Abstract
Free full text

Human Cyclin K, a Novel RNA Polymerase II-Associated Cyclin Possessing Both Carboxy-Terminal Domain Kinase and Cdk-Activating Kinase Activity
Abstract
The gene coding for human cyclin K was isolated as a CPR (cell-cycle progression restoration) gene by virtue of its ability to impart a Far− phenotype to the budding yeast Saccharomyces cerevisiae and to rescue the lethality of a deletion of the G1 cyclin genes CLN1, CLN2, and CLN3. The cyclin K gene encodes a 357-amino-acid protein most closely related to human cyclins C and H, which have been proposed to play a role in regulating basal transcription through their association with and activation of cyclin-dependent kinases (Cdks) that phosphorylate the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase II (RNAP II). Murine and Drosophila melanogaster homologs of cyclin K have also been identified. Cyclin K mRNA is ubiquitously expressed in adult mouse and human tissues, but is most abundant in the developing germ cells of the adult testis and ovaries. Cyclin K is associated with potent CTD kinase and Cdk kinase (CAK) activity in vitro and coimmunoprecipitates with the large subunit of RNAP II. Thus, cyclin K represents a new member of the “transcription” cyclin family which may play a dual role in regulating Cdk and RNAP II activity.
Cyclins were first identified in the eggs of marine invertebrates as proteins whose amounts increased during interphase and then abruptly dropped at each mitosis (17). Cyclins are absolutely required for the kinase activity of cyclin-dependent kinases (Cdks). The first Cdk gene was identified in Saccharomyces cerevisiae as a temperature-sensitive mutant in the cell division cycle, cdc28 (23). Subsequently, CDC28 was found to encode the catalytic subunit of a protein kinase called p34 (3) that was the homolog of the Schizosaccharomyces pombe cdc2+ gene (39). In these yeasts, p34cdc28/cdc2 controls two critical cell-cycle transitions, the G1-S transition and G2-mitosis (M) transition (39, 45).
Since these initial discoveries in yeasts and invertebrates, the number and functional diversity of Cdks and the cyclins that regulate them have grown considerably. Several of the human Cdks and cyclins were isolated based on their ability to rescue mutations in their yeast counterparts (16, 30, 31, 37) or by sequence similarity (35). The key regulators of the G1 progression in mammalian cells include three D-type cyclins (D1, D2, and D3), which bind to and regulate either Cdk4 or Cdk6, and cyclin E, which associates later in G1 with Cdk2 (reviewed in reference 52). Human Cdk1 (also known as Cdc2) in association with cyclin B is required at the G2-M-phase transition (13, 14, 27, 33).
In addition to the accumulation and destruction of cyclins, Cdks are both positively and negatively regulated by phosphorylation. Full activation of Cdks requires the phosphorylation of a highly conserved threonine residue in the T-loop (T161 in Cdc2). A candidate for this Cdk-activating kinase (CAK) is itself composed of a Cdk, Cdk7, and a cyclin, cyclin H (20, 54). Recently, a CAK from budding yeast, called Cak1 or Civ1 (25, 59), was identified that is unrelated to Cdk7. This brings into question whether Cdk7-cyclin H is actually acting as CAK in vivo and whether there are additional CAK activities in mammals.
In addition to their important role as essential regulators of the cell-division cycle, cyclins and Cdks have been shown to participate in other seemingly unrelated cellular processes. Most revealing was the isolation of cyclin H-Cdk7 and its yeast homolog, Ccl1-Kin28, as subunits of the basal transcription repair factor TFIIH (18, 49). Another cyclin-Cdk pair, Srb11-Srb10 (32), and its human homolog, cyclin C-Cdk8, are also components of the RNA polymerase II (RNAP II) holoenzyme (34, 46, 57). The yeast RNAP II holoenzyme is a large, stable complex composed of RNAP II; a subset of general transcription factors, including TFIIB, TFIIF, and TFIIH; and a group of proteins called the Srbs. Srbs function as mediators between the RNAP II holoenzyme and promoter-selective factors, which results in the activation or repression of transcription.
The regulation imparted by the two known cyclin-Cdk components of the RNAP II holoenzyme is thought to occur through the RNAP II largest subunit’s essential carboxyl-terminal domain (CTD) (1, 38, 64), which consists of multiple heptapeptide repeats with the consensus sequence YSPTSPS (reviewed in reference 41). Multiple kinases phosphorylate the CTD in vivo (7, 29, 44), a modification that is likely to be important for transcription. In S. cerevisiae, three distinct cyclin-Cdk complexes that phosphorylate CTD have been characterized: Ccl1-Kin28 (18), Srb11-Srb10 (32, 58), and Ctk2-Ctk1 (55). Null alleles of KIN28 result in a decrease in the abundance of all mRNA species (8). The Ccl1-Kin28 kinase complex is present in budding yeast TFIIH (8, 61) and phosphorylates CTD but not Cdc28 in vitro. SRB10 (SSN3) and SRB11(SSN8) were isolated as suppressors of a CTD truncation mutation (32) and an snf1 mutation (5, 26). Mutations in SRB10 or SRB11 cause global defects in class II gene expression and affect the ability of cells to respond to transcription regulators in vivo (26). CTK2, a C-type cyclin, and CTKK1, a CTD kinase, form a complex in vivo, although a role in transcription has not been demonstrated (55). In addition, several related cyclin-Cdk complexes have been identified in S. pombe, but their roles in transcription or Cdk activation are unknown (4, 11, 21, 36).
The precise regulatory roles of the cyclin-kinase components of the RNAP II holoenzyme have not been elucidated. Recent genetic and biochemical evidence suggests that transcription is initiated in vivo by an RNAP II holoenzyme consisting of RNAP II; a subset of general transcription factors, including TFIIH; and a mediator subcomplex that includes the Srb proteins cyclin C and Cdk8. The mediator complex has been proposed to provide the critical connection between upstream signal transduction cascades and promoter-specific transcriptional initiation and elongation. Recently, cyclin H-Cdk7 as part of the TFIIH complex has been shown to stimulate the N-terminal activation function AF-1 of retinoic acid receptor α1 (RARα1) (47). Thus, phosphorylation by the TFIIH-associated Cdk7 could provide a regulatory interface between RNAP II and transregulatory factors. How phosphorylation of CTD controls RNAP function is unknown. It may be that the cyclical phosphorylation and dephosphorylation of the CTD are required for multiple steps in a single transcription cycle. Alternatively, individual Cdk complexes could phosphorylate different parts of the CTD, thereby regulating different steps in transcription. If cyclin Cdks mediate transcriptional activation for different transcription factors, as has been implied for cyclin H-Cdk7 and AF-1, then these cyclin-Cdk complexes may be tissue-specific regulators of gene expression during development.
Many questions remain concerning the roles of cyclin-Cdks in activating other Cdks as CAKs and their newly discovered role in regulation of gene expression as components of the RNAP II holoenzyme. The true identity of the human CAK has not been established, and the physiological roles of cyclin-Cdks in regulating gene expression have not been elucidated. Here, we describe the discovery of a new human cyclin present in RNAP II complexes, designated cyclin K, which is associated with both a CTD kinase and CAK activity in vitro. Cyclin K is only the second cyclin, after cyclin H, to display both CAK and CTD kinase activity in vitro.
MATERIALS AND METHODS
Cell culture and cell lines.
All cell lines described in this study, including the retinal pigment epithelium (RPE) cell line (gift of Alberta Davis), were maintained in growth medium consisting of Dulbecco’s modified Eagle’s medium (DMEM) with high concentrations of glucose, 2 mM glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), and 5% heat-inactivated fetal calf serum in a 5% CO2 incubator.
Antibodies.
Polyclonal antibodies against full-length cyclin K were prepared by immunizing rabbits with a recombinant glutathione S-transferase (GST)-cyclin K fusion protein produced either in bacteria or in insect cells. Antibodies were affinity purified on hemagglutinin (HA)-cyclin K (35 mg/ml) coupled to an Affigel 10 column (Bio-Rad), released with a pH shock (100 mM glycine [pH 2.4]), and neutralized with 1 M Tris (pH 8.0). These antibodies (α-KFL) were concentrated to 2 mg/ml by centrifugation through Centricon-30 columns. For anti-cyclin K peptide antibodies (α-Kpep), a 16-amino-acid C-terminus peptide that was conserved between mice and humans was generated, coupled to keyhole limpet hemocyanin, and injected into rabbits. Rabbit α-Kpep antisera were affinity purified by using the peptide coupled to activated CH Sepharose 4B, eluted, and concentrated as described above.
Chromosomal mapping.
Human cyclin K cDNA was used as a probe to obtain two independent genomic P1 clones (Genome Systems, Inc.), which were fluorescently labeled and used for in situ hybridization on mitotic chromosomes (Baylor FISH Core Facility). To further sublocalize cyclin K, CEPH mega YACs (748G11, 935B5, 884G2, 878G10, 840C11, and 777B4), which span the D14S13 locus of 14q32, were examined for the presence of cyclin K gene coding sequences by using intronic PCR primers (30F, gag tgg tgc tga aca cgt ggt; and 25R, atc aac att tga aca tgt aca) which amplify a 151-bp fragment containing exon 5. The positive YACs (777B4, 935B5, 884G2, and 74G11) had the presence of cyclin K confirmed by Southern blot hybridization.
In vitro binding assays.
Cdk1 to -8 and cyclin K were in vitro transcribed and translated with TnT reticulocyte lysate (Promega). Radiolabeled translation products were then added to 30 μl of either GST-cyclin K or GST-glutathione beads and incubated in NETN buffer (20 mM Tris [pH 7.5], 120 mM NaCl, 0.5% Nonidet P-40, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 20 U of aprotinin per ml, 1 mM NaF) at 4°C for 2 h. Batch assays were washed three times with NETN buffer and run on sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis (SDS-PAGE) gels.
Baculovirus infections.
GST-cyclin K was subcloned into XbaI-BamHI-digested pVL1392 (Pharmingen, Inc.). Recombinant Autographa californica nuclear polyhedrosis viruses were then generated by using the BaculoGold transfection system (Pharmingen, Inc.) and amplified.
Metabolic labeling and immunoprecipitations.
Cells were incubated in serum-free DMEM without methionine for 30 min and then labeled with a 200-mCi/ml mixture of [35S]methionine and [35S]cysteine in serum-free DMEM without methionine for 4 h at 37°C. Cells were lysed in cyclin K lysis buffer (50 nM HEPES [pH 7.50], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1% Tween 20) containing 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 20 U of aprotinin per ml, 10 mM β-glycerophosphate, 1 mM NaF, and 0.1 mM sodium orthovanadate. Immunoprecipitations were performed for 6 h at 4°C. Immunoprecipitated proteins (IPs) were washed five times with 1 ml of 400 mM NaCl NETN buffer and separated on 10% polyacrylamide gels containing SDS, and proteins were visualized by fluorography. Two microliters (2 μg) of affinity-purified α-Kpep antibodies or 5 μl of normal rabbit serum (NRS) was used to immunoprecipitate lysates corresponding to one 10-cm-diameter petri dish (approximately 500 μg). One microgram of cyclin K peptide was used to inhibit 2 μg of anti-cyclin K antibody. This represents an approximately 80-fold molar excess of peptide over antibody recognition sites.
Immunoblotting.
For immunoblots from whole lysates, cell pellets were washed in phosphate-buffered saline, lysed in 100 mM Tris (pH 6.8)–2% SDS–100 mM DTT, sonicated, and boiled. A total of 60 μg of each cell lysate was then electrophoresed through SDS-PAGE (10% polyacrylamide) gels and transferred to Immobilon-P membrane (Millipore). Blots were blocked with 5% milk in Tris-buffered saline with 0.2% Tween 20 (TBST) and incubated for 1 h at room temperature with primary antibody diluted 1:1,000. Blots were washed three times for 20 min each, incubated in secondary antibody (Amersham) at a 1:5,000 dilution in milk-TBST for 2 h, washed again, and developed with the Amersham ECL (enhanced chemiluminescence) reagents.
For immunoprecipitations, α-Kpep antibodies were covalently cross-linked to protein A-Sepharose beads at a concentration of 2 mg of total immunoglobulin per ml of swollen beads, by using 20 mM dimethyl pimelimidate in 200 mM borate (pH 9.0). Beads were washed with 100 mM glycine (pH 2.5) to remove any antibodies that had not been cross-linked. NRS was also coupled separately, as a negative control. A total of 100 μl of beads was used for each IP from 500 μg of cell lysate. IP beads were washed four times with wash buffer containing 250 mM NaCl, 20 mM Tris (pH 7.5), and 0.5% Nonidet P-40 and immunoblotted as described above.
CTD phosphorylation assay.
Twenty microliters of protein A-Sepharose beads covalently cross-linked with crude α-Kpep antibodies and 2 μg of affinity-purified α-Kpep or NRS were used for IPs from selected cell lines and washed again in cyclin K-CTD kinase buffer containing 2.5 mM MnCl2, 10 mM MgCl2, 50 mM Tris (pH 7.4), and 5 mM DTT. For peptide competition, 10 μg of cyclin K peptide was preincubated with 20 μl of α-Kpep–protein A-coupled beads (40 μg of crude α-Kpep), or 1 μg of cyclin K peptide was preincubated with 2 μg of α-Kpep for 1 h at 4°C (a 40-fold molar excess of peptide). CTD kinase assays were performed by adding 50 μl of a mix that contained cyclin K CTD kinase buffer, 100 ng of GST-CTD, 5 μM ATP, and 0.05 mCi of [γ-32P]ATP (3,000 Ci/mmol; New England Nuclear) to the α-Kpep or NRS IP complexes, and these mixtures were then incubated at room temperature for 1 h. Complexes were then boiled in Laemmli sample buffer and electrophoresed on SDS-PAGE (8% polyacrylamide) gels.
In vitro CAK associated with cyclin K.
RPE and WI-38 cells were lysed, and immunoprecipitations were performed with 2 μg each of affinity-purified α-Kpep antibodies, anti-Cdk7 antibodies (Santa Cruz Biotechnology, Inc.), and NRS antibodies. Bacterially expressed and purified cyclin A and HA-tagged Cdk2 (0.1 μg of each) were incubated for 20 min at room temperature with 20 μl (out of 100 μl) of cyclin K IPs, cyclin K IPs preincubated with peptide, Cdk7 IPs, or NRS IPs in a total volume of 100 μl, containing 25 mM HEPES, 10 mM MgCl2, 1 mM ATP, 1 mM DTT (pH 7.4), 1 mM phenylmethylsulfonyl fluoride, 0.1 mM Na3VO4, 2 μg of aprotinin per ml, and 1 μg of leupeptin per ml. After activation, the complexes were assayed for H1 kinase activity by the addition of 30 μl of histone H1 kinase mix (5 μg of histone H1 [Boehringer Mannheim], 50 mM ATP, 1 μCi of [α-32P]ATP, 25 mM HEPES, 10 mM MgCl2, 1 mM DTT [pH 7.4]). After 10 min at room temperature, the reactions were stopped by addition of SDS sample buffer, and the mixtures were electrophoresed by SDS-PAGE (10% polyacrylamide). Dried gels were autoradiographed for 10 to 20 min.
Isolation of mouse cyclin K cDNA.
PCR primers directed toward the N terminus (5F, ATG AAG GAG AAT AAA GAA AAT) and an internal region (8R, GAT TTC AAA TTT GCA CAA ACG) of human cyclin K were used to amplify a 660-bp fragment from a mouse breast epithelial cDNA library in pACTII (65). This PCR product was subcloned into pCRII (Invitrogen) and used as a probe to screen a total mouse testis cDNA library to obtain a full-length mouse cyclin K cDNA.
In situ hybridizations.
In situ hybridizations of mouse embryos collected at 9, 10.5, 12.5, and 15 days postcoitus were performed as described previously (43). Sections are from C57 Black embryos and were subjected to in situ hybridization with the mouse cyclin K 35S-labeled riboprobe.
RESULTS
Identification of CPR4, which can function as a G1 cyclin in S. cerevisiae and can cause a Far− phenotype in yeast.
A screen was performed for human cDNAs that could overcome activation of the mating pheromone pathway when expressed in the yeast strain SY2227. SY2227 contains STE4 under the control of the GAL1 promoter and a FUS1-HIS3 fusion. The presence of galactose induces STE4, which acts in a dominant fashion to titrate out Gpa1 and activate the mating pathway and cell cycle arrest. We sought to isolate CPR (cell cycle progression restoration) cDNAs that could specifically overcome the growth arrest due to the mating pheromone without interfering with the signal transduction pathway activating FUS1 gene expression, as was previously shown for the far1 mutation (6) (Fig. (Fig.1A).1A). A human HepG2 cDNA library under GAP control in pAB23BXN (50) was introduced into SY2227. Transformants were selected on plates containing synthetic complete medium lacking histidine and uracil (SC-his-ura) supplemented with 2% galactose and 5 mM 3-aminotriazol to ensure high-level FUS1 expression (48). Under these conditions, normal Ura+ SY2227 cells are arrested because of the activation of the mating pathway. Of two million transformants, 420 colonies survived selection, and 29 imparted the Cpr+ phenotype upon reintroduction into SY2227 (Fig. (Fig.1B).1B). These fell into 13 classes based on DNA sequence analysis (15).
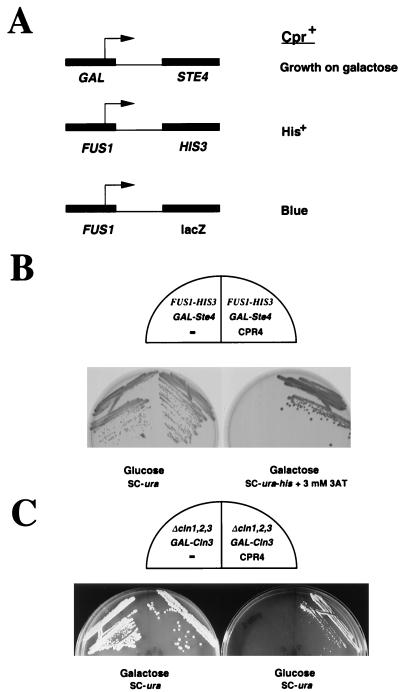
Isolation of human cyclin K by a functional selection strategy with yeast cells. (A) Selection scheme for isolation of CPR genes. Galactose induces high levels of the Gα subunit Ste4, which binds to and inhibits the Gα subunit Gpa1, thus freeing Gα for signaling and activation of cell cycle arrest and transcription. To eliminate selection of cDNAs that interfere with the upstream signal transduction cascade, CPR gene-harboring yeast cells are required to grow on galactose while simultaneously maintaining an active FUS1-HIS3 gene, thus ensuring the activity of the transcriptional branch. The level of activity of the FUS1 promoter is quantified with a FUS1-lacZ reporter. (B) Yeast strain SY2227 (15) expressing CPR4 (human cyclin K) is resistant to Ste4 overproduction by GAL-STE4 without interference of FUS-HIS3 expression. SY2227 strains harboring either pCPR4 or pUN80 (a URA3 control plasmid lacking CPR4/cyclin K) were streaked onto SC plates lacking uracil with 2% glucose as a carbon source (SC-ura) or SC plates lacking histidine and uracil (SC-ura-his) and containing 3 mM 3-aminotriazol (3AT) and 2% galactose. The 3-aminotriazole is used to select for expression of the FUS1-HIS3 fusion. (C) CPR4/cyclin K rescues the lethality of a yeast strain with cln1, cln2, and cln3 deletions. The yeast strain Y145 has a deletion of cln1, cln2, and cln3 and is kept alive by an integrated GAL-CLN3 gene. On galactose medium, CLN3 is transcribed and the cells live. On glucose, the GAL promoter is shut off and the cells fail to grow. Y145 expressing CPR4/cyclin K is able to grow on glucose, while the same strain harboring a control plasmid, pUN80, fails to grow on glucose.
One mechanism by which a gene might impart a Far− phenotype to yeast is to bypass the G1 cyclin requirements at START. Mutations that stabilize Cln3 (DAF1-1) are mating factor resistant (10). To test whether any of the CPR genes was rescued via this mechanism, each CPR gene was examined for its ability to rescue the lethality associated with a cln1 cln2 cln3 triple mutant that is kept alive by CLN3 under control of the GAL promoter. This strain grows in the presence of galactose but fails to grow in glucose. Only one human cDNA, CPR4, rescued this conditional lethality (Fig. (Fig.1C).1C). A characterization of the remaining 12 CPR genes is presented elsewhere (15).
CPR4 encodes a novel human cyclin, designated cyclin K.
Sequencing of CPR4 revealed that it encoded a new human cyclin that was only distantly related to the other members of the cyclin family. Because of its sequence divergence, CPR4 constituted a new subtype of cyclin (Fig. (Fig.2A2A and B). Although there is as yet no human cyclin J, because a Drosophila gene has recently been named “cyclin J” (19), we elected to call this gene “cyclin K.”
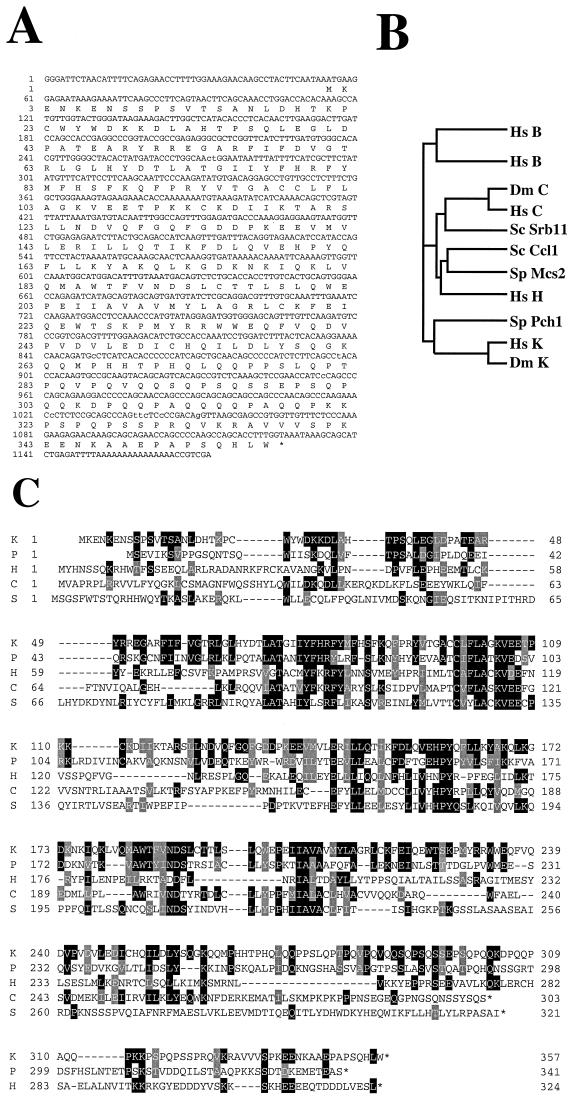
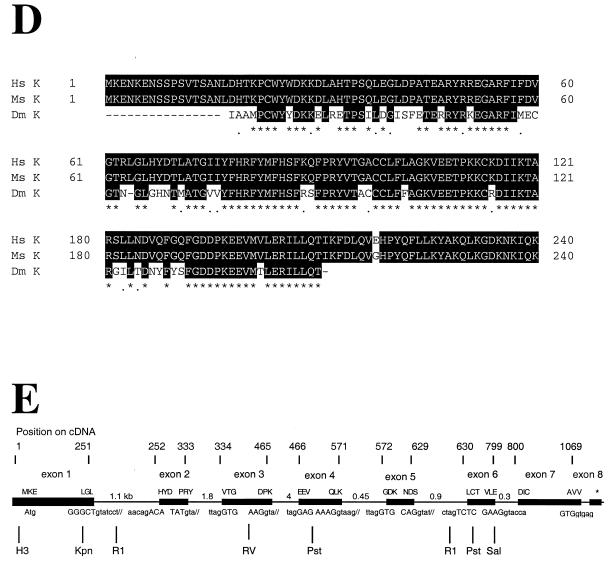
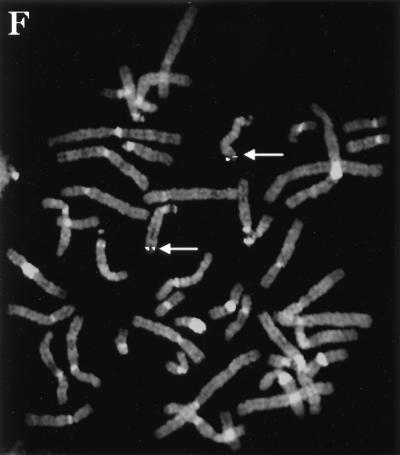
The cyclin K gene encodes a new class of metazoan cyclin that is related to cyclins C and H. (A) cDNA and protein sequence of the CPR4 gene, which encodes a new human cyclin, designated cyclin K. (B) An evolutionary tree of the cyclin C, H, and K subfamilies from humans and yeast. Hs, human; Dm, D. melanogaster; Sc, S. cerevisiae; Sp, S. pombe. Protein sequence similarities between all cyclin types were compared by using the program Pileup from the GCG (Genetics Computer Group, Madison, Wis.) sequence analysis package. The vertical lines indicate divergence; the horizontal lines indicate the relative time between each occurrence. (C) Homology alignment with human cyclins C and H and yeast cyclins Pch1 and Srb11. Black boxes indicate identities, and gray boxes indicate similarities. K, human cyclin K; P, S. pombe Pch1; H, human cyclin H; C, human cyclin C; S, S. cerevisiae Srb11. (D) Cyclin box alignment of human, mouse, and Drosophila cyclin K. The GenBank accession numbers for the cyclin K genes are as follows: human, AF060515; mouse, AF060517; and Drosophila, AF060516. (E) Genomic structure of human cyclin K. Human P1 clones containing cyclin K were subcloned into pBluescript KS II+ and sequenced. (F) The human cyclin K gene is located on chromosome 14q32. In situ hybridization was performed on mitotic chromosomes by using fluorescently labeled cyclin K P1 clones. Arrows indicate the localization of cyclin K on chromosome 14 at 14q32.
Cyclin K’s 1.1-kb cDNA encoded a 357-amino-acid open reading frame with a predicted molecular mass of 41.6 kDa (Fig. (Fig.2A).2A). Among the human cyclins, cyclin K is most similar to human cyclin C, with which it shares 22% identity and 44% similarity overall and 28% identity and 53% similarity within the cyclin box (Fig. (Fig.2B2B and C). Among the yeast cyclins, cyclin K is most similar to S. pombe Pch1, with which it shares 26% identity and 51% similarity overall and 34% identity and 58% similarity within the cyclin box (Fig. (Fig.2B2B and C). This high degree of similarity is also shared with an S. cerevisiae cyclin C homolog, Srb11 (32). Pch1 is an essential C-type cyclin whose function is unknown (21). SRB11 mutants were identified as suppressors of the Cs phenotype of partial truncation mutants in CTD (58), and alleles of SRB11 have been isolated in three other screens for mutations involved in the regulated repression of transcription (ssn8, are1, and ume1 mutants). ssn8 mutants derepress catabolite-repressed genes (26), are1 mutants cause partial loss of α2-mediated repression of a-specific genes in α-cells (62), and ume3 mutants relieved the repression of genes normally expressed only during meiosis (56).
We have also cloned mouse cyclin K and have found that it shares 99% amino acid identity (Fig. (Fig.2D)2D) with human cyclin K. In addition, examination of the database revealed an expressed sequence tag (EST) (AA201937) encoding a Drosophila melanogaster cyclin K homolog with significant conservation with human cyclin K (Fig. (Fig.22D).
Cyclin K resides within the refined USH1A locus.
The chromosomal location and genomic organization of human cyclin K were determined. The human cyclin K genomic locus was isolated on a P1 clone, and sequence analysis revealed that it is comprised of eight coding exons (Fig. (Fig.2E).2E). Fluorescent in situ hybridization with the cyclin K genomic P1 clones revealed that cyclin K mapped to 14q32, a region that harbors several human disease loci, including Usher syndrome type 1A (USH1A) (Fig. (Fig.2F)2F) (28). Usher syndrome is an autosomal recessive disease that is a frequent cause of combined deafness and blindness in humans (53, 60) and is marked by severe to profound congenital hearing loss, absent vestibular response, and progressive pigmentary retinopathy. The genetic mapping of USH1A has been further refined to the D14S13 locus, with a location score in log base 10 (lod) of 4.90 (28). Cyclin K genomic PCR primers were used to show that cyclin K was present on overlapping YACs (CEPH mega YACs 935B5, 777B4, and 884G2) that comprise this locus, which is approximately 800 kb (15a), demonstrating that cyclin K is tightly linked to this disease gene. However, mutations in cyclin K have not been detected in USH1A families (15a).
Cyclin K is ubiquitously expressed.
A cyclin K probe recognized a 2.3-kb RNA transcript present in all mouse and human adult tissues examined (Fig. (Fig.3A),3A), with the highest level of expression observed in the testis. Additional 2.9- and 1.4-kb transcripts were also detected in some tissues, most prominently in the testis and thymus.
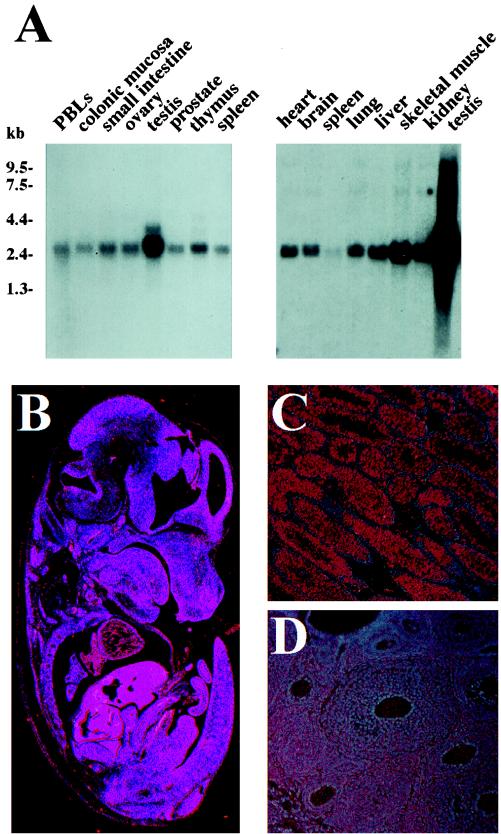
Expression pattern of cyclin K mRNA. (A) Northern blots containing poly(A)+ RNA (2 μg per lane) from the indicated tissues were probed with full-length human cyclin K cDNA as described in Materials and Methods. PBLs, peripheral blood lymphocytes. Exposure times were 1 h. (B) Sagittal section from a 15.5-day-postcoitus C57 Black embryo that was subjected to in situ hybridization with the mouse cyclin K 35S-labeled riboprobe. (C) Transverse section through an adult mouse testis. (D) Transverse section through an adult mouse ovary.
Two rabbit polyclonal antibodies were generated against a GST-cyclin K fusion protein that was expressed and purified from Escherichia coli (α-KFL). Crude α-KFL sera recognized subnanogram quantities of bacterially expressed HA-cyclin K on Western blots (data not shown). Recombinant cyclin K was expressed as an HA-tagged fusion protein (HA-cyclin K) in bacteria and ran as a 50-kDa protein on SDS-PAGE (data not shown). Western blotting of cell extracts from several mammalian cell lines with affinity-purified α-KFL revealed that the cyclin K protein migrates as doublet with a predominant 54-kDa species and a less-abundant 52-kDa species (Fig. (Fig.4A,4A, HepG2 lane). Furthermore, cyclin K abundance was variable among cell lines. Cyclin K was present in high levels in a HepG2 cell line and an RPE cell line, but was at much lower levels in the U-2 OS and WI-38 cell lines (Fig. (Fig.4A).4A).
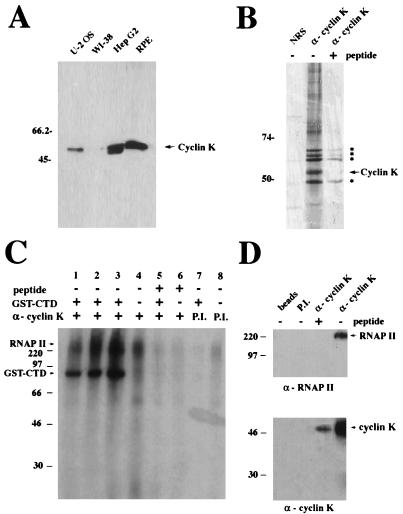
Human cyclin K is associated with a potent CTD kinase activity. (A) Western blot analysis of human cell lysates probed with antibodies (α) to cyclin K. Sixty micrograms of protein from each of the following whole-cell extracts was probed with affinity-purified α-Kpep antibodies: U-2 OS, osteogenic sarcoma; WI-38, human diploid fibroblast; HepG2, hepatocellular carcinoma; and RPE (gift of A. Davis). (B) Immunoprecipitation of endogenous cyclin K protein from the [35S]methionine-labeled RPE cell line. Two microliters (2 μg) of affinity-purified α-Kpep antibodies or 5 μl of NRS was used to immunoprecipitate protein from lysates (500 μg). One microgram of cyclin K peptide (80-fold molar excess) was used to inhibit 2 μg of α-Kpep antibody. Asterisks correspond to bands associated with the cyclin K immunoprecipitation. (C) GST-CTD in vitro kinase activity associated with cyclin K. RPE whole-cell extracts (100 μg) were immunoprecipitated with either affinity-purified α-Kpep (lanes 1 to 6) or preimmune (P.I.) sera (lanes 7 and 8) that had been preincubated with (lanes 5 and 6) or without (lanes 1, 2, 3, 4, 7, and 8) peptide competitor, and the resultant complexes were used for in vitro kinase assays with (lanes 1, 2, 3, 5, and 7) or without (lanes 4, 6, and 8) the addition of a bacterially produced GST-CTD fusion protein. (D) Association of cyclin K with RNAP II in RPE. Immunoprecipitations were performed with 100 μg of RPE cell extract by using 20 μl each of protein A-Sepharose beads, protein A-Sepharose beads coupled with preimmune (P.I.) sera, and protein A-Sepharose beads covalently cross-linked with crude α-Kpep antibodies, with or without preincubation with cyclin K peptide competitor. Western blots were then performed with anti-RNAP II (ARNA3) antibodies (upper panel) and α-KFL antibodies (lower panel) as probes.
Expression of cyclin K in adult tissues and during mouse embryonic development.
Human and mouse tissue Northern blots revealed that cyclin K’s message was much more abundant in the testis than in all other adult tissues examined (Fig. (Fig.3A).3A). Cyclin K’s mRNA expression was examined in the adult mouse testis by in situ hybridization of histological sections of the mouse testis and ovary with 35S-labeled antisense and sense mouse cyclin K riboprobes. Cyclin K transcripts were clearly detectable throughout the seminiferous epithelium, but were not detectable in the peritubular cells or the interstitial tissue between adjacent tubules, which harbors the Leydig cells (Fig. (Fig.3C).3C). Determination of the exact stage of the germ cells expressing cyclin K will require further expression studies, performed in conjunction with staining with known cell-specific markers.
Histological sections of adult mouse ovaries were also analyzed for cyclin K expression (Fig. (Fig.3D).3D). Cyclin K message was highly abundant in the developing oocyte. Thus, cyclin K is highly expressed in the developing germ cells of both sexes.
The expression pattern of cyclin K during mouse embryogenesis was examined. Cyclin K was detectable throughout a 12.5-day embryo (Fig. (Fig.3B).3B). Upon further inspection, cyclin K appeared more abundant in several tissues and cell types, including the eye and cochlea (data not shown). Within the ear, cyclin K was highly expressed in the crista ampullaris of the semicircular canals (data not shown). Within the eye, the mRNA was most abundant in the single-cell layer that comprises the RPE, in agreement with the RPE protein blot.
Cyclin K does not associate with Cdk1 to Cdk8 in vitro.
To examine possible Cdk partners for cyclin K, in vitro binding assays using bacterially expressed HA-cyclin K or insect cell-purified GST-cyclin K failed to show a significant association of cyclin K with a battery of in vitro-translated Cdks (Cdk1 to -8) (data not shown). Because of the sequence similarity among cyclins C, H, and K, further attempts were made to detect an association of cyclin K with Cdk7 and Cdk8. Insect cells (Sf9) coinfected with baculovirus GST-cyclin K and HA-Cdk7 failed to show appreciable association of these proteins by glutathione bead chromatography or HA immunoprecipitation (data not shown). Because Cdk7 associates with cyclin H with the assistance of the RING finger protein Mat1 (12), these cyclin K binding assays were also performed with both Cdk7 and Mat1, but still failed to show appreciable association (36a). Therefore, cyclin K does not appear to associate with Cdk1 to -8 in vitro and is likely to associate with one of the less-characterized Cdks in vivo. Immunoprecipitation of cyclin K from 35S-labeled cells by using the antipeptide antibody revealed the presence of five highly labeled bands in the 48- to 65-kDa range that were competed with the cyclin K peptide (Fig. (Fig.4B).4B). One or more of these is likely to be the Cdk subunit responsible for cyclin K’s kinase activity.
Insect cell-expressed GST-cyclin K is associated with a potent CTD kinase.
Insect cell GST-cyclin K purified by glutathione affinity was used to search for candidate substrates for cyclin K-associated kinases, by using GST glutathione-affinity extracts as a negative control. Bacterial GST-cyclin K incubated with HeLa extracts was also employed. Both insect cell and bacterially produced GST-cyclin Ks failed to display kinase activity toward candidate substrates, including histone H1, phosphorylase a, phosphorylase b, myelin basic protein (MBP), or GST-Rb, but insect cell-derived GST-cyclin K, but not GST alone, copurified with a potent kinase activity toward a GST-CTD fusion protein (data not shown).
Cyclin K is associated with a potent RNAP II-CTD kinase in RPE cells.
To explore the possibility that cyclin K may have an association with RNAP II and kinase activity towards it, the RPE cell line, which expressed relatively high levels of cyclin K, was chosen for biochemical analysis. α-Kpep IP kinase assays, with histone H1 and GST-CTD as substrates, were performed in parallel with WI-38 and RPE cell extracts. Cyclin K IP Western blots were hampered by the fact that cyclin K migrates at 54 kDa, which overlaps immunoglobulin G migration. For this reason, α-Kpep was covalently cross-linked to protein A-Sepharose beads and used for IP Western blots. A GST-CTD kinase activity was detected in IPs from the RPE cell line but not in IPs from the WI-38 extracts (data not shown) or IPs with NRS (Fig. (Fig.4C).4C). This activity correlates with the relative abundance of cyclin K in these cells. A phosphorylated band, at approximately 90 kDa, is present in cyclin K IPs from RPE cells (Fig. (Fig.4C,4C, lanes 1 to 3) but is not detectable in reactions lacking the GST-CTD substrate (lanes 4, 6, and 8), IPs preincubated with antigenic peptide in molar excess (lane 5), or IPs with preimmune sera (lane 7). Therefore, cyclin K IPs from RPE cells are associated with a potent CTD kinase. This activity is not present in cyclin K IPs from WI-38 cells, which have low levels of cyclin K protein (Fig. (Fig.4A).4A). These data suggest that cyclin K may have a Cdk partner with CTD kinase activity.
Cyclin K is associated with the large subunit of RNAP II in RPE cells.
In addition to the 90-kDa GST-CTD phosphorylated band, additional phosphorylation bands of 220 and 240 kDa were present and were found only in the immune complexes from RPE cells (Fig. (Fig.4C,4C, lanes 1 to 4). The size of these phosphoproteins, as well as the knowledge that the related cyclins C and H are in complexes with RNAP II, suggested that this substrate might be RNAP II. RNAP II exists in two major forms: RNAP IIa, which migrates with an apparent molecular mass of 220 kDa in SDS-PAGE gels; and a form with additional phosphates on the CTD, RNAP IIo, which migrates at approximately 240 kDa. To test whether RNAP II was associated with human cyclin K, α-Kpep IPs were probed with a monoclonal antibody directed against an internal motif of RNAP II (ARNA3). A 220-kDa band corresponding to RNAP IIa was observed in the cyclin K IPs and was not present in control lanes (Fig. (Fig.4D).4D). This band directly overlapped the 220-kDa phosphoprotein band observed in the kinase reaction. The ARNA3 antibody failed to recognize the 240-kDa band of RNAP IIo, which is consistent with prior characterizations of this antibody (24).
Cyclin K is a CAK in vitro.
When cyclin H is associated with Cdk7, the kinase complex is able to phosphorylate either the C-terminal domain of the largest subunit of RNAP II (CTD) or Cdk2. To test whether cyclin K also exhibits this dual capacity, immunoprecipitations of cyclin K from RPE cell extracts were performed, and the resulting complexes were used for kinase assays against bacterially produced Cdk2 complexed with cyclin A (9). Cyclin K IPs exhibited a potent kinase activity toward Cdk2 (data not shown), and this phosphorylation results in the activation of Cdk2 kinase activity toward a histone H1 substrate (Fig. (Fig.5A).5A). Cyclin K IPs from WI-38 cells displayed significantly less CAK activity, consistent with the lower levels of cyclin K in WI-38 cells (Fig. (Fig.5A).5A). Cyclin K IPs preincubated with peptide (+pep) competitor failed to exhibit significant levels of this CAK activity, and CAK activity is not present in IPs when NRS is used (Fig. (Fig.5A).5A). To demonstrate that this activity is working through phosphorylation of T161 on Cdk2, the Cdk2 (T161A) mutant was used as a substrate in the CAK assays. Cdk2 proteins, but not Cdk2 (T161A) mutants, were capable of being activated as histone H1 kinases by cyclin K IPs (Fig. (Fig.5B).5B). Thus, like cyclin H, cyclin K is associated with an in vitro kinase activity toward both RNAP II-CTD and Cdk2 (CAK).
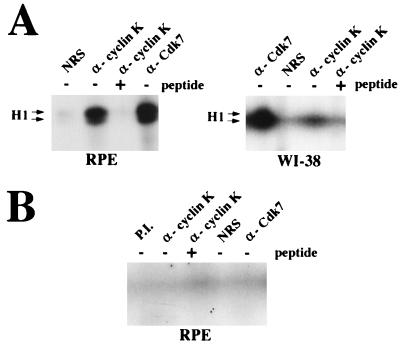
Cyclin K is a CAK in vitro. (A) Cyclin K is associated with a CAK activity towards cyclin A-Cdk2. Immunoprecipitations were performed from RPE and WI-38 cell lysates by using 2 μg each of NRS, affinity-purified α-Kpep antibodies, affinity-purified α-Kpep antibodies preincubated with peptide competitor, and anti-Cdk7 (Santa Cruz Biotechnology, Inc.) antibodies as described in Materials and Methods. IPs were incubated with 0.1 μg of bacterially expressed and purified cyclin A, HA-Cdk2, and ATP. After activation, the complexes were assayed for H1 kinase activity with [γ-32P]ATP as previously described (9), and reaction products were electrophoresed by SDS-PAGE (10% polyacrylamide) and visualized by autoradiography. (B) The cyclin K-associated CAK activity requires T161 of Cdk2. CAK assays were performed as described for panel A with IPs from RPE cell extracts, except that instead of HA-Cdk2, the T161A mutant of HA-Cdk2 was used.
DISCUSSION
In this study, we have identified a novel cyclin, cyclin K, that is associated with RNAP II and a kinase that phosphorylates a GST-CTD fusion protein in vitro. Cyclin K-associated CTD activity is detected in cyclin K IPs as well as baculovirus-produced cyclin K. In addition, the amount of active CTD kinase correlates with the levels of cyclin K in the cell lines tested. Furthermore, the fact that cyclin K is bound to RNAP II strengthens the likelihood that the CTD of RNAP II is a physiologically relevant substrate for the cyclin K-kinase complex. These data suggest that cyclin K and the unidentified cyclin K-associated kinase behave similarly to cyclin H-Cdk7, a kinase component of TFIIH involved in the regulation of RNAP II.
The role of this CTD phosphorylation with respect to transcriptional initiation or postinitiation processes has not been elucidated. It has been demonstrated for Drosophila heat shock genes and certain other loci in vivo that an initiated but paused RNAP II is form IIA (hypophosphorylated), while actively elongating polymerase is form IIo (hyperphosphorylated) (40, 63). Thus, these loci are likely to require CTD phosphorylation for elongation, not initiation. More recently, the human immunodeficiency virus type 1 Tat protein, which is known to activate postinitiation phases of transcription, has been shown to associate with CTD kinases unrelated to TFIIH, which phosphorylate CTD in vitro (24, 42). This phosphorylation is thought to increase the processivity of RNAP II (42). Moreover, E1A and VP16 have been shown to interact with a CTD kinase that is suspected to be cyclin C-Cdk8 (22). In addition, overexpression of cyclin C is able to stimulate transcription at certain promoters (51). Therefore, cyclin K could participate in either promoter-specific activation or transcript-specific elongation by analogy to known CTD kinases.
Cyclins C, H, and K are candidates for molecules involved in relaying extracellular growth or differentiation signals to the transcription apparatus. The binding and phosphorylation of the transactivator RARα1 by cyclin H-Cdk7 in TFIIH result in the stimulation of activation function (47). These data do not explain how the apparently ligand-independent activation of RARα1 through phosphorylation by cyclin H-Cdk7 could be connected to the ligand dependence of transactivation of RARα1 by retinoic acid. However, this finding points to a role in specific signal transduction. If these CTD kinases act in signal transduction cascades to regulate RNAP, one might anticipate tissue-specific patterns of expression that correlate with target gene activation. Cyclin K, which shows very high levels of expression in specific tissues, such as the testes, could be involved in activating tissue-specific sets of genes used in those tissues at high levels.
In addition, we have shown that a cyclin K-associated kinase phosphorylates Cdk2-cyclin A in vitro, resulting in Cdk2 activation towards histone H1 as a substrate. A possible caveat is that the CAK activity present in cyclin K IPs may be due to an associated kinase with CAK activity that is not the direct Cdk binding partner of cyclin K, a possibility that cannot be eliminated at this time. The level of the cyclin K-associated CAK activity correlates with the level of cyclin K protein in the cell lines tested, consistent with the notion that the activity is due to cyclin K. Without knowledge of the kinase partner for cyclin K and the reconstitution of the active cyclin K kinase complex, it will not be possible to definitively prove that the associated activity is due to cyclin K. These caveats aside, cyclin K is the second human cyclin after cyclin H to exhibit both RNAP II CTD kinase activity and CAK activity in vitro. However, the in vivo relevance of both cyclin H’s and cyclin K’s associated CAK activity has not been established.
Human cyclins can be divided into three groups, which also seem to reflect their in vivo function. Group I consists of cyclins A, B, D1, D2, D3, E, and F, which have proven roles in cell cycle regulation. Group II consists of cyclins C, H, and, now, K, which have proposed roles in transcriptional regulation. Finally, group III consists of cyclins G1, G2, and I, which are likely to have roles distinct from either cell cycle or transcriptional regulation (2). Here, we have described the identification and characterization of a new member of group II, cyclin K. Our genetic and biochemical evidence suggests that cyclin K plays a specialized, cell-type-specific role in regulating RNAP II via CTD phosphorylation and, in conjunction with its yeast Cpr+ (Far−) phenotype, is also a candidate for integrating cell growth with cell cycle progression in metazoans. Moreover, cyclin K and its unidentified kinase partner are capable of activating Cdk2 (CAK) in vitro. Testing the hypotheses concerning the possible roles of cyclin K in RNAP II and Cdk regulation will require identification of its kinase partner and interference with cyclin K function in vivo.
ACKNOWLEDGMENTS
We thank Wade Harper and members of the Elledge lab for comments, helpful discussions, and reagents. We also thank the Baylor FISH Core Facility and Vesna Jurecic for mapping CPR4. We thank E. Nigg for communicating results before publication.
M.C.E. is supported by the National Institutes of Health Medical Scientist Training Program at Baylor College of Medicine. This work was supported by grant GM44664 from the Public Health Service to S.J.E. S.J.E. is a PEW Scholar in Biomedical Sciences and an Investigator of the Howard Hughes Medical Institute.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.18.7.4291
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc109013?pdf=render
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/content/full/18/7/4291
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/18/7/4291
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/18/7/4291
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Cyclin-dependent kinases: Masters of the eukaryotic universe.
Wiley Interdiscip Rev RNA, e1816, 17 Sep 2023
Cited by: 3 articles | PMID: 37718413 | PMCID: PMC10909489
Review Free full text in Europe PMC
Coming of Age: Targeting Cyclin K in Cancers.
Cells, 12(16):2044, 11 Aug 2023
Cited by: 0 articles | PMID: 37626854 | PMCID: PMC10453554
Review Free full text in Europe PMC
The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control.
Cells, 11(19):3058, 29 Sep 2022
Cited by: 3 articles | PMID: 36231024 | PMCID: PMC9563267
Review Free full text in Europe PMC
Targeting CDK9 for Anti-Cancer Therapeutics.
Cancers (Basel), 13(9):2181, 01 May 2021
Cited by: 38 articles | PMID: 34062779 | PMCID: PMC8124690
Review Free full text in Europe PMC
Topology-driven protein-protein interaction network analysis detects genetic sub-networks regulating reproductive capacity.
Elife, 9:e54082, 09 Sep 2020
Cited by: 8 articles | PMID: 32901612 | PMCID: PMC7550192
Go to all (59) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (3)
- (1 citation) ENA - AA201937
- (1 citation) ENA - AF060515
- (1 citation) ENA - AF060517
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants.
Plant J, 24(1):11-20, 01 Oct 2000
Cited by: 44 articles | PMID: 11029700
Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II.
Mol Biol Cell, 7(4):505-513, 01 Apr 1996
Cited by: 50 articles | PMID: 8730095 | PMCID: PMC275905
The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis.
Plant Cell, 16(11):2954-2966, 14 Oct 2004
Cited by: 48 articles | PMID: 15486101
The cyclin C/Cdk8 kinase.
Prog Cell Cycle Res, 2:197-204, 01 Jan 1996
Cited by: 8 articles | PMID: 9552396
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (3)
Grant ID: R01 GM044664
Grant ID: R37 GM044664
Grant ID: GM44664





