher-level interactions in an even further reduced data set.
To date, very few publications have incorporated interaction testing of GWA data. This is perhaps not surprising as GWA studies have naturally focussed on single-locus testing in the first instance. Curtis 110 performed pairwise tests of association at 396,591 markers using 541 subjects (cases and controls) from a genomewide study of Parkinson's disease. He found no significant epistatic interactions, possibly because of the small sample size and/or because of the interaction test employed (which might have been more powerful if restricted to cases alone). Gayan et al. 15 used the same data set to perform two-locus interaction testing via their interaction-detection approach known as ‘Hypothesis Free Clinical Cloning’ (HFCC). This approach involves testing for association (while allowing for interaction) under a set of pre-specified fully penetrant disease models, with the tests performed within several different subgroups of the data (considered as ‘replication groups’). For the Parkinson's analysis, each subgroup consisted of approximately 90 cases and 90 controls, which seems a remarkably small sample size for this kind of analysis; not surprisingly, little consistency between results was found when the analysis was repeated using different partitions of the data. Emily et al. 60 reported four significant cases of epistasis in the WTCCC data using an approach that narrows the search space based on experimental knowledge of biological networks.
Given the large number of GWA studies that have recently or are currently being performed, it is clear that, for many, genomewide interaction testing will be the natural next step following single-locus testing. We await with interest the results of these analyses.
Supplementary Material
SupplTable1
SupplText1
SupplText2
Acknowledgements
Support for this work was provided by the Wellcome Trust (Grant reference 074524). I thank Jeff Barrett for assistance with interpretation of the WTCCC Crohn's results, and the WTCCC for making their data freely available. Thanks also to Jason Moore for useful discussions concerning data-mining methods in general and MDR in particular, and to Kevin Keen for pointing out the origins of the term ‘epistasis’.
Glossary
| Data-mining | The process of extracting hidden patterns and potentially useful information from large amounts of data. |
| Machine learning | The ability of a program to learn from experience — that is, to modify its execution on the basis of newly acquired information. A major focus of machine learning research is to automatically produce models (rules and patterns) from data. Hence, machine learning is closely related to fields such as data-mining, pattern recognition and statistics. |
| Bayesian model selection | A statistical approach for selecting (choosing between) models by incorporating both prior distributions for parameters of the models and the observed experimental data. |
| Maximum likelihood | A statistical approach that is used to make inferences about the combination of parameter values that gives the greatest probability of obtaining the observed data. |
| Saturated | A term for a statistical model that is as full as possible (‘saturated’) with parameters. Such a model is sometimes useful as it serves as a benchmark to quantify how well a simpler model (one with fewer parameters) fits the data. |
| Penetrance | The probability of displaying a particular phenotype (e.g. succumbing to a disease) given that one has a specific genotype. |
| Marginal effects | The average effects (e.g. penetrances) of a single variable, averaged over the possible values taken by some other variable(s). These could be calculated, for one locus of a two-locus system, say, as the average of the two-locus penetrances, averaged over the three possible genotypes at the other locus, using the relevant population genotype frequencies for both loci |
| Logistic regression model | A statistical model that is used when the outcome is binary in nature. Relates the log odds of the probability of an event to a linear combination of predictor variables. |
| Multinomial regression | A statistical approach, similar to logistic regression, that is used when the outcome takes one of several possible categorical values. |
| Information theory | A branch of applied mathematics involving the quantification of information. |
| Confounding | A phenomenon whereby the measure of association between two variables is distorted because other variables, associated with both variables of interest, are not controlled for in the calculation. |
| Empirical Bayes procedure | A hierarchical model in which the hyperparameter is not a random variable but is estimated by some other (often classical) means. |
| Information theory | A branch of applied mathematics involving the quantification of information |
| Entropy | A key measure used in information theory, that quantifies the uncertainty associated with a random variable. For example, a variable indicating the outcome from a throw of a fair coin (2 equally likely outcomes) will have less entropy than a variable indicating the outcome from a roll of a die (6 equally likely outcomes). |
| Permutation | An approach often used in hypothesis testing. In this approach, an empirical distribution of a test statistic is obtained by permuting the original sample many times and re-calculating the value of the test statistic in each permuted data set. Each permuted sample is considered to be a sample of the population under the null hypothesis. |
| Multiple testing | An analysis in which multiple independent hypotheses are tested. If a large number of tests are performed, the significance level (p-value) of any particular test must be interpreted in the light of this fact, as the overall combined probability of making a type I error will increase. |
| Bonferroni correction | The simplest correction of individual p-values for multiple-hypothesis testing: pcorrected = 1 − (1 − puncorrected)n, where n is the number of hypotheses tested. This formula assumes that the hypotheses are all independent, and simplifies to pcorrected = npuncorrected when npuncorrected <<1. |
| Q-Q plot | A quantile-quantile (Q-Q) plot is a diagnostic plot that can be used to compare the distribution of observed test statistics with the distribution expected under the null. Those tests that lie significantly above the line of equality between observed and expected quantiles are considered significant in the context of the number of tests performed. |
| High-dimensional data | Data containing information on a very large number of variables, albeit possibly measured in a small number of subjects or replicates. |
| Cross-validation | A technique that involves partitioning a data set into smaller sub-samples, performing an analysis in one sub-sample and using the other sub-sample to measure or validate how well the analysis has performed. To reduce variability, multiple rounds of cross-validation are often performed using different partitions of the data and the validation results are averaged over the rounds. |
| Overfitting | The phenomenon whereby a complex model might provide a good fit to the current data set, but is in fact ‘over’ fitted to the random quirks in that paticular data set, and thus does not provide such good generalizabililty to future data sets as would a simpler model. |
| Frequentist | A statistical approach for testing hypotheses by assessing the strength of evidence for the hypothesis provided by the data. |
| Burn-in period | In Markov chain Monte Carlo (MCMC) analysis, a period at the start of the computation in which the values taken by the parameters are ignored (thrown away) for the purposes of constructing the posterior distribution. |
| Compositional, statistical and functional epistasis | Three different forms of epistasis as described by Phillips 20. Compositional epistasis refers to the blocking of one allelic effect by an allele at another locus. Statistical epistasis refers to the average effect of substitution of alleles at combinations of loci, with respect to the population average genetic background. Functional epistasis refers to the molecular interactions that proteins and other genetic elements have with one another. |
| Bootstrap samples | These are data sets obtained by taking a random sample of the original data, usually with replacement. One then applies the same analysis as was applied to the real data. This is repeated many times, allowing one to assess the variability in results incurred due to random sampling. |
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrg2579
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2872761?pdf=render
Citations & impact
Impact metrics
Article citations
Multicategory Survival Outcomes Classification via Overlapping Group Screening Process Based on Multinomial Logistic Regression Model With Application to TCGA Transcriptomic Data.
Cancer Inform, 23:11769351241286710, 08 Oct 2024
Cited by: 0 articles | PMID: 39385930 | PMCID: PMC11462568
Association of SNP-SNP interactions of surfactant protein genes with severity of respiratory syncytial virus infection in children.
Physiol Genomics, 56(10):691-697, 02 Sep 2024
Cited by: 0 articles | PMID: 39222066
Cluster effect for SNP-SNP interaction pairs for predicting complex traits.
Sci Rep, 14(1):18677, 12 Aug 2024
Cited by: 0 articles | PMID: 39134575 | PMCID: PMC11319716
Identification of interactions between genetic risk scores and dietary patterns for personalized prevention of kidney dysfunction in a population-based cohort.
Nutr Diabetes, 14(1):62, 14 Aug 2024
Cited by: 0 articles | PMID: 39143076 | PMCID: PMC11325018
Biomedical literature mining: graph kernel-based learning for gene-gene interaction extraction.
Eur J Med Res, 29(1):404, 02 Aug 2024
Cited by: 0 articles | PMID: 39095899 | PMCID: PMC11297645
Go to all (705) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 11 of 11)
- (4 citations) dbSNP - rs4471699
- (2 citations) dbSNP - rs931058
- (1 citation) dbSNP - rs2532292
- (1 citation) dbSNP - rs447169
- (1 citation) dbSNP - rs434157
- (1 citation) dbSNP - rs2076756
- (1 citation) dbSNP - rs12150547
- (1 citation) dbSNP - rs17574824
- (1 citation) dbSNP - rs10824773
- (1 citation) dbSNP - rs17689882
- (1 citation) dbSNP - rs17650381
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
SNPHarvester: a filtering-based approach for detecting epistatic interactions in genome-wide association studies.
Bioinformatics, 25(4):504-511, 19 Dec 2008
Cited by: 75 articles | PMID: 19098029
Detecting purely epistatic multi-locus interactions by an omnibus permutation test on ensembles of two-locus analyses.
BMC Bioinformatics, 10:294, 17 Sep 2009
Cited by: 25 articles | PMID: 19761607 | PMCID: PMC2759961
Detecting epistatic SNPs associated with complex diseases via a Bayesian classification tree search method.
Ann Hum Genet, 75(1):112-121, 01 Dec 2010
Cited by: 3 articles | PMID: 21121902 | PMCID: PMC3178841
Detecting epistasis in human complex traits.
Nat Rev Genet, 15(11):722-733, 09 Sep 2014
Cited by: 210 articles | PMID: 25200660
Review
Funding
Funders who supported this work.
Wellcome Trust (1)
Development of statistical methodology for detection and characterisation of genetic factors in complex disease.
Prof Heather Cordell, University of Cambridge
Grant ID: 074524