Abstract
Free full text

Structure of a human ASF1a/HIRA complex and insights into specificity of histone chaperone complex assembly
Abstract
Human HIRA, ASF1a, ASF1b and CAF-1 are evolutionally conserved histone chaperones that form multiple functionally distinct chromatin assembly complexes, with roles linked to diverse nuclear process, such as DNA replication and formation of heterochromatin in senescent cells. We report the crystal structure of an ASF1a/HIRA heterodimer and a biochemical dissection of ASF1a's mutually exclusive interactions with HIRA and the p60 subunit of CAF-1. The HIRA B-domain forms an antiparallel β-hairpin that binds perpendicular to the strands of the β-sandwich of ASF1a, via β-sheet, salt-bridge and van der Waals contacts. The N- and C-terminal regions of ASF1a and ASF1b determine the different affinities of these two proteins for HIRA, by contacting regions outside the HIRA B-domain. CAF-1 p60 also employs B-domain-like motifs for binding to ASF1a, thereby competing with HIRA. Together, these studies begin to define the molecular determinants of assembly of functionally diverse macromolecular histone chaperone complexes.
The basic repeating unit of eukaryotic chromatin is the core nucleosome. Each core nucleosome consists of a histone (H3/H4)2 heterotetramer, two histone (H2A/H2B) heterodimers and about 147 base pairs of DNA. During chromatin assembly, histone chaperones donate histones to DNA1. In human cells, the deposition of the histone H3/H4 complex involves four histone chaperones, Chromatin Assembly Factor 1 (CAF-1), HIstone Regulatory Homolog A (HIRA), and two Anti-Silencing Factor paralogs (ASF1a and ASF1b).
CAF-1, an evolutionarily conserved heterotrimeric chaperone comprised of p150, p60 and p48 subunits, mediates DNA synthesis-coupled chromatin assembly in S-phase1,2. CAF-1 is also required for chromatin assembly coupled to DNA repair3–8, formation of specialized chromatin structures, such as transcriptionally silent chromatin at telomeres, rDNA yeast mating loci, and plant gene loci3,9–12, and for proper pericentromeric chromatin structure in yeast13.
The functions of the HIRA histone chaperone are substantially different to CAF-1. HIRA is the human ortholog of two proteins known to silence histone gene expression and create transcriptionally silent heterochromatin in yeast, flies, plants and humans14–18. In yeast, Hir1 and Hir2 also contribute to formation of pericentromeric chromatin structure13. However, the Hir-mediated silencing pathway is genetically separable from the CAF-1 pathway16, and HIRA does not appear to participate in DNA replication or repair coupled chromatin assembly19,20. Instead, in vitro and during decondensation of the sperm pronucleus in flies, HIRA specifically deposits a histone variant, histone H3.3, in a DNA replication and repair independent manner19,20 21.
The ASF1 proteins share functions with both the CAF-1 and HIRA histone chaperones16,22,23. In human cells, there are two ASF1 paralogs, ASF1a and ASF1b, which are highly conserved at the N-terminal core domains but more diverged at their C-terminal tails (Supplementary Fig. 1a and 1b)24. ASF1a, but not ASF1b, physically interacts with HIRA and this complex drives formation of specialized domains of facultative heterochromatin, called senescence-associated heterochromatin foci (SAHF), in senescent human cells18,20. Both ASF1 proteins also interact with CAF-1 p6022,25,26, and these interactions are thought to contribute to DNA replication and repair-coupled chromatin assembly26–29. One role of ASF1 proteins in these complexes is to bind to the histone H3/H4 complex30,31. Of note, the binding of HIRA and CAF-1 to ASF1 proteins appears to be mutually exclusive20. This exclusivity, as well as the specific binding of HIRA to ASF1a but not ASF1b, is likely to be governing principles that dictate the functional specialization of the CAF-1 and HIRA-containing chaperone complexes. Therefore, we set out to define the molecular basis of this specificity.
Previously, we showed that HIRA specifically recognizes the N-terminal core domain of ASF1a via an evolutionarily conserved stretch of amino acids, called the B-domain18,32. Now, by mutagenesis, biochemical and X-ray crystallographic approaches, we have characterized this interface. We show that the HIRA B-domain forms a β-hairpin that interacts extensively with the β-sandwich structure of the ASF1a N-terminal core domain through β-sheet, salt bridge and van der Waals interactions. We also define the molecular basis of HIRA's specificity for ASF1a, over ASF1b. Finally, we show that the CAF-1 p60 C-terminus utilizes HIRA B-domain-like motifs to recognize the same HIRA-binding surface on ASF1a, explaining why HIRA and CAF-1 p60 bind mutually exclusively to ASF1a, to mediate distinct chromatin regulatory activities.
RESULTS
A minimum HIRA fragment that binds to ASF1a
Wild-type human HIRA is a polypeptide of 1017 amino acids. Previously, we showed that residues 421–729 are sufficient for binding to the evolutionarily conserved N-terminal core domain of ASF1a, residues 1–155 (Supplementary Fig. 1a), and for repression of histone gene expression and formation of SAHF in human cells 18,33. This ASF1a-interacting domain of HIRA contains a stretch of about 37 amino acids that is evolutionarily conserved in HIRA and its orthologs and named the B-domain (defined as residues 439–47532)(Fig. 1a). The B-domain is necessary for binding to ASF1a, repression of histone gene expression and formation of SAHF18,33. To date, the B-domain has not been described outside of the HIRA family, although the p60 subunit of CAF-1 shares some homology with this domain (Fig. 1b).
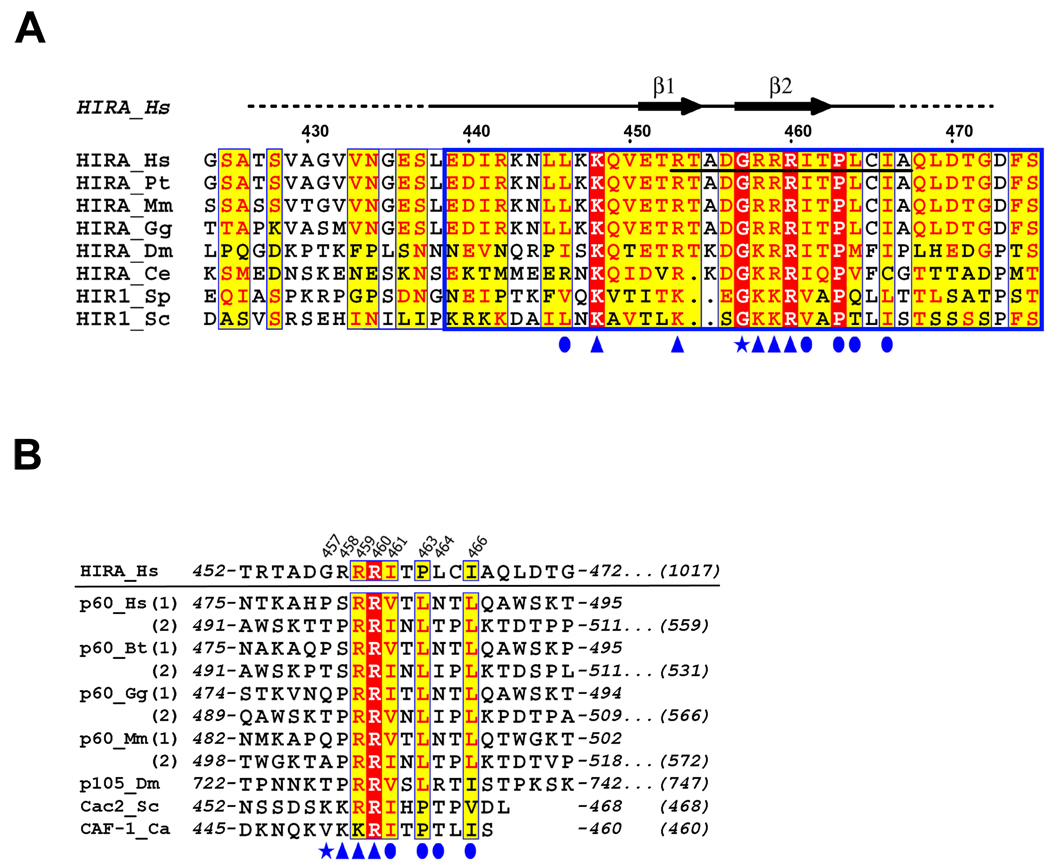
(a) The extended B-domain of HIRA homologues are aligned with strictly conserved (identical) residues shaded in red and conserved residues shaded in yellow, with the minimal HIRA peptide required for ASF1a binding underlined. The blue box indicates the core B-domain, defined previously by sequence analysis and functional studies18,32,33. The residue numbering and the secondary structure for human HIRA are indicated above the sequence alignment. Ordered regions of HIRA in the crystal structure are indicated by the solid black line above the alignment and disordered regions are indicated with a dashed line. Residues of HIRA that make contacts to ASF1a are indicated with blue symbols (arrowhead - salt bridge interaction; oval - hydrophobic interaction; star - structural and/or non-salt bridge hydrogen bonding). Abbreviations for various organisms are Bt, B. Taurus; Ca, C. albicans; Ce, C. elegans; Dm, D. melanogaster; Gg, G. gallus; Hs, H. sapiens; Mm, M. musculus; Pt, P. troglodytes; Sc, S. cerevisiae; and Sp, S. pombe;
(b) Sequence alignment of the B-domain-like motifs of human CAF-1 p60 and its homologues from other species. For the proteins that have two motifs, both of them are given separately with (1) and (2) designations. The human HIRA B-domain is shown on the top for comparison. Residues of HIRA that make contacts to ASF1a are numbered on the top and indicated below the alignment using the same symbols described in (A). Starting and ending residue numbers are marked on the sequence, with the total number of residues in the full-length proteins indicated at the end of the sequences. Abbreviations for various organisms are as in (A).
To determine which region of the HIRA B-domain makes critical contacts with ASF1a, we made a series of scanning mutations, deleting 7–10 amino acids of the B-domain at a time. The HA-tagged HIRA proteins were tested for binding to ASF1a in vitro. The two innermost deletions (of residues 449–458 and 459–468), encompassing the most highly conserved residues of the B-domain (Fig. 1a), had the most debilitating effect on binding to ASF1a, whereas the two outermost deletions had a more modest effect (Fig. 2a).
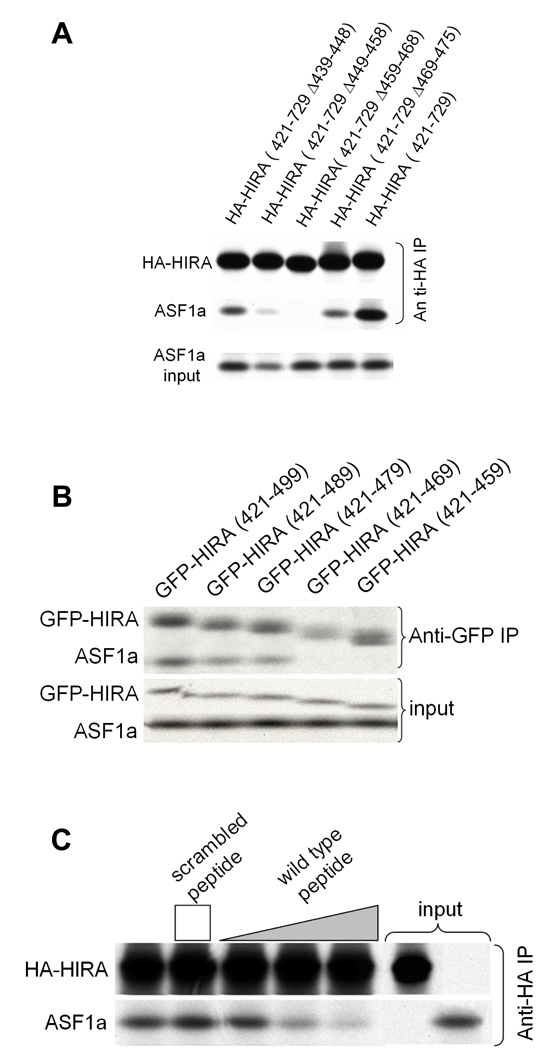
(a) Binding of in vitro translated 35S-labeled HA-HIRA(421–729) and the indicated B-domain deletion mutants to 35S-labeled Myc-ASF1a.
(b) Binding of in vitro translated 35S-labeled GFP-HIRA(421–729) and the indicated C-terminal deletion mutants to 35S-labeled Myc-ASF1a.
(c) Binding of in vitro translated 35S-labeled HA-HIRA(421–729) to 35S-labeled Myc-ASF1a in the presence of 0, 1, 10 and 50µg of the wild-type HIRA peptide, RTADGRRRITPLCIA, or 50µg of the scrambled peptide, GRAARITPRDTLRCI. The height of the square and triangle indicates relative peptide concentrations.
Having identified the regions of the B-domain that are necessary for binding to ASF1a, we next set out to define the minimum region of HIRA that is sufficient for binding. A series of HA-tagged deletion mutants derived from HIRA(421–729) was tested for binding to ASF1a. As shown in Supplementary Fig. 2, C-terminal deletions as far as residue 509 all retained binding to ASF1a, although HA-HIRA(421–509) bound with reduced efficiency compared to HA-HIRA(421–729). An additional series of C-terminal deletions was made, this time fused at their N-terminus to GFP. As shown in Fig. 2b, binding was retained in GFP-HIRA(421–479) but undetectable in GFP-HIRA(421–469). We conclude that the minimum region of HIRA that retains binding to ASF1a lies within amino acids 421–479, underscoring the role of the B-domain in ASF1a binding32 (Fig. 1a).
Next, we asked whether a short peptide spanning the most conserved amino acids of the B-domain was sufficient to interact with ASF1a, by testing its ability to compete with the binding of HA-HIRA(421–729) to ASF1a. Specifically, we tested residues 453–467 of wild-type HIRA, RTADGRRRITPLCIA, and a sequence-scrambled version, GRAARITPRDTLRCI, as a control. The wild-type peptide, but not the scrambled peptide, competed-out the interaction between ASF1a and HA-HIRA(421–729) (Fig. 2c). Together, these results show that the B-domain is both necessary and sufficient for interaction with ASF1a.
Structure of the ASF1a/HIRA complex
Having defined the minimal region of HIRA require for Asf1a interaction, we set out to determine the X-ray crystal structure of an ASF1a/HIRA complex. Towards this goal, we coexpressed the HIRA B-domain (427–472) and the ASF1a N-terminal core domain (1–154) in E. coli and purified the tightly associated heterodimeric complex to homogeneity. Crystals were obtained in the space group P6522, containing two ASF1a/HIRA complexes per asymmetric unit. The structure of the complex was determined by molecular replacement using the crystal structure of the N-terminal core domain of yeast Asf1p34 as a search model. This produced a clear solution for the two ASF1a molecules in the asymmetric unit and excellent density for the two ASF1a-bound HIRA fragments (Fig. 3a). One HIRA fragment could be confidently built into the electron density map from residues 446 to 466 and the other from residues 449 to 464 (Fig. 3b). We presume that the remainder of the HIRA fragments is disordered. The final model was refined to 2.7 Å resolution with excellent refinement statistics (Table 1).
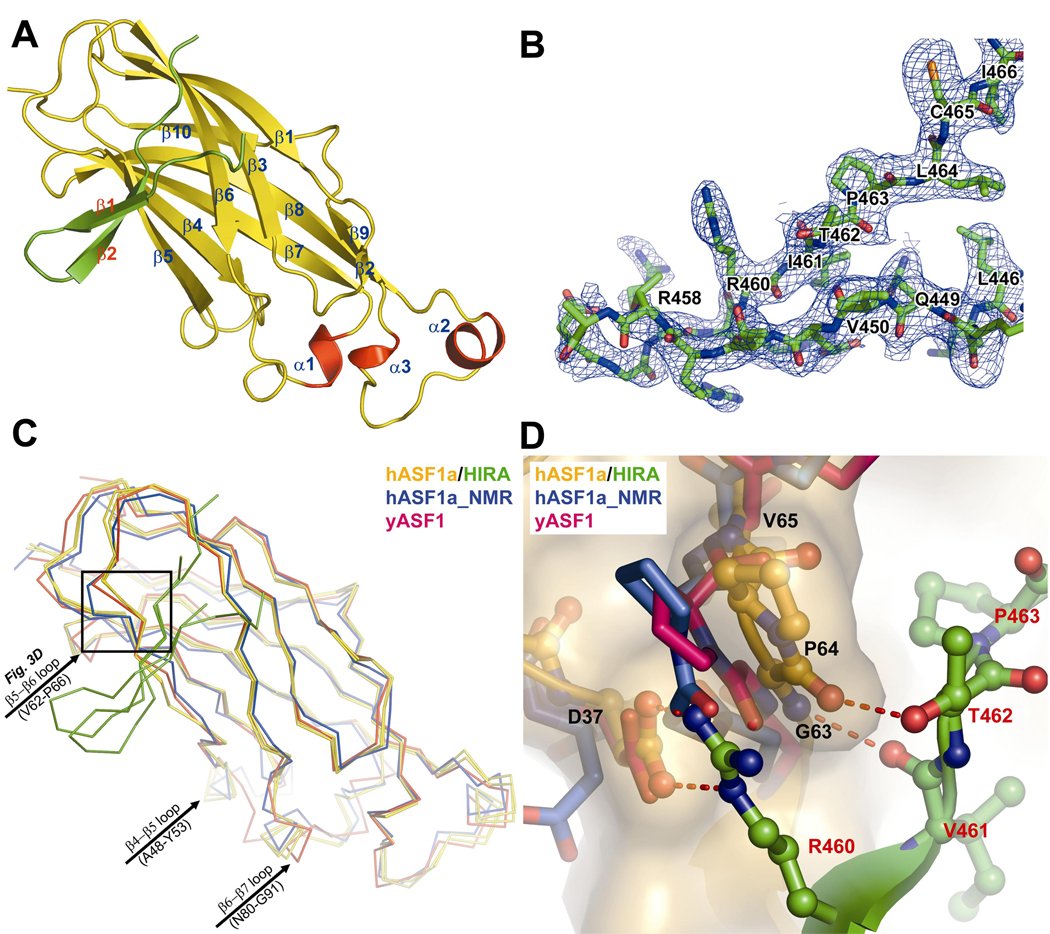
(a) Ribbon representation of the ASF1a/HIRA complex. ASF1a is in yellow, with the three helices in red and the bound HIRA fragment in green.
(b) Section of the composite-omit electron density map (contoured at 1.0 σ) of the ASF1a/HIRA complex crystal, showing a region of the HIRA fragment.
(c) Cα superposition of the two ASF1a/HIRA complexes (ASF1a in yellow and HIRA green) within the asymmetric unit, with the nascent structures of yeast Asf1p (red) and human ASF1a (blue). The black box defines the close-up region in (d).
(d) A close-up view of the superposition of the structures in (c) around the ASF1a VGP (residues 62–64) triplet. Only one complex of the two ASF1aN/HIRA complexes in the asymmetric unit cell, highlighted with a ball-and-stick model, is displayed for clarity. The color-coding is as described in (c) with CPK coloring and hydrogen bonds shown. Fig. 3, 4a4a and and77 were created with Pymol (DeLano W.L.; www.pymol.org).
Table 1
Data Collection and Refinement Statistics
| Human ASF1a/HIRA complex | |
|---|---|
| Data Collection | |
| Wavelength (Å) | 0.980 |
| Space Group | P6522 |
| Cell Dimensions | |
 a, b, c (Å) a, b, c (Å) | 116.2, 116.2, 167.6 |
 α, β, γ (°) α, β, γ (°) | 90.0, 90.0,120.0 |
| Resolution (Å) | 25–2.7 (2.8−2.7) |
| Rmerge | 0.037 (0.315) |
| I/σI | 42.6 (5.3) |
| Completeness (%) | 100 (100) |
| Redundancy | 8.4 (8.6) |
| Refinement | |
| Resolution (Å) | 2.7 |
| No. reflections | 19,106 |
| Rwork / Rfree | 22.9/26.9 |
| No. atoms | |
 Proteins(ASF1a/HIRA) Proteins(ASF1a/HIRA) | 2,784 (2482/302) |
 Water Water | 130 |
| B-factors | |
 Proteins(ASF1a/HIRA) Proteins(ASF1a/HIRA) | 53.1 (53.1/53.0) |
 Water Water | 49.4 |
| R.m.s deviations | |
 Bond lengths (Å) Bond lengths (Å) | 0.0067 |
 Bond angles (°) Bond angles (°) | 1.421 |
Values in parentheses are for the highest-resolution shell
The two ASF1a N-terminal core domains in the asymmetric unit (Cα RMSD 0.45Å) adopt the same elongated immunoglobulin-like β sandwich fold (Fig. 3a) previously described for the nascent structures of human ASF1a31 and yeast Asf1p34, with Cα RMSD of 1.1 Å relative to either structure (Fig. 3c). However, variation between the free and HIRA-bound ASF1 polypeptides is observed at two less conserved loops (Fig. 3c), and interestingly, also at the highly conserved β5–β6 loop at the HIRA interface (Fig. 3d). In particular, it appears that the β5–β6 turn of free ASF1a, especially around Gly63 and Pro64, is in a conformation that interferes with HIRA binding, but adjusts to accommodate the HIRA B-domain upon complex formation.
Each of the two bound HIRA polypeptides in the asymmetric unit forms a β-hairpin containing two strands of 4 and 6 residues with a 2-residue tight turn and loop extensions N- and C-terminal to the β-strands (Fig. 3b). In particular, residues Glu451–Thr454 and Gly457–Ile461 form a two-stranded anti-parallel β-sheet that is stabilized by 5 intra-molecular main-chain and 4 side-chain hydrogen bonds, including a bidentate salt-bridge between residues Glu451 and Arg459 (Fig. 4a). Gly457 at the β-hairpin turn also adapts unique dihedral angles to facilitate two sets of hydrogen bonds between the backbone of Gly457 and residues Arg453 and Thr454 that stabilize the β-hairpin structure. The high degree of conservation of residues that mediate the intra-molecular interactions within the β-hairpin of the HIRA B domain, and in particular the strict conservation of Gly457 (Fig. 1a), suggests that this structure may be preformed prior to ASF1a binding.
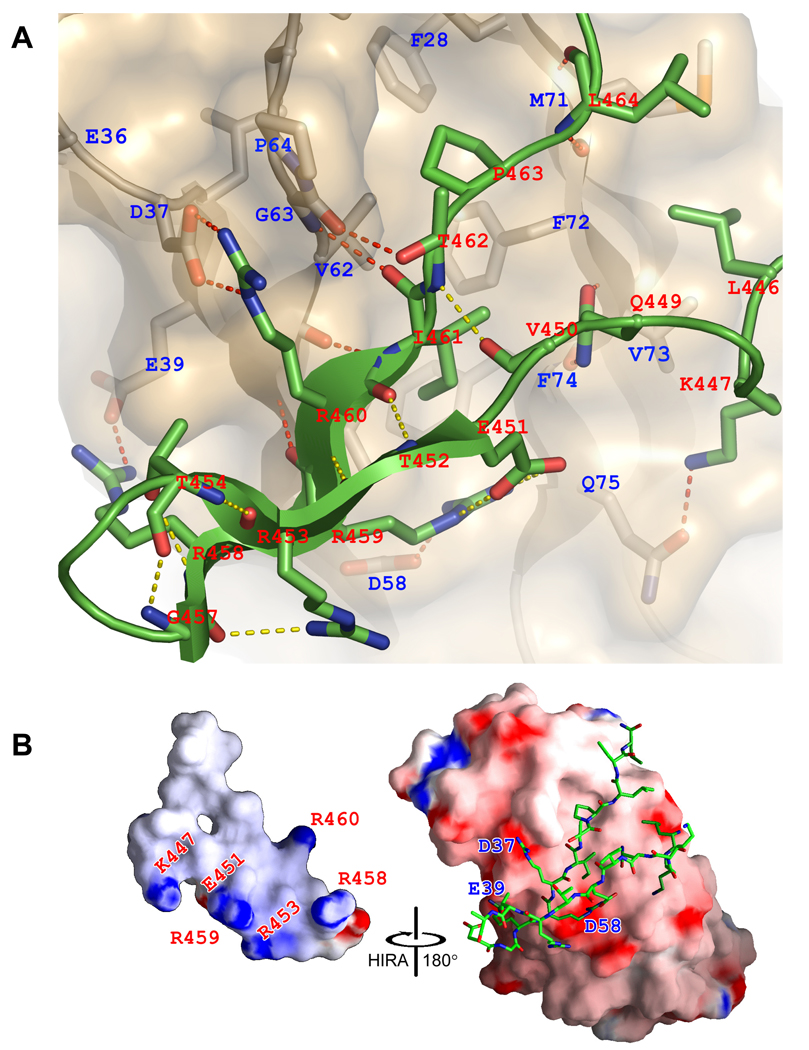
(a) Close-up view of the ASF1a/HIRA interface. ASF1a is shown as a gold ribbon in combination with a semi-transparent surface representation and HIRA as a green ribbon. Residues from both molecules that mediate hydrogen-bonds (including salt-bridges) and hydrophobic interactions are highlighted as sticks with CPK coloring. Hydrogen bonds are shown with dashed lines (yellow for intra-molecular and red for inter-molecular).
(b) Surface electrostatic representation of ASF1a and HIRA moieties in the complex. For clarity, the HIRA fragment in the complex is shown only as sticks with CPK coloring (right), with the HIRA surface shown in an open-book format (left). Residues involved in salt-bridge interaction are indicated on the corresponding surface. This figure was made with GRASP41.
The B-domain of HIRA binds along the edge and perpendicular to the strands of the β-sandwich of the ASF1a N-terminal core domain and makes extensive interactions with ASF1a, burying 652Å2 of HIRA surface (Fig. 3a). Both ASF1a/HIRA complexes in the asymmetric unit show similar contacts despite the different HIRA conformations of residues around the β-hairpin turn (Fig. 3c). The ASF1a-binding site for HIRA is a shallow hydrophobic cleft that is primarily lined by residues Val60, Val62, Val65, Pro66, Phe72, and Phe74 from the β5–β6 region and residue Phe28 and Leu38 from the β3 and β4 strands, all involved in van der Waals interactions with residues Ile461, Pro463 and Leu464 of the HIRA B-domain (Fig. 4a). There is also a cluster of salt bridges formed between the acidic residues Glu39, Asp58 and Asp37 of ASF1a and the basic residues Arg458, Arg459 and Arg460 of HIRA, respectively (Fig. 4b). Finally, residues Leu61–Gly63 of ASF1a (β5) form β-sheet interactions with residues Arg459–Ile461 (β2) of HIRA. The loops before and after the β-hairpin are both secured onto ASF1a by hydrogen bonds and hydrophobic interactions (Fig. 4a). Taken together, it appears that HIRA residues Arg458–Ile466, within the minimum ASF1a-binding peptide of HIRA (residues Arg453–Ala467) identified above, play the most critical role in ASF1a interaction. Most of the interacting ASF1a and HIRA residues involved in this interaction are either strictly or highly conserved within both proteins (Fig. 1a and Supplementary Fig. 1a), highlighting the importance of these interactions.
Biochemical characterization of the ASF1a/HIRA interface
Equipped with detailed structural insight into the ASF1a/HIRA interface, we then used targeted mutagenesis and in vitro binding assays to verify the functional significance of individual interactions between ASF1a and HIRA. We first examined the importance of the salt bridge interactions. As can be seen in Fig. 5a, alanine substitution of either the second or third arginine in Arg458–Arg460 of HIRA that interacts with D58 and D37 of ASF1a, completely abolishes binding.
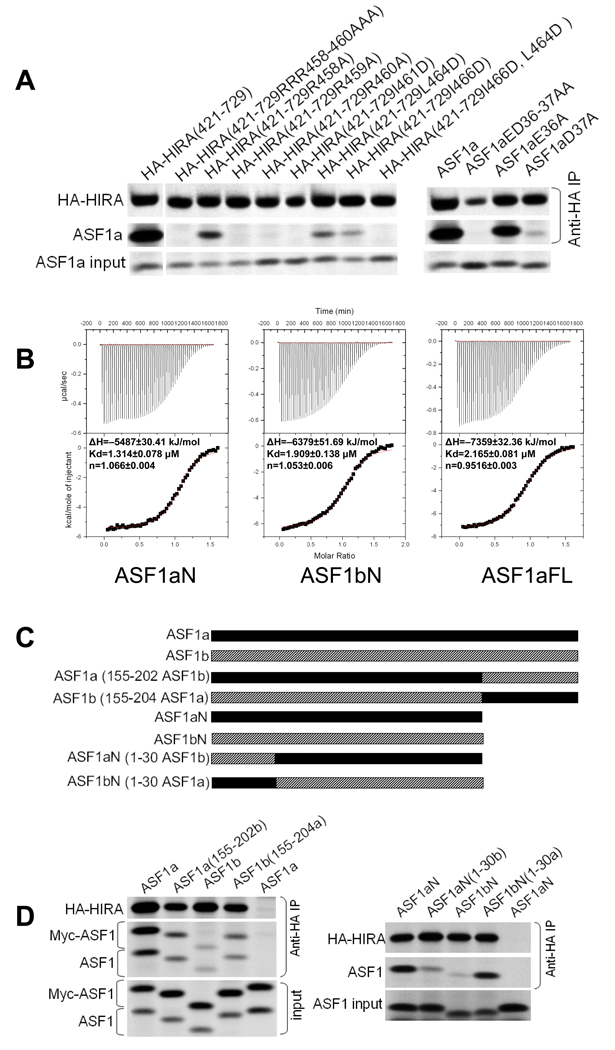
(a) Left- Binding of in vitro translated 35S-labeled HA-HIRA(421–729) or the indicated mutants to 35S-labeled Myc-ASF1a. Right- Binding of in vitro translated 35S-labeled HA-HIRA(421–729) to 5S-labeled Myc-ASF1a or the indicated mutants.
(b) Representative data from isothermal titration calorimetry titrations of HIRA B-domain peptide (residues 453–467) titrated into ASF1aN (residues 1–154), ASF1bN (residues 1–154), and ASF1aFL (full length, residues 1–204). The area under each injection spike (upper section) is integrated and fitted by using nonlinear least-squares regression analysis (lower section). The derived enthalpies (ΔH), dissociation constant (Kd) and stoichiometry (n) are indicated for each of the titrations. This figure was prepared using Origin 5.0.
(c) Schematic of the protein chimaeras used in (d).
(d) Left - Binding of in vitro translated 35S-labeled HA-HIRA(421–729) to the indicated 35S-labeled ASF1 proteins and their C-termini (155–202) swapped chimaeras. Right - Binding of in vitro translated 35S-labeled HA-HIRA(421–729) to the indicated 35S-labeled ASF1 proteins and their N-termini (1–30) swapped chimaeras.
In addition, alanine substitution of the first arginine, Arg458, partially blocks binding. Correspondingly, we probed the contribution of the respective interacting residues in ASF1a. Previously, we showed that double substitution of the two acidic residues Glu36 and Asp37 with alanine residues abolishes binding to HIRA(421–729)18. Consistent with the structural finding that only Asp37 is involved in salt-bridge formation, the single substitution Asp37A abolishes binding to HIRA(421–729), while E36A has little effect (Fig. 5a). Taken together, these results reveal that the charge interactions between highly conserved residues HIRA-Arg460 and ASF1a-Asp37, and HIRA-Arg459 and ASF1a-Asp58, are crucial for complex formation (Fig. 1a and Supplementary Fig. 1a).
The role of the hydrophobic interactions between ASF1a and HIRA was also evaluated. Fig. 5a shows that mutation of HIRA residue Ile461, one of two hydrophobic residues within the hydrophobic cleft of ASF1a, to aspartic acid abolishes the interaction between HIRA(421–729) and ASF1a. In addition, mutation of either Leu464D or Ile466D in HIRA(421–729) partially blocks ASF1a binding, and mutation of both together completely abolishes binding. Together, these findings agree with our structural findings that the hydrophobic cleft of ASF1a compliments Ile461 and Pro463 of HIRA, consistent with the strict evolutionary conservation of these two residues (Fig. 1a). A L464D/I466D double mutant also disrupts ASF1a binding, suggesting that, although less conserved, this part of HIRA still plays a role (Fig. 5a).
In an earlier study, we showed that mutation of the strictly conserved triplet Val-Gly-Pro to Aal-Ala-Ala (residues 62–64), located in the β5–β6 loop region of ASF1a, completely abolishes HIRA binding 34. The structural comparison of the complex with free ASF1 proteins as described above suggested that the unique conformation and relative flexibility of this loop region is critical for proper ASF1a/HIRA interactions. This mutation, largely by eliminating the unique conformations of Gly63 and cis-Proline 64, not only disrupts direct hydrogen bonding with HIRA, but also causes potential steric occlusion of other contacts, such as the interaction between Asp37 of ASF1a and Arg460 of HIRA (Figs. 3d and 4a).4a). Taking all our results together, we conclude that a combination of β-sheet, electrostatic and hydrophobic interactions stabilize the interaction between the ASF1a N-terminal core and the HIRA B domain.
Specificity of HIRA for ASF1a over ASF1b
In light of the earlier observations by us and Nakatani and coworkers that HIRA preferentially associates with ASF1a over ASF1b, in vitro and in vivo18,20, we were surprised that each of the ASF1a residues involved in HIRA B-domain binding are strictly conserved in ASF1b (Supplementary Fig. 1a). This suggests that interactions between the ASF1a N-terminal core domain and the HIRA B-domain are not responsible for HIRA's discrimination between ASF1a and ASF1b. To test this, we measured binding of the recombinant N-terminal core domains of ASF1a and ASF1b to the minimal HIRA B-domain peptide (residues 453–467), using isothermal titration calorimetry (ITC). We also tested the full-length ASF1a protein, to evaluate the contribution of the C-terminal region of ASF1a to HIRA B-domain binding. We were unable to prepare the full-length recombinant ASF1b protein in soluble form for ITC. The ITC studies show that the HIRA peptide binds with comparable affinity to each of the ASF1 proteins, with dissociation constants in the range of 1.3–2.1 µM (Fig. 5b). These results confirm that HIRA does not distinguish between ASF1a and ASF1b through the interaction between the HIRA B-domain and the ASF1 N-terminal core domain. It further reveals that the C-terminal region of ASF1a does not influence binding of the HIRA B-domain.
The ITC and structural studies together suggest that differential binding of ASF1a and ASF1b to HIRA might be mediated by regions outside of the HIRA B-domain and/or regions outside of the ASF1a core domain region that makes HIRA B-domain interactions. Previously, we showed that simultaneous substitution into ASF1b of two regions of ASF1a that are relatively poorly conserved in ASF1b, residues 31–37 and the C-terminal 50 residues, potentiated binding of ASF1b to HIRA18. However subsequent studies revealed that substitution of residues 31–37 alone does not potentiate binding (data not shown). To further investigate the role of the non-conserved C-terminal tails of ASF1a and ASF1b (Supplementary Fig. 1b) in HIRA binding, we tested binding of HA-tagged HIRA(421–729) to ASF1 chimeras in which the C-terminal tail regions of ASF1a and ASF1b were swapped (Fig. 5c). As can be seen in Fig. 5d (left panel), swapping the C-terminal tails between ASF1a and ASF1b decreases binding of ASF1a but increases binding of ASF1b. Based on these results, we conclude that, compared to the ASF1b C-terminal tail, the ASF1a C-terminus potentiates binding to HIRA.
Interestingly, in these HIRA binding studies, we observed that the ASF1a N-terminal core domain lacking the C-terminus (ASF1aN), the same region of ASF1a cocrystallized with HIRA, bound better to HIRA(421–729) than the corresponding fragment of ASF1b (ASF1bN) (Figs. 5c and 5d, left panel). These results, coupled with our ITC studies showing that the N-terminal core domains of ASF1a and ASF1b bound indistinguishably to the HIRA B-domain alone, suggest that another region of HIRA might bind to the N-terminal core domain of ASF1a. Of interest, the N-terminal 30 residues of ASF1a and ASF1b are less conserved than the rest of the core domain (Supplementary Fig. 1a) and lie in the vicinity of the HIRA B-domain binding surface (see Fig. 7a). To test whether this part of ASF1a mediates additional contacts with HIRA outside of the B-domain and thus contributes to HIRA's specificity for ASF1a, we prepared chimeric proteins in which the N-terminal 30 residues were swapped between the ASF1a and ASF1b core domains and tested them for binding to HIRA(421–729). These experiments showed that ASF1bN carrying the N-terminal 30 residues of ASF1a binds HIRA(421–729) much better than native ASF1bN (Fig. 5d, right panel). Conversely, ASF1aN carrying the N-terminal 30 residues of ASF1b binds much worse to HIRA(421–729) than native ASF1aN. Based on these results, we conclude that the N-terminal 30 residues of ASF1a and ASF1b also contribute to the binding specificity for HIRA. Also, as shown in Fig. 5d (right panel), ASF1aN and ASF1bN(1–30 ASF1a) have similarly high binding affinity for HIRA(421–729), while ASF1bN and ASF1aN(1–30 ASF1b) have similarly low binding affinities, suggesting that residues 31–155 of ASF1a and ASF1b do not make direct contributions to relative HIRA binding specificity.

(a) Surface representation of the ASF1a/HIRA complex, with residues 1–30 of ASF1a highlighted in blue. The color-code highlights the sequence variation between ASF1a and ASF1b as indicated in the color bar. The HIRA fragment bound to ASF1a is also shown in green surface representation for reference.
(b) Hydrophobic residues (in yellow) are mapped onto a ribbon representation of ASF1a, with the cluster of hydrophobic residues at the histone H3 binding site31 shown as a space-filling model. The HIRA fragment bound to ASF1a is shown as a green surface representation for reference and the published NMR data is used to model a histone H3 C-terminal helix (in red) onto the ASF1a surface. The relative orientations between the left and right panels are indicated.
(c) A schematic model for ASF1a and ASF1b association with HIRA, CAF-1 p60 and histone H3. See text for details.
In sum, our binding studies show that HIRA’s discrimination between ASF1a and ASF1b depends on part(s) of the HIRA protein outside of the B-domain and both the N-terminal 30 and C-terminal 50 residues of the ASF1 proteins.
B-domain-like motifs in CAF-1 p60
Although ASF1a interacts with both HIRA and CAF-1 p60, it does not simultaneously copurify with both proteins20. Therefore, we hypothesized that CAF-1 p60 might bind ASF1a in an analogous fashion to HIRA, using a B-domain-like motif. To test this, we searched CAF-1 p60 homologues for the consensus HIRA B-domain motif, GRRRIXPLXI, which provides the critical binding determinants for ASF1a. This search identified either one or two B-domain-like motifs in the C-terminal tail of each of CAF-1 p60 homologue in the sequence database (Fig. 1b). Interestingly, a previous report showed that the C-terminal region of the Drosophila CAF-1 p60 homolog (p105), which also harbors this motif, mediates ASF1 binding25.
Despite their overall homology in the B-domain and B-domain-like regions, a very noticeable difference between HIRA and the CAF-1 p60 homologues is the lack of conservation of the first and, to a lesser extent, the second of the three conserved arginine residues in HIRA (residues 458–460) (Fig. 1a and 1b), that make important salt-bridge interactions with ASF1a in the complex (Fig. 4a). To assess the consequence of these changes, a series of mutations was generated in the 458–Arg-Arg-Arg–460 triplet of the HIRA B-domain and tested for ASF1aN binding. As shown in Fig. 6a, single or double arginine to lysine mutations within residues 458–460 do not result in a marked perturbation of ASF1a binding, while a triple arginine to lysine change results in a more dramatic decrease. As shown in Fig. 5a and 6a,6a, when Arg458 is mutated to alanine, to mimic the less polar residue found in the B-domain-like motifs in CAF-1 p60, the HIRA B-domain still maintains a relatively high level of ASF1aN binding. However, when the R458A mutation is combined with either the R459K or R460K mutations, ASF1a binding is substantially impaired, albeit not abolished (Fig. 6a). A combination of the R458A mutation with the R459K, R460K double mutation completely abolishes ASF1aN binding (Fig. 6a), indicating that maintaining at least one arginine (instead of lysine) at either position Arg459 or Arg460 is critical for ASF1aN binding. These results suggest that the B-domain like motifs of CAF-1 p60 and its homologues could mediate binding of the corresponding proteins to ASF1aN.


(a) Binding of the HIRA B-domain and selected mutants as GST fusion proteins to purified ASF1aN inputs.
(b) Binding of affinity purified human CAF-1 p60 and yeast Cac2 as GST fusion proteins to purified ASF1aN proteins.
(c) Binding of affinity purified CAF-1 p60 GST fusion proteins and their selected mutants to purified ASF1aN proteins.
(d) Physical interactions between ectopically expressed CAF-1 p60 and ASF1a proteins in human U2OS cells as tested by immunoprecipitation-western blot analysis. Left – Interaction between HA-tagged CAF-1 p60 wild-type or truncation mutant p60(1–481) and Myc-tagged wild-type ASF1a. Right – Interaction between HA-tagged CAF-1 p60 wild-type or mutant p60(RR482AA) or p60(RR498AA) with Myc-tagged wild-type ASF1a.
(e) Binding of bacterially expressed and affinity purified HIRA (427–472) and CAF-1 p60 (376–559) GST fusion proteins to purified ASF1aN, ASF1bN, ASF1a full-length or ASF1aN(D37A) mutant proteins.
(f) Disruption of CAF-1 p60 (469–559)/ASF1aN and HIRA (427–472)/ASF1aN complexes by a HIRA (453–467) peptide. In (b), (c), (e) and (f), only the upper (intact) band of each individual GST fusion protein was used for evaluation of binding to ASF1a because of partial proteolysis of the fusion proteins during expression and purification.
To directly test whether the two B-domain-like motifs in human CAF-1 p60 (within residues 480–490 and 496–506) mediate ASF1a binding, various CAF-1 p60 C-terminal fragments were tested for binding to the ASF1a N-terminal core domain. As shown in Fig. 6b, CAF-1 p60 fragments containing both B-domain-like motifs (residues 376–509, 376–559, 469–509 and 469–559) interact with ASF1a, while a polypeptide with both motifs deleted (residues 376–481) shows very weak binding. A fragment harboring only the first B-domain-like motif (residues 376–496) binds with slightly reduced affinity, relative to the fragments harboring both motifs. ASF1a pull-down experiments with the yeast CAF-1 p60 homologue, Cac2p, showed that this protein also interacts with ASF1a, via a conserved B-domain-like motif within residues 457–468 (Fig. 6b). Taken together, these studies show that human and yeast CAF-1 p60 bind to the conserved ASF1a core domain through their evolutionarily conserved C-terminal B-domain-like motifs.
To verify that ASF1a interacts with both HIRA and CAF-1 p60 through the B-domain motifs and to establish the relative importance of the first and second B-domain-like motifs in CAF-1 p60, we carried out ASF1a pull-down studies with the shortest CAF-1 p60 fragment (469–509) that contains both motifs, harboring mutations of the arginine residues in both motifs, Arg483 and Arg499, that are equivalent to the critical Arg460 in HIRA. As shown in Fig. 6c, the R483A/R499A double mutation in CAF-1 p60 also abolishes the ASF1a interaction, while a comparable double mutation to lysine markedly compromises the interaction. Pull-down experiments with single R483A and R499A mutations of CAF-1 p60 revealed similar residual ASF1a binding capacities (Fig. 6c), indicating that either B-domain-like motif of CAF-1 p60 can bind to ASF1a in vitro in the context of this minimal CAF-1 p60 fragment.
To test if other regions of CAF-1 p60 participate in ASF1a binding and to investigate whether both B-domain-like motifs of CAF-1 p60 contribute to ASF1a binding in the intact full-length CAF-1 p60 protein and in vivo, we carried out co-immunoprecipitation assays employing epitope-tagged CAF-1 p60 proteins ectopically expressed in U2OS cells. Specifically, we employed native full length CAF-1 p60, full length proteins containing RR to AA mutations in either the first or second B-domain-like motifs and a fragment harboring the N-terminal WD40 repeats, but not the two C-terminal B-domain-like motifs (residues 1–481). As shown in Fig. 6d (left panel), while wild-type CAF-1 p60 exhibits strong binding activity, CAF-1 p60(1–481) is completely inactive in ASF1a binding, indicating that the N-terminal WD40 repeats are not sufficient for ASF1a binding and that the C-terminal region, harboring the two B-domain-like motifs is critical. As shown in Fig. 6d (right panel), intact CAF-1 p60 harboring the RR to AA mutations in the second B-domain-like motif binds to ASF1a with wild-type efficiency, while mutation of the first B-domain-like motif completely abolishes binding. Taking all our results together, we conclude that in the context of the full-length CAF-1 p60 protein in vivo the first B-domain-like motif of CAF-1 is primarily responsible for ASF1a binding.
We next compared CAF-1 p60 binding to ASF1a and ASF1b, using a fragment of CAF-1 p60 missing only the N-terminal WD40 repeats (residues 376–559), since these repeats are neither necessary nor sufficient for ASF1a binding (Fig. 6b and 6d, left panel). As shown in Fig. 6e, CAF-1 p60(376–559) binds equally well to ASF1aN, ASF1bN and full length ASF1a, consistent with earlier findings that CAF-1 p60 associates with both ASF1a and ASF1b in vivo20.
Additional binding experiments using the ASF1aN(D37A) mutant, a mutant which does not bind to HIRA (Fig. 5a), were also carried out. These experiments reveal that the ASF1aN(D37A) mutant does not bind to CAF-1 p60 (376–559), nor to the HIRA B-domain, indicating that CAF-1 p60 makes interactions with the ASF1a and ASF1b core domains that are analogous to interactions of the HIRA B-domain with ASF1a.
These data suggest that the HIRA B-domain and the B-domain-like motifs of CAF-1 p60 bind to the ASF1a core domain in a similar and, therefore, mutually exclusive fashion. To directly test this, we purified ASF1aN/HIRA(427–472) and ASF1aN/CAF-1 p60(469–559) complexes and mixed them with increasing concentrations of HIRA peptide (453–467) (0, 1 and 5 mM). As shown in Fig. 6f, the HIRA peptide displaces the majority of the CAF-1 p60(469–559) fragment, and moderately displaces the HIRA B-domain (427–472) from ASF1a. An unrelated peptide did not displace either HIRA or CAF-1 p60 at the highest concentration (Fig. 6f). This confirms that HIRA and CAF-1 p60 bind mutually exclusively to the N-terminal core of ASF1a.
DISCUSSION
Here we have presented structural and functional data to define the essential interaction interface between two histone chaperones, HIRA and ASF1a, which function as a complex in vivo. In the process, we discovered that the B-domain, previously defined only in human HIRA and its orthologs, is conserved as an ASF1 binding motif in CAF-1 p60 proteins from yeast to human. Moreover, we found that HIRA's specific binding to ASF1a, but not ASF1b, is dictated by flanking sequences outside of the core interaction domains of both proteins. The structure of the ASF1a/HIRA complex reveals that the N-terminal 30 residues of ASF1 drapes across the long axis of the elongated ASF1a core domain and, as shown in Fig. 7a, is in position to interact with regions N- and C-terminal to the HIRA B-domain that would run into this region of ASF1a. Without structural information on the C-terminal tail region of ASF1a, which we show is also involved in HIRA binding, we are unable to assign its relative positioning to the core domain upon HIRA binding, but we propose that it may be adjacent to the ASF1aN/HIRA-B domain interface, as schematized in Fig. 7c. Thus, the B-domain of HIRA defines a conserved ASF1 binding motif whose specificity is modified by other sequences within both the ASF1 protein and HIRA itself.
We identified B-domain-like motifs in CAF-1 p60 that mediate an interaction with ASF1a that is mutually exclusive with the HIRA interaction, suggesting that both HIRA and CAF-1 compete for ASF1a association in vivo to form distinct chromatin regulatory activities. We further verified that although both B-domain-like motifs of p60 contain the sequence determinants for ASF1a association, only the first motif is involved in ASF1a association in the context of the full length CAF-1 p60 protein in vivo, suggesting a 1:1 stoichiometry of the ASF1a/CAF-1 p60 complex, similar to that of the ASF1a/HIRA complex. However, our sequence comparisons, binding assays, and peptide displacement studies also indicate that CAF-1 p60 B-domain has a lower binding affinity for ASF1aN than does the HIRA B-domain (Figs. 6a–f), probably due to variations at residues equivalent to HIRA R458, I461, and L464, which are involved in important charge or hydrophobic interactions with ASF1a.
The physiological relevance of our finding that ASF1a binds to both HIRA and CAF-1 p60 through the same domain is reinforced by additional cellular studies recently reported by Sanematsu et al.35. In this study, the authors reported that an ASF1a(E36A,D37A) mutant, when stably expressed in a conditional ASF1a-knockout chicken DT40 cell line, failed to interact with CAF-1 p60. These authors also showed in vitro that ASF1a(E36A,D37A) does not bind to wild-type HIRA. We previously showed in vitro and in vivo that ASF1a(E36A,D37A) and a HIRA mutant lacking the B-domain both fail to interact with their wild-type cognate binding partners18.
A recent NMR chemical shift analysis of ASF1a, in the absence and presence of the histone H3 C-terminal helix, identified the histone H3 binding surface of ASF1a as a hydrophobic surface on the β4-loop and β7 strand of ASF1a, a result confirmed by mutagenesis studies31. Based on these results, it appears that the histone H3/H4 complex and HIRA bind on opposite faces of ASF1a, as modeled in Fig. 7b. This is consistent with mutational data showing that the D37R,E39R double mutation that disrupts the ASF1a/HIRA interaction does not affect the interaction between ASF1a and histone H331 (our unpublished data). A schematic model of the ASF1a interaction with HIRA, CAF-1 p60 and histone H3 that incorporates our findings is presented in Fig. 7c. Key features of this model are that 1) both HIRA and CAF-1 p60 use analogous B-domains to compete for the same binding surface on ASF1a; 2) HIRA regions outside of the B-domain participate in ASF1a binding via interaction with the N- and C-terminal fragments of ASF1a, potentiating HIRA’s specificity for ASF1a over ASF1b, while CAF-1 p60 binds comparably to ASF1a and ASF1b; 3) the B-domain binding surface of ASF1 proteins lie away from their histone H3/H4 binding surface but nonetheless may communicate through other regions of ASF1.
Future structural and biochemical studies, comparable to those reported here, would uncover additional principles that guide the assembly and activity of multi-subunit histone chaperone complexes. In sum, we have begun to define the basic principles that govern the assembly of functionally distinct multi-subunit histone chaperone complexes. These principles, together with additional targeting and regulatory subunits, no doubt contribute to the cell's ability to target functionally specific histone chaperone complexes to chromatin at the correct time and place.
METHODS
Molecular cloning and protein expression
Genes encoding full-length protein and N-terminal core domain (residues 1–154) of both human ASF1a and ASF1b were cloned into a modified pET Duet vector (Novagen) with a TEV-cleavable 6His-tag. The HIRA B-domain (residues 427–472) and various CAF-1 p60 and Cac2 constructs were cloned into a modified pCDF Duet vector (Novagen) with a TEV-cleavable N-terminal GST fusion. The appropriate cDNA fragments coding for in vitro translated HIRA and ASF1 proteins were amplified by PCR and subcloned into pcDNA3 (Invitrogen) or pSG5 (Stratagene). Selected point mutations described in the text were introduced by site directed mutagenesis using the QuikChange mutagenesis kit according to the manufacturer’s protocol (Invitrogen®) or by two step mutagenic PCR and sequenced to confirm mutations.
Recombinant proteins were expressed in BL21-Gold (DE3) cells at 37°C for 3 hours or at 18°C overnight. ASF1a/HIRA and ASF1a/CAF-1 p60 complexes were produced by coexpression. After standard GST- or 6His-tagged protein affinity purifications, proteins were cleaved by TEV protease when necessary and then further purified by ion exchange and gel filtration chromatography. The HIRA(453–467) peptide was synthesized using FMOC on solid-phase, purified using reversed-phase HPLC, and confirmed by mass spectrometry (Wistar Institute, Proteomics Core Facility).
Isothermal Titration Calorimetry (ITC) assays
ASF1 proteins (at 0.025 mM) and HIRA peptide (at 0.2 mM) were dialyzed against PBS-βME (1 × PBS, 5 mM β-mecaptoethanol) prior to ITC analysis. HIRA peptide was titrated into ASF1a proteins at 15°C with 56 injections (5 µl/each), with other details essentially as previously described36.
In vitro binding assays
Selected GST fusion proteins expressed in 5 ml E. coli cultures were purified by and retained on 100 µl glutathione sepharose beads and washed 5 times with PBS-βME. Samples were quantitated by SDS-PAGE analysis before the addition of excess stoichiometric amounts of ASF1aN or the D37A mutant. Binding reactions were incubated at 4°C for 2 hours with gentle rotation before washing 3 times with PBS-βME, followed by two more washes using PBS-βME with addition of 160 mM NaCl and 0.05% Tween 20. Samples from the last wash were resolved by SDS-PAGE and visualized by coomassie blue staining.
35S-labeled in vitro translated proteins were derived from pcDNA3 or pSG5 based plasmids using the Promega Transcription and Translation kit, according to the manufacturer’s instructions. Binding assays and peptide competition experiments were performed as described previously37. Anti-HA (12CA5, Santa Cruz) and anti-myc antibodies (9E10, Santa Cruz) were from the indicated supplier.
For B-domain binding competition experiments, ASF1aN/HIRA(427–472) and ASF1aN/CAF-1 p60 (469–559) complexes were prepared as described above. Four aliquots of about 100 ug of each complex were retained on 100 ul of glutathione beads in PBS-βME and an increasing amount of the HIRA B-domain peptide was added to each aliquot to a final volume of 0.5 ml and incubated at 4°C for 3 hours. Beads were washed 3 times with PBS-βME and analyzed by SDS-PAGE to evaluate the displacement of ASF1a from the beads that was caused by the competitive binding of HIRA(453–467) peptide to ASF1aN previously bound to either CAF-1 p60(469–559) or HIRA(427–472).
Transfections and immunoprecipitations
Human U2OS cells were cultured and transfected with pcDNA3 (Invitrogen) based plasmids encoding epitope tagged CAF-1 p60 and ASF1a proteins as described previously18. Immunoprecipitations and western blots with anti-HA (Y11, Santa Cruz) and anti-Myc (9E10, Santa Cruz) antibodies were also performed as described previously18.
Crystallization, data collection, structure determination and refinement
Crystals of the human ASF1aN/HIRA(427–472) complex were grown by hanging drop vapor diffusion at room temperature and were obtained by mixing 2 µl of a 0.5 mM protein complex solution (in 20 mM Hepes pH7.0, 150 mM NaCl and 5 mM β-ME) with 2 µl of reservoir solution containing 1.6 M NaH2PO4/K2HPO4 at pH 5.6, and equilibrating over 1.0 ml of reservoir solution. Crystals were fully grown within two weeks to a typical size of 0.3mm×0.3mm×0.2mm, and cryoprotected for data collection by step-wise transfer into reservoir solutions supplemented with 15–25% glycerol (v/v). Diffraction data from the crystals was obtained at beamline X6A at the National Synchrotron Light Source at Brookhaven National Laboratory. All data was processed with the HKL 2000 suite (HKL Research, Charlottesville, VA). The structure was solved by molecular replacement with the program PHASER38, using protein residues 1–154 from the yeast Asf1 protein structure34 as a search model. The structure was refined by simulated annealing, torsion angle dynamics, and B factor adjustments in CNS39 with iterative manual adjustments of the model and placement of solvent molecules using the program O40. The final model was checked for errors with composite-simulated annealing omit maps, and a final round of refinement resulted in a model with excellent refinement statistics and geometry (Table 1). For the two HIRA fragments in the asymmetric unit, residues 427–445 and 467–472, and 427–448 and 465–472 are missing, respectively. The two ASF1a molecules are relatively well defined, with only some side-chain density missing for some residues in loop regions.
ACKNOWLEDGEMENTS
We would like to thank M. Allaire for assistance with data collection. This work was supported by NIH grants to R.M. and P.D.A, a Leukemia and Lymphoma Society Scholar Award to P.D.A and a grant from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health awarded to the Wistar Institute. Part of this research was conducted on beamline X6A at the National Synchrotron Light Source at Brookhaven National Laboratory, which is supported by the U.S. Department of Energy under contract No.DE-AC02-98CH10886. Beamline X6A is funded by NIH/NIGMS under agreement Y1 GM-0080-03.
Footnotes
The coordinates of the ASF1a/HIRA complex structure has been deposited with the Protein Data Bank (PDB) with access code 2I32.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
AUTHOR CONTRIBUTIONS
Y.T. and M.V.P. designed and carried out experiments reported in the manuscript, and prepared manuscript figures and text; K.Z., M.G., A.C. and R.D. carried out preliminary studies that led to experiments reported in the manuscript; P.D.A and R.M. designed and supervised experiments and prepared manuscript text. All authors read and approved the submitted manuscript.REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/nsmb1147
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2933817
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
HIRA complex deposition of histone H3.3 is driven by histone tetramerization and histone-DNA binding.
J Biol Chem, 300(9):107604, 24 Jul 2024
Cited by: 0 articles | PMID: 39059488 | PMCID: PMC11388340
Structure of the Hir histone chaperone complex.
Mol Cell, 84(14):2601-2617.e12, 25 Jun 2024
Cited by: 2 articles | PMID: 38925115
Replisome Proximal Protein Associations and Dynamic Proteomic Changes at Stalled Replication Forks.
Mol Cell Proteomics, 23(5):100767, 13 Apr 2024
Cited by: 0 articles | PMID: 38615877
Regulation of endogenous retroviruses in murine embryonic stem cells and early embryos.
J Mol Cell Biol, 15(8):mjad052, 01 Jan 2024
Cited by: 2 articles | PMID: 37604781 | PMCID: PMC10794949
Review Free full text in Europe PMC
Independent and Complementary Functions of Caf1b and Hir1 for Chromatin Assembly in Tetrahymena thermophila.
Cells, 12(24):2828, 13 Dec 2023
Cited by: 0 articles | PMID: 38132148 | PMCID: PMC10741905
Go to all (127) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 2I32View structure
-
(1 citation)
PDBe - 6HisView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells.
Mol Cell Biol, 29(3):758-770, 24 Nov 2008
Cited by: 82 articles | PMID: 19029251 | PMCID: PMC2630694
Identification of an ubinuclein 1 region required for stability and function of the human HIRA/UBN1/CABIN1/ASF1a histone H3.3 chaperone complex.
Biochemistry, 51(12):2366-2377, 16 Mar 2012
Cited by: 24 articles | PMID: 22401310 | PMCID: PMC3320765
Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex.
Mol Cell Biol, 31(19):4107-4118, 01 Aug 2011
Cited by: 63 articles | PMID: 21807893 | PMCID: PMC3187368
A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function.
J Mol Biol, 429(13):1924-1933, 19 Nov 2016
Cited by: 22 articles | PMID: 27871933 | PMCID: PMC5438300
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIA NIH HHS (2)
Grant ID: P01 AG031862
Grant ID: P01 AG031862-010001
NIGMS NIH HHS (4)
Grant ID: R01 GM060293
Grant ID: R01 GM060293-01
Grant ID: Y01 GM000080
Grant ID: Y1 GM-0080-3





