Abstract
Free full text

A role for cdk9-cyclin k in maintaining genome integrity
Abstract
Cyclin-dependent kinase 9 (CDK9), with its cyclin T regulatory subunit, is a component of the positive transcription elongation factor b (P-TEFb) complex, which stimulates transcription elongation and also functions in co-transcriptional histone modification, mRNA processing and mRNA export. CDK9 also binds to cyclin K but the function of this CDK9-cyclin K complex is less clear. Others and we have recently shown that CDK9 functions directly in maintaining genome integrity. This activity is restricted to CDK9-cyclin K. Depletion of CDK9 or its cyclin K but not cyclin T regulatory subunit impairs cell cycle recovery in response to replication stress and induces spontaneous DNA damage in replicating cells. CDK9-cyclin K also interacts with ATR and other DNA damage response and DNA repair proteins. CDK9 accumulates on chromatin and limits the amount of single-stranded DNA in response to replication stress. Collectively, these data are consistent with a model in which CDK9 responds to replication stress by localizing to chromatin to reduce the breakdown of stalled replication forks and promote recovery from replication arrest. The direct role of CDK9-cyclin K in pathways that maintain genome integrity in response to replication stress appear to be evolutionarily conserved.
Introduction
The precise replication of the genome and the continuous surveillance of its integrity are essential for cell survival and the avoidance of various diseases, including cancer. The genome is constantly exposed to environmental and endogenous genotoxic insults that challenge DNA replication. To cope with this challenge, the replication stress response (RSR), a subset of the DNA damage response (DDR), coordinates diverse DNA repair and cell cycle checkpoint pathways. The RSR is critical for the prevention of cancer by acting as a barrier against genomic instability and tumorigenesis.1,2 In human pre-malignant lesions, aberrant DNA replication induces activation of the RSR, thus mobilizing cell cycle arrest, DNA repair or apoptosis. Mutations in the RSR promote the survival and proliferation of genetically unstable cells eventually leading to cancer.
One component of the RSR is the ATR (ATM and Rad3-related) checkpoint kinase, which primarily responds to single-stranded DNA (ssDNA) generated by processing of double-strand breaks (DSB) or at stalled replication forks.3 ATR function is essential to stabilize stalled replication forks and promote recovery. Disruption of ATR causes early embryonic lethality in mice,4 and cells lacking ATR have defects associated with DNA replication,5 chromosomal instability and expression of fragile sites of mammalian chromosomes,6 specific regions in the genome that are particularly prone to breakage under conditions of replicative stress. ATR senses stalled replication forks as a consequence of fork uncoupling. When DNA polymerases stall, the MCM replicative helicases continue DNA unwinding ahead of the replication fork, leading to the generation of ssDNA, which is then bound by the single-stranded binding protein RPA.7 This association initiates the checkpoint response and may allow ATRIP to recruit ATR to sites of ssDNA.5,8,9
Efficient activation of ATR requires at least several other factors. The ssDNA-RPA complex recruits the RAD17 clamp loader10 which then loads the RAD9-HUS1-RAD1 (9-1-1) clamp complex onto DNA.11 RPA also contains a protein interaction domain that binds several checkpoint proteins including ATRIP and RAD9 to promote the assembly of checkpoint signaling complexes.12 Phosphorylated RAD9 recruits topoisomerase II beta binding protein 1 (TopBP1), which binds and activates ATR.13–16 Once activated, ATR phosphorylates numerous substrates including the CHK1 kinase that helps to disperse the signal. The ATR-dependent phosphorylation of CHK1 is dependent on claspin, a mediator protein. We recently completed an unbiased loss of function genetic screen in human cultured cells to identify additional components of the RSR.17 One of the proteins we identified in the screen was CDK9.
CDK9 is a serine/threonine kinase that is a member of the cyclin-dependent kinase (CDK) family. Unlike most CDKs, which function in regulating cell cycle transitions, CDK9, like CDK7 and CDK8, has been linked predominantly with transcription, although it also plays a role in co-transcriptional histone modification, pre-mRNA processing, mRNA export and co-activation of HIV replication.18–20 CDK9 forms a heterodimer with a regulatory subunit, cyclin T1, T2a, T2b or K, to form the main component of the P-TEFb complex, which stimulates transcription elongation by phosphorylating the carboxyl-terminal domain (CTD) of RPB1, the largest subunit of RNA polymerase II as well as the negative transcription elongation factors—negative elongation factor (NELF) and DRB sensitivity inducing factor (DSIF). CDK9 is expressed broadly in all types of human tissues and its expression along with that of its regulatory T-type cyclins does not vary throughout the cell cycle.21 The cell cycle-dependent expression of cyclin K is not known. CDK9 is present in two isoforms in mammalian cells, CDK9-42 and CDK9-55, which are named for their molecular weights and differ only by the addition of a 117 amino acid sequence fused in frame to the amino-terminal end of the CDK9-42 sequence,22 although the functional difference between the two isoforms is not clear. Both CDK9 isoforms can form heterodimers with different cyclin T and cyclin K regulatory subunits, thus forming eight distinct p-TEFb complexes. While all of the CDK9/cyclin complexes can phosphorylate the CTD of RPB1 in vitro, only CDK9-cyclin T1 can interact with the HIV transcriptional activator, Tat, to promote HIV replication23 and only CDK9-cyclin T2a can bind to MyoD to promote myogenic transcription.24 The function of CDK9-cyclin K has been less clear. Cyclin K was first identified as a protein that could rescue the lethality of deleting the G1 cyclin proteins, Cln1, Cln2 and Cln3 in S. cerevisiae and was found to have CDK activating kinase (CAK) activity towards CDK2 in vitro.25 Cyclin K interacts with CDK9 in vitro and in vivo,26,27 and the CDK9/cyclin K complex can activate transcription only when tethered to RNA but not DNA.27 Cyclin K expression is also activated transcriptionally by the p53 tumor suppressor in response to DNA damage by adriamycin, ultraviolet (UV) light and ionizing radiation (IR).28
Function for CDK9-Cyclin K in the Replication Stress Response
Using a RNA interference screen to identify genes required for recovery following a transient high dose of hydroxyurea (HU), an agent that stalls replication forks by inhibiting ribonucleotide reductase and thus depleting nucleotides required for DNA synthesis, we recently identified CDK9 as a HU sensitivity gene.17 We further showed that depletion of CDK9 in cells impairs cell cycle recovery following a transient pulse of HU or aphidicolin, an inhibitor of DNA polymerase α and δ, and that this deficit can be complemented by expressing exogenous wild-type CDK9 (42 kDa isoform) but not kinase dead CDK9. Depletion of CDK9 in cells also induced phosphorylation of γH2AX, a marker for DNA double-strand breaks, in the absence of exogenous damage. The implication is that in the absence of exogenous damage, replication forks may stall and collapse when CDK9 is silenced, leading to the formation of DNA double-strand breaks. Indeed, an increase in phosphorylation of γH2AX was seen in cells co-labeled with cyclin A, a marker for cells in S and G2 phase, and with BrdU, a marker for cells in S phase, suggesting that depletion of CDK9 induces DNA damage in replicating cells. Further single-molecule experiments will be needed to specifically examine whether fork collapse is the cause of this damage. A similar induction in phosphorylation of γH2AX was also recently reported following silencing of the 55 kDa isoform of CDK9,29 further validating CDK9 as a genome maintenance protein. Additional studies will be necessary to detemine if the two CDK9 isoforms are both independently required for genome maintenance or may have partial redundancy.
To determine which regulatory subunit works with CDK9 in the RSR, we also examined cell cycle recovery after a replication challenge of HU or aphidicolin in cells silenced for cyclins T1, T2 and K. Depletion of cyclin K but not cyclin T1 or T2 impaired cell cycle recovery, suggesting that cylin K is the regulatory subunit that works with CDK9 in the RSR. Similar to CDK9, depletion of cyclin K also induced phosphorylation of γH2AX in the absence of exogenous damage. These findings demonstrate that CDK9-cyclin K is required for genome maintenance activities and has different biological activities to CDK9-cyclin T.
Direct Role for CDK9-Cyclin K in Maintaining Genome Integrity
Previous studies have shown that CDK9 may selectively regulate the expression of a restricted subset of genes instead of the expression of most genes by RNA polymerase II.30 Given CDK9's well characterized role in promoting transcription elongation, is it possible that the RSR phenotypes observed following CDK9 silencing are an indirect result of transcriptional silencing of a subset of genes involved in DNA replication or the DNA damage response? We performed a genome-wide expression analysis in cells depleted of CDK9 but did not find significant upregulation of downregulation or obvious DNA replication or DNA damage response genes, which could explain our loss of function phenotypes. Although we cannot rule out the possibility that CDK9 may affect the expression of a previously uncharacterized DNA replication or DNA damage response gene, our findings suggest that transcriptional changes may not account for the RSR defects seen in our cells. Definitive evidence will require generation of a separation of function CDK9 mutant incapable of stimulating RNA polymerase II elongation but still able to mediate replication stress responses. As cyclin K and cyclin T1 have structurally different binding surfaces to CDK9,31,32 it may also be possible to generate a CDK9 mutant, which can bind to cyclin K but not cyclin T.
We performed co-immunopreciptation studies and found that CDK9 interacts in a complex or complexes with ATR, ATRIP and the mediator protein claspin. Significantly, ATR also complexes with cyclin K but not cyclin T1 or T2, consistent with the role of cyclin K as the key regulatory subunit of CDK9 in the RSR. These interactions are not mediated by DNA as they are resistant to treatment with nuclease and are not regulated by replication stress as they are unchanged following treatment with HU. An interaction between the 55 kDa isoform of CDK9 with the non-homologous end-joining repair protein, Ku70, was also recently reported in reference 29. This interaction was likewise not DNA dependent as it was preserved after treatment with nuclease. Ku70 did not interact in a complex with cyclin T1 or T2 and the authors were unable to determine if it interacted with cyclin K due to high background. Still, these data lend support of a direct role for CDK9 in genome maintenance that is independent of its interaction with cyclin T.
Functional Conservation in Yeast
The role of CDK9 in maintaining genome integrity may be conserved in yeast. CDK9 has two homologues in S. cerevisiae, Bur1 and Ctk1,33 although Ctk1 may be more closely homologous with mammalian CDK12.34 Bur1 was recently found to interact with Rfa1 and Rfa2, the S. cerevisiae homologues of mammalian RPA, in a direct but DNA dependent manner.35 Deletion of the Bur1 carboxyl-terminus, which mediates its interaction with Rfa1, causes sensitivity to both HU and methyl methansulfonate (MMS), which stalls replication forks by akylating DNA. Deletion of Bur2, the cyclin partner of Bur1, also causes sensitivity to MMS and cisplatin.36,37 Mutations of the amino or carboxyl terminus of Bur1 induce spontanenous Rad52 foci even in the absence of exogenous damage, consistent with the role of Bur1 in preventing the breakdown of stalled replication forks. A bur1 mutant also suppressed the HU sensitivity of a mutant mec1, the S. cerevisiae homologue of mammalian ATR, suggesting that Bur1 acts upstream of Mec1. The authors also performed transcriptional genome wide profiling of a Bur1 mutant but did not find significant upregulation or downregulation of genome maintenance genes.
Bur1 also genetically interacts with Mrc1, Csm3, Tof1 and Sgs1.38 Mrc1, the S. cerevisiae homologue of mammalian claspin, Tof1 and Sgs1, a RecQ helicase with similarity to human BLM and WRN, help to stabilize DNA polymerase at stalled replication forks. Both Mrc1 and Sgs1 also regulate the checkpoint kinase Rad53.39,40 Likewise, Ctk1 is synthetically sick with Dia2, Dun1 and Srs2, synthetically lethal with Mre11, Rad50, Rad51, Rad52, Rad54, Rad55, Rad6 and Xrs2, and genetically interacts with Rfa2 and Rpa34.41,42 Dia2 is associated with the replisome and regulates replication fork progression.43,44 Rad6 regulates PCNA and acts on single-stranded gaps left behind newly started replication forks.45,46 Mre11, Rad50 and Xrs2 help to stabilize the replisome at stalled forks.47 Dun1 and Rad6 both regulate the ribonucleotide reductase inhibitor Sml1.48,49 Srs2, Mre11, Rad50, Rad51, Rad52, Rad54 and Rad55 all function in the repair of DNA double-strand breaks. A mutation in the T-loop of S. pombe CDK9 also causes UV hypersensitivity.50 These data point towards an evolutionarily conserved function of CDK9 in the replication stress response.
Regulation of CDK9 by Replication Stress
Proteins which function in the DNA damage response are often regulated by DNA damage. Although we found no evidence that the kinase activity of CDK9 is regulated by replication stress, we did observe an increase in chromatin bound CDK9 in HU-treated cells. Moreover, the amount of chromatin bound RPA70 is increased in CDK9 depleted cells in response to HU, suggesting that CDK9 limits the amount of ssDNA that is available for RPA70 binding. These findings are consistent with a model in which CDK9 responds to replication stress by localizing to chromatin to reduce the breakdown of stalled replication forks. The regulation of CDK9 by replication stress may thus be similar to that of ATR, which is regulated by localization but not by intrinsic kinase activity.
We propose a model in which CDK9 functions in distinct pools, one in which CDK9 interacts in a complex with cyclin T1 or cylin T2 to regulate transcription and one in which CDK9 interacts in a complex with cyclin K to directly regulate the breakdown of stalled replication forks (Fig. 1). Given CDK9's interaction in a complex with ATR, ATRIP and claspin, where might CDK9 function in this pathway? In contrast to Bur1, which interacts with Rfa1 and has been suggested to function upstream of Mec1, depletion of CDK9 in human cells does not affect ATR dependent phosphorylation of CHK1 or MCM2, implying that CDK9 likely does not function upstream of ATR. CDK9 depletion in cells does not impair CHK1 autophosphorylation but does induce stabilization of CDC25A, a phosphorylation target of CHK1, which is involved in the G2/M checkpoint. These data imply that CDK9 may function downstream of CHK1 or perhaps in a parallel pathway to regulate CDC25A stability. Indeed CDK9 does contain multiple CHK1 consensus R-X-X-S/T and ATR consensus SQ/TQ substrate motifs, which are evolutionarily conserved. Intriguingly, some of these motifs are found in the kinase activation domain of CDK9.
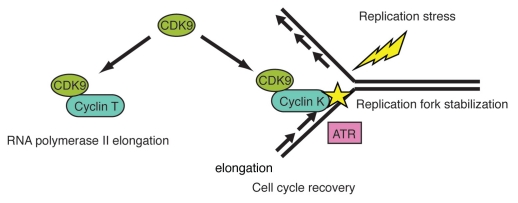
Proposed model for CDK9-cyclin K in the Replication Stress Response. CDK9 has multiple functions. With cyclin T, CDK9 functions in promoting RNA polymerase II elongation. In response to replication stress, CDK9 with cyclin K is mobilized to chromatin where it functions to prevent the breakdown of stalled replication forks and promote cell cycle recovery. CDK9-cyclin K interacts in a complex with ATR and other DNA damage response and repair proteins but likely functions downstream or in a parallel pathway to ATR and CHK1.
The specific function of CDK9 in the RSR remains to be determined. We have shown that the kinase function of CDK9 is essential for mediating recovery following replication arrest. While we cannot rule out the possibility that it may be controlling a transcriptional RSR program through RNA polymerase II phosphorylation, our negative microarray gene expression data, lack of involvement of CDK9-CCNT complexes and interaction of CDK9-CCNK with components of the RSR suggest a more direct effect. Further insight into CDK9's precise mechanism in the RSR will ultimately require identification of its substrates that mediate its genome maintenance activities.
Acknowledgements
This work was supported by a National Cancer Institute grant (R01CA136933) to D.C., the Vanderbilt Center in Molecular Toxicology (P30ES000267), Vanderbilt Ingram Cancer Center and Vanderbilt Institute for Clinical and Translational Research (UL1024975), a Department of Defense Breast Cancer Research Program Era of Hope Postdoctoral Award to D.S.Y., and a National Cancer Institute grant (K08CA143902) to D.S.Y., who is a Georgia Cancer Coalition Distinguished Cancer Scholar.
References
Articles from Cell Cycle are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.4161/cc.10.1.14364
Read article for free, from open access legal sources, via Unpaywall:
https://www.tandfonline.com/doi/pdf/10.4161/cc.10.1.14364?needAccess=true
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4161/cc.10.1.14364
Article citations
Discovery of small molecule degraders for modulating cell cycle.
Front Med, 17(5):823-854, 01 Oct 2023
Cited by: 1 article | PMID: 37935945
Review
The novel CDK9 inhibitor, XPW1, alone and in combination with BRD4 inhibitor JQ1, for the treatment of clear cell renal cell carcinoma.
Br J Cancer, 129(12):1915-1929, 26 Oct 2023
Cited by: 0 articles | PMID: 37884683 | PMCID: PMC10703862
The impact and outcomes of cancer-macrophage fusion.
BMC Cancer, 23(1):497, 01 Jun 2023
Cited by: 1 article | PMID: 37264310 | PMCID: PMC10236829
Discovery and characterization of a specific inhibitor of serine-threonine kinase cyclin-dependent kinase-like 5 (CDKL5) demonstrates role in hippocampal CA1 physiology.
Elife, 12:e88206, 25 Jul 2023
Cited by: 10 articles | PMID: 37490324 | PMCID: PMC10406435
PROTAC'ing oncoproteins: targeted protein degradation for cancer therapy.
Mol Cancer, 22(1):62, 30 Mar 2023
Cited by: 16 articles | PMID: 36991452 | PMCID: PMC10061819
Review Free full text in Europe PMC
Go to all (35) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cyclin-dependent kinase 9-cyclin K functions in the replication stress response.
EMBO Rep, 11(11):876-882, 08 Oct 2010
Cited by: 69 articles | PMID: 20930849 | PMCID: PMC2966956
Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription.
J Biol Chem, 274(49):34527-34530, 01 Dec 1999
Cited by: 136 articles | PMID: 10574912
Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages.
Curr HIV Res, 1(4):395-404, 01 Oct 2003
Cited by: 49 articles | PMID: 15049426
Review
Cyclins that don't cycle--cyclin T/cyclin-dependent kinase-9 determines cardiac muscle cell size.
Cell Cycle, 2(2):99-104, 01 Mar 2003
Cited by: 26 articles | PMID: 12695656
Review
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: R01CA136933
Grant ID: K08 CA143902
Grant ID: K08CA143902
Grant ID: R01 CA136933
Grant ID: R01 CA136933-04
NIEHS NIH HHS (2)
Grant ID: P30ES000267
Grant ID: P30 ES000267
PHS HHS (1)
Grant ID: UL1024975
 1 and
1 and 



