Abstract
Free full text

Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells
Abstract
Recent studies have demonstrated that the Ten-eleven translocation (Tet) family proteins can enzymatically convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). While 5mC has been studied extensively, little is known about the distribution and function of 5hmC. Here we present a genome-wide profile of 5hmC in mouse embryonic stem (ES) cells. A combined analysis of global 5hmC distribution and gene expression profile in wild-type and Tet1-depleted ES cells suggests that 5hmC is enriched at both gene bodies of actively transcribed genes and extended promoter regions of Polycomb-repressed developmental regulators. Thus, our study reveals the first genome-wide 5hmC distribution in pluripotent stem cells, and supports its dual function in regulating gene expression.
Mammalian genomes are chemically modified by DNA cytosine methylation, an inheritable epigenetic mark that is implicated in many biological and pathological processes, including gene regulation, genomic imprinting, X-chromosome inactivation, suppression of transposable elements, and tumorigenesis (Cedar and Bergman 2009; Ooi et al. 2009). Genome-wide studies of mammalian DNA methylation have shown that 5-methylcytosine (5mC) is widely distributed across the genome in a nonrandom manner (Weber et al. 2007; Fouse et al. 2008; Meissner et al. 2008; Lister et al. 2009). In conjunction with other epigenetic modifications, 5mC can regulate accessibility of the DNA to transcription factors and chromatin regulators, thereby contributing to gene regulation and cellular differentiation.
Recent studies have uncovered 5-hydroxymethylcytosine (5hmC) as the sixth base of the genome, and that the Ten-eleven translocation (Tet) family proteins is responsible for the generation of 5hmC from 5mC in mammalian cells (Kriaucionis and Heintz 2009; Tahiliani et al. 2009; Ito et al. 2010). This new discovery raises the possibility that 5hmC may function as another epigenetic mark by altering chromatin structure or contributing to the recruitment or exclusion of other DNA-binding proteins that affect transcription. Recent reports have shown that 5hmC is relatively enriched in several cell types, including mouse embryonic stem (ES) cells and certain neuronal cells (Kriaucionis and Heintz 2009; Tahiliani et al. 2009; Globisch et al. 2010; Szwagierczak et al. 2010). Expression and functional analysis have further demonstrated that Tet1, the founding member of the Tet family, is highly expressed in mouse ES cells, and depletion of Tet1 results in impairment of ES cell self-renewal and maintenance (Ito et al. 2010). Consistent with the essential role of Tet1 in ES cells, we showed recently that Tet1 occupies regulatory regions of both pluripotency-related genes and Polycomb group (PcG) protein-repressed developmental regulators (Wu et al. 2011). However, little is known about the genomic distribution of 5hmC, dependence of 5hmC on Tet1 occupancy, and the regulatory function of 5hmC on transcription. Here we report the first genome-wide map of 5hmC occupancy in mouse ES cells. The comparison of 5hmC distribution with other epigenetic marks and global expression profile provides evidence for a role of 5hmC in both transcriptional activation and repression.
Results and Discussion
Genome-wide distribution of 5hmC in mouse ES cells
To test the specificity of 5hmC antibodies in immunoprecipitating unmethylated, methylated, and hydroxymethylated synthetic DNA, we optimized the amount of input DNA and found that affinity-purified polyclonal antibodies (Active Motif) for 5hmC specifically immunoprecipitated 5hmC-containing, but not 5mC- or C-containing, DNA under denaturing conditions (Supplemental Fig. S1A). We then tested the ability of both rabbit polyclonal (Active Motif) and rat monoclonal (Diagenode) antibodies in immunoprecipitation of genomic DNA at three Tet1-bound targets (Nanog, Tcl1, and Sox17) determined by Tet1 chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) (Wu et al. 2011). Quantitative PCR (qPCR) analysis indicated that both antibodies could readily immunoprecipitate 5hmC-modified genomic DNA, and 5hmC polyclonal antibodies showed slightly higher immunoprecipitation efficiency as compared with the monoclonal antibody (Supplemental Fig. S1B,C).
To determine global distribution of 5hmC, we further tested 5hmC antibody-based immunoprecipitation combined with chromosome-wide tiling microarrays (Supplemental Fig. S2A). We found that both antibodies could consistently detect peaks of 5hmC at defined genomic regions, and that the 5hmC profile was different from that of 5mC at both gene and probe levels (Supplemental Fig. S2A–C), suggesting that the 5hmC antibody is specific. Since the majority of Tet1-binding sites in mouse ES cells are within nonrepetitive, gene-rich genomic regions (Wu et al. 2011), we mapped 5hmC distribution using whole-genome tiling microarrays that covered the entire nonrepetitive portion of the mouse genome. A total of 91,913 genomic regions enriched with 5hmC were identified with high confidence (Supplemental Table S1). Nearly 60% of 5hmC peaks were found to be within gene bodies of annotated RefSeq genes (Fig. 1A), suggesting that 5hmC is also preferentially associated with gene-rich regions of the genome (Supplemental Fig. 2A). We note that a recent study profiling the 5hmC distribution in mouse cerebellum using a different method has come to a similar conclusion (Song et al. 2011). Further analysis showed that most 5hmC-enriched regions were associated with moderate CpG density (Fig. 1B). Thus, 5hmC antibody-based DNA immunoprecipitation (hMeDIP) provides a simple and specific tool to investigate the genomic distribution and function of 5hmC in mammalian cells.
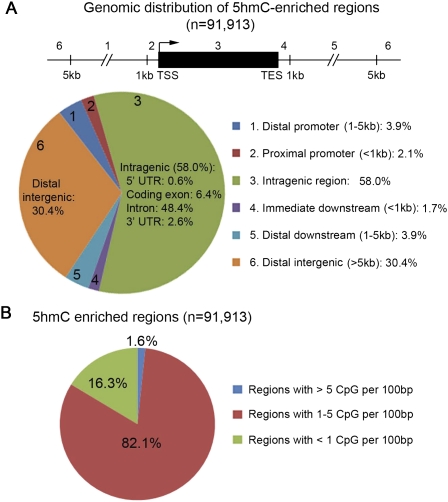
Genomic distribution of 5hmC in mouse ES cells. (A) Genomic distribution of 5hmC-enriched regions ([−log10 peak P-value] > 2.3) relative to University of California at San Diego RefSeq genes (NCBI build 36). The genome-wide 5hmC occupancy was determined by whole-genome tiling microarray analysis. (B) Proportion of 5hmC-enriched regions with different CpG densities. Note that 5hmC is enriched at genomic regions with moderate-density CpG dinucleotides.
Tet1 is required for maintaining 5hmC levels at defined genomic regions
We next mapped the 5hmC peaks to all annotated RefSeq genes in the mouse genome. We found that 5hmC was preferentially enriched in Tet1-bound genes as compared with Tet1-unbound genes (Fig. 2A), consistent with the known enzymatic activity of Tet1. Further analyses of 5hmC peaks within regions flanking transcriptional start sites (TSSs) of annotated genes indicated that 5hmC levels were high in genomic regions flanking CpG-rich proximal promoters and within Tet1-bound CpG-poor promoters (Supplemental Fig. S3A). Mapping of 5hmC peaks to Tet1-bound sites also supported the observation that 5hmC tended to be more enriched in genomic regions with medium levels of CpG density (Supplemental Fig. S3B,C). In fact, 5hmC appeared to be excluded from Tet1-binding sites with high CpG density (Supplemental Fig. S3C). The lack of high levels of 5hmC within these CpG-rich regions may possibly be explained by two nonmutually exclusive mechanisms: (1) Tet1 is capable of rapidly hydrolyzing 5mC into 5hmC, which in turn is converted to unmethylated cytosine by yet-to-be-identified downstream enzymes within CpG-rich regions. (2) High levels of trimethylated H3K4 (H3K4me3) within CpG-rich regions prevent efficient binding of the de novo DNA methyltransferase complex Dnmt3a2/Dnmt3b/Dnmt3l in mouse ES cells to these DNA sequences (Ooi et al. 2007), and thus inhibit accumulation of 5mC (Supplemental Fig. S3C).
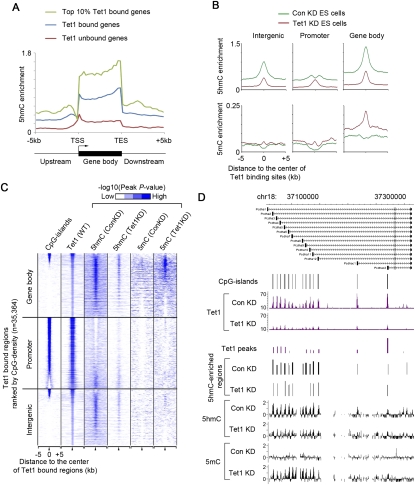
Tet1 is required for maintaining 5hmC levels at defined genomic regions in ES cells. (A) Distribution of 5hmC relative to all annotated genes in ES cells. Averaged 5hmC enrichment (measured by −log10 peak P-value) in 200-base-pair (bp) bins upstream of/downstream from gene bodies or at 5% intervals within the gene body is shown along the transcription units from 5 kb upstream of TSSs to 5 kb downstream from the transcriptional end sites (TESs). Note that 5hmC levels are generally enriched in Tet1-bound genes as compared with Tet1-unbound genes. (B) Changes in 5hmC and 5mC enrichment (measured by −log10 peak P-value) in response to Tet1 knockdown are shown for Tet1-bound regions associated with different genomic features (gene body, intergenic region, and promoter [2 kb flanking TSSs]). (C) Heat map representation of CpG islands and occupancy of Tet1, 5mC, and 5hmC in mouse ES cells at all Tet1-enriched regions (5 kb flanking the center of Tet1 peaks). The heat map is rank-ordered by CpG density of genomic regions within 500 bp flanking the center of Tet1 peaks. The enrichment of 5hmC and 5mC was determined by whole-genome tiling microarrays. Tet1-bound regions at gene bodies, promoters, and intergenic regions are shown separately. The enrichment of Tet1 binding was determined previously by ChIP-seq analyses (Wu et al. 2011). All average binding was measured by −log10 (peak P-values) in 200-bp bins and are shown by color scale. The following color scales (white, no enrichment; blue, high enrichment) are used for 5hmC, 5mC, and Tet1, respectively: [0, 2], [0, 0.5], and [0, 50]. The presence of CpG islands is displayed in color (blue, present; white, absent). (D) Tet1 occupancy and changes in 5hmC/5mC levels are shown for a group of representative Tet1 targets (Pcdha gene cluster on chr18) in control (Con) and Tet1 knockdown (Tet1 KD) ES cells. Tet1 ChIP-seq data in control knockdown (Con KD) and Tet1 knockdown (Tet1 KD) are shown in read counts, with the Y-axis floor set to 0.2 read per million reads. 5hmC and 5mC levels are shown as log2 ratios of immunoprecipitation/input (IP/input).
To further investigate the dependence of 5hmC distribution on Tet1 occupancy, we performed lentivirus-mediated knockdown of Tet1 in mouse ES cells (Ito et al. 2010). The results shown in Figure 2B demonstrate that knockdown of Tet1 resulted in reduced 5hmC levels at Tet1 regions throughout the genome that include Tet1-bound promoters, gene bodies, and intergenic regions. Many Tet1-binding sites, particularly those at CpG-rich proximal promoters, did not show a decrease in 5hmC level in the absence of Tet1 proteins (Fig. 2B,C; Supplemental Fig. S4A,B). This is likely due to a lack of 5hmC enrichment at these sites in wild-type mouse ES cells (Fig. 2C; Supplemental Fig. S3C). However, an increase in 5mC levels was still frequently observed within both promoter and nonpromoter Tet1-binding sites (Fig. 2B–D; Supplemental Fig. S4A), indicating that Tet1 has a role in maintaining a DNA hypomethylated state at these sites. Taken together, these results indicate that Tet1 is required for establishing a defined genomic pattern of 5hmC, and also for initiating an enzymatic cascade to maintain CpG-rich gene promoters in a DNA hypomethylated state.
5hmC is enriched in both Tet1-activated and Tet1-repressed genes
Previous genome-wide analyses have shown that distribution of DNA methylation in the genome may also be regulated by histone modifications (Ooi et al. 2007; Schlesinger et al. 2007; Mohn et al. 2008). For example, H3K4me3 at proximal promoters generally regulates DNA methylation levels in a negative fashion (Weber et al. 2007; Meissner et al. 2008), whereas H3K36me3 within actively transcribed gene bodies is positively correlated with the presence of high levels of DNA methylation (Ball et al. 2009). To further investigate the relationship between Tet1, 5hmC, and major histone modifications at Tet1-bound genes, we cross-referenced the 5hmC profile with published genome-wide occupancy of major histone modifications. As Tet1 can bind to both actively transcribed genes and Polycomb repression complex 2 (PRC2)-repressed developmental regulators (positive for Ezh2 and H3K27me3) (Wu et al. 2011), we analyzed these two groups of Tet1-bound genes separately (Fig. 3A). This analysis revealed that 5hmC was relatively more enriched at intragenic regions (Fig. 3B,C), particularly at the 3′ end of the gene body for actively transcribed Tet1-only targets (e.g., Rest in Fig. 3C), similar to the transcription elongation mark H3K36me3 (Mikkelsen et al. 2007). In contrast, enrichment of 5hmC was more prominent at extended promoter regions—including both upstream of and downstream from TSSs (Fig. 3B,C)—of Tet1/PRC2-cobound targets (e.g., Lhx2 in Fig. 3C). Thus, 5hmC enrichment at Tet1-bound genes may contribute to maintenance of both the transcriptionally active and inactive chromatin states by functionally interacting with distinct histone modifications and their associated proteins.
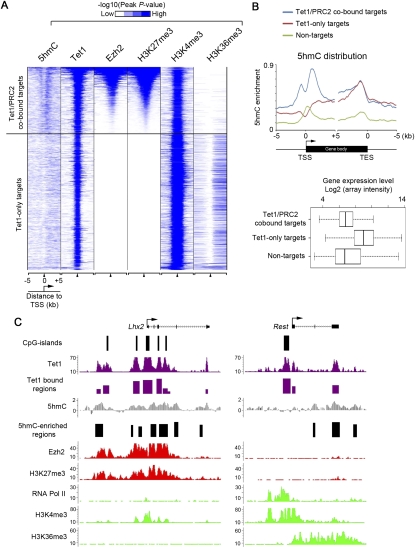
5hmC is enriched in both repressed (bivalent, Tet1/PRC2-cobound) and actively transcribed (Tet1-only) genes. (A) Heat map representation of genomic regions with enriched 5hmC, binding profile of Tet1 and Ezh2, and major histone modifications (H3K4me3, H3K27me3, and H3K36me3) (Mikkelsen et al. 2007) in mouse ES cells at all Tet1 target genes (5 kb flanking TSSs). The heat map is rank-ordered from genes with the highest H3K27me3 enrichment to no H3K27me3 within 5-kb genomic regions flanking TSSs. The enrichment of 5hmC and 5mC was determined by whole-genome tiling microarrays. The enrichment of Tet1, H3K4me3, H3K27me3, and H3K36me3 binding was determined previously by ChIP-seq analyses (Mikkelsen et al. 2007; Wu et al. 2011). All average binding was measured by −log10 (peak P-values) in 200-bp bins and is shown by color scale. The following color scales (white, no enrichment; blue, high enrichment) are used for 5hmC/5mC, Tet1/H3K27me3/H3K36me3, and H3K4me3, respectively: [0, 2], [0, 50], and [0, 100]. The presence of CpG islands is displayed in color (blue, present; white, absent). (B) Average distribution profiles of 5hmC enrichment are shown for Tet1/PRC2-cobound targets, Tet1-only targets, and nontargets. Averaged expression levels of these three groups of genes are shown in the bottom panels (measured by log2 values of expression microarray signals). (C) Shown are profiles of Tet1 (Wu et al. 2011), 5hmC, Ezh2 (Ku et al. 2008), RNA polymerase II (Seila et al. 2008), and major histone modification (Mikkelsen et al. 2007) occupancy at two representative Tet1 targets: a Tet1/PRC2-cobound target (Lhx2), and a Tet1-only target (Rest promoter). ChIP-seq data in mouse ES cells are shown in read counts, with the Y-axis floor set to 0.2 read per million reads.
Relationship between 5hmC distribution and chromatin occupancy of pluripotency-related transcription factors and other genomic features
The fact that DNA methylation can affect the binding of many DNA-binding proteins to their target sequences raises the possibility that 5hmC may also be involved in regulating the protein–DNA interactions. To investigate this potential relationship in mouse ES cells, we mapped 5hmC microarray signals to previously determined binding sites of a set of proteins important for pluripotency (e.g., Nanog, Sox2, and Oct4) (Chen et al. 2008). In contrast to a general depletion of 5mC at DNA–protein interaction sites, we observed a relative enrichment of 5hmC toward the site of most DNA-binding proteins (Supplemental Fig. S5). Previous analysis of DNA methylation in human ES cells using whole-genome bisulfite sequencing suggests that 5mC in a non-CpG context, but not CpG DNA methylation, is greatly depleted from binding sites of transcription factors related to pluripotency (Lister et al. 2009). Since bisulfite treatment cannot discriminate 5mC from 5hmC (Huang et al. 2010; Jin et al. 2010), bisulfite sequencing may overestimate the 5mC levels at these binding sites. Indeed, specific antibody-based immunoprecipitation analysis of 5mC and 5hmC in mouse ES cells indicated that 5mC was generally depleted from DNA–protein interaction sites, whereas 5hmC was relatively enriched at these sites (Supplemental Fig. S5).
We next analyzed a set of genomic features defined by histone modifications or sequence-specific DNA-binding proteins, including H3K4me3-enriched promoter regions, methylated H3K4 (H3K4me1)-enriched enhancers, Ctcf-marked insulators, and H3K36me3-enriched transcribed intragenic regions (Mikkelsen et al. 2007; Chen et al. 2008; Meissner et al. 2008). Except for H3K36me3-enriched regions, 5mC was, in general, depleted from these genomic features (Supplemental Fig. S5), consistent with the notion that DNA methylation negatively regulates most DNA–protein interactions. In contrast, average signal profiles of 5hmC showed a relative enrichment at promoters, enhancers, transcribed regions, and insulators (Supplemental Fig. S5). Notably, the general enrichment of 5hmC at H3K4me3 peaks indicates that the absence of 5hmC at a subset of CpG-rich, H3K4me3-enriched proximal promoters is probably due to the existence of additional regulatory factors of Tet1 activity or proteins capable of rapidly converting 5hmC into unmethylated cytosine at these sites. Furthermore, the observed enrichment of 5hmC and concomitant depletion of 5mC at enhancer or insulator sequences may therefore contribute to maintaining a more accessible chromatin structure for binding of enhancer proteins (e.g., p300) and Ctcf to these sites. Enrichment of both 5hmC and 5mC at actively transcribed regions marked by high levels of H3K36me3 suggests a transcriptional link between these two marks (Supplemental Fig. S5).
5hmC has different distribution profiles at active and repressed genes
To investigate a potential role of 5hmC in transcriptional regulation, we examined the relationship between 5hmC distribution and the global gene expression profile. This analysis showed that 5hmC was relatively enriched within intragenic regions of genes transcribed at high and medium levels (Fig. 4A, blue and red; Supplemental Fig. S6, blue and red), as well as promoter regions of transcriptionally inactive genes (Fig. 4A, green; Supplemental Fig. S6, green). Further analysis of 5hmC distribution on Tet1-bound genes ranked by their expression levels also supported a potential role of 5hmC in both transcriptional activation and repression (Fig. 4B). Interestingly, 5hmC was enriched at promoters of both Tet1/PRC2-cobound (Supplemental Fig. S7, blue) and Tet1-only targets that were expressed at low levels (Supplemental Fig. S7, purple) in mouse ES cells, suggesting that promoter 5hmC may function as a general repressive mark (Supplemental Fig. S7). To investigate further how 5hmC distribution may contribute to Tet1-dependent gene expression, we compared the 5hmC profiles between control and Tet1-depleted ES cells on Tet1-repressed and Tet1-activated targets. We found that 5hmC levels were decreased at both groups of Tet1 targets (Supplemental Fig. S8). A decrease in 5hmC was more pronounced at promoter regions and the 5′ end of intragenic regions on Tet1-repressed targets, whereas a depletion of intragenic 5hmC was evident for Tet1-activated targets. Taken together, these results indicate that, similar to 5mC, 5hmC may play a complex role in transcriptional regulation, depending on its location (Wu et al. 2010). Our analysis of 5hmC distribution in mouse ES cells suggests that promoter and gene body 5hmC may preferentially contribute to gene repression and activation, respectively.
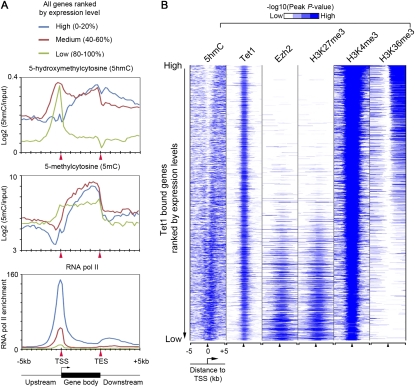
Relationship between 5hmC enrichment and gene expression in mouse ES cells. (A) Distribution of 5hmC, 5mC, and RNA polymerase II at genes expressed at different levels in ES cells. Enrichment of 5hmC and 5mC was measured by raw log2 ratios of immunoprecipitation/input (IP/input) and MEDME-corrected values of log2 ratios, respectively. (B) Heat map representation of genomic regions with enriched 5hmC, binding profile of Tet1 and Ezh2, and major histone modifications in mouse ES cells at all Tet1 target genes (5 kb flanking TSSs). The heat map is rank-ordered by gene expression levels of Tet1-bound genes. All average binding was measured by −log10 (peak P-values) in 200-bp bins and is shown by color scale. The following color scales (white, no enrichemnt; blue, high enrichment) are used for 5hmC, Tet1/Ezh2/H3K27me3/H3K36me3, and H3K4me3, respectively: [0, 2], [0, 50], and [0, 100].
In summary, our studies have presented a genome-wide map of 5hmC in mouse ES cells. Systematic comparison of 5hmC distribution, Tet1 occupancy, and major histone modifications indicate that 5hmC may be involved in establishing and maintaining chromatin structure for both actively transcribed genes and PcG-repressed developmental regulators. We also provide initial evidence indicating that 5hmC may contribute to both transcriptional activation and repression in a context-dependent manner. Collectively, these results and the demonstration of a simple antibody-based approach in genome-wide 5hmC mapping have set the stage for further understanding the functions of Tet family proteins and 5hmC in development and disease.
Materials and methods
Genome-wide and locus-specific 5hmC analysis (hMeDIP)
To immunoprecipitate 5hmC, genomic DNA was sequentially digested with proteinase K and RNase A and purified by phenol-chloroform extraction. Purified genomic DNA was sonicated to 200–1000 base pairs (bp) and heat-denatured (10 min, 95°C). An aliquot of sonicated genomic DNA was saved as input. Five micrograms of fragmented genomic DNA was immunoprecipitated with 5 μg of rat 5hmC Ab (Diagenode, catalog no. MAb-633HMC) or rabbit 5hmC Ab (Active Motif, catalog no. 39791) overnight at 4°C in a final volume of 500 μL of immunoprecipitation buffer (10 mM sodium phosphate at pH 7.0, 140 mM NaCl, 0.05% Triton X-100). The DNA–antibody mixture was incubated with 30 μL of protein G Dynabeads (Invitrogen) for 2 h at 4°C and washed three times with 1 mL of immunoprecipitation buffer. The beads were then treated with proteinase K for at least 3 h at 55°C, and the methylated DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. For whole-genome DNA tiling microarray analysis, immunoprecipitated 5hmC-containing DNA from control or Tet1-depleted ES cells was cohybridized with input DNA to mouse whole-genome tiling microarrays (NimbleGen). Locus-specific hMeDIP–qPCR was performed similarly using nondenatured genomic DNA, and immunoprecipitated DNA was analyzed on an ABI 7300 system (Applied Biosystems) using SYBR Green (Invitrogen). Primer sequences are listed in Supplemental Table S2.
To evaluate the immunoprecipitation efficiency of 5hmC antibodies (Active Motif) for synthetic DNA (949 bp; Zimo Research), 25 pg of unmethylated, methylated, or hydroxymethylated DNA was diluted in 480 μL of 1× TE buffer. DNA was heat-denatured for 10 min at 95°C, and chilled for 5 min on ice. 5hmC immunoprecipitation was performed as described above. qPCR was carried out with primers H/me-1-F (AGGTGGAGGAAGGTGATGTC) and H/me-1-R (ATAAACCGAACCGCTACACC).
Whole-genome tiling microarray analysis
For whole-genome DNA tiling microarray analysis of 5hmC distribution, immunoprecipitated and input DNA was prepared from both control and Tet1-depleted ES cells and amplified using a whole-genome amplification kit (Sigma). Probe labeling, amplification, hydridization, data extraction, and analysis were performed as described previously (Wu et al. 2011).
For identification of probes associated with significant levels of 5hmC, a nonparametric one-sided Kolmogorov-Smirno (KS) test was used. Briefly, from the scaled log2 ratio data, a fixed-length window (750 bp) was placed around each consecutive probe, and the one-sided KS test was applied to determine whether the probes were drawn from a significantly more positive distribution of intensity log ratios than those in the rest of the array. The resulting score for each probe was the [−log10 P-value] from the windowed KS test around that probe. Peak data files were generated from the P-value data files using NimbleScan version 2.5. Peaks within 500 bp of each other were merged. For calculating the absolute 5mC levels in control knockdown and Tet1 knoockdown ES cells, the MEDME program (Pelizzola et al. 2008) was used to correct the nonlinear relationship between microarray signals and genomic CpG density. To visualize 5mC and 5hmC distributions in the Cisgenome browser (Ji et al. 2008), probe-level smoothing (log2 ratios of probes within 1 kb are averaged) was performed for each probe. To calculate the peak distribution, averaged 5hmC or 5mC enrichment (measured by [−log10 peak P-value]) was binned to 200-bp intervals within genomic regions 5 kb upstream of and downstream from TSSs or transcriptional end sites (TESs) of annotated RefSeq genes. Heat maps were generated and visualized using Cluster3 and Java Treeview, respectively. 5mC and 5hmC whole-genome tiling microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE26833 (Wu et al. 2011) and GSE27613, respectively.
Constructs and antibodies
All of the constructs and antibodies used in this study were described previously (Ito et al. 2010; Wu et al. 2011).
ChIP-seq, gene expression profiling, and data analysis
ChIP-seq experiments and data analysis for Tet1 were described previously (Wu et al. 2011). ChIP-seq data sets of H3K4me3, H3K27me3, H3K36me3 (Mikkelsen et al. 2007), Ezh2 (Ku et al. 2008), and RNA polymerase II (Seila et al. 2008) were obtained from previous publications and reanalyzed in MACS using identical parameters (except statistical cutoff was set to P-value < 10−5). ChIP-seq sequencing read counts for each ChIP-seq experiments were binned into 400-bp windows at 100-bp steps along the genome and visualized in the Cisgenome browser (Ji et al. 2008). To assign ChIP-seq enriched regions to genes, RefSeq genes were downloaded from the UCSC Table Browser (May 2010). For all data sets, genes with enriched regions within 5 kb of their TSSs were called bound. Gene expression profiling analysis of control and Tet1-depleted mouse ES cells was carried out using the Affymetrix GeneChip Mouse Genome 430 2.0 array. Tet1 ChIP-seq and gene expression microarray data have been deposited in the Gene Expression Omnibus under accession number GSE26833 (Wu et al. 2011).
Mouse ES cell culture, Tet knockdown, and qPCR
Mouse E14Tg2A ES cells were cultured in feeder-free conditions (Ito et al. 2010). Control and Tet1 knockdown cell preparation and qPCR verification were described previously (Wu et al. 2011).
Acknowledgments
We thank Brian Abraham and Iouri Chepelev for help with data transfer, Jinzhao Wang for FACS sorting, and Susan Wu for critical reading of the manuscript. This work was supported by NIH grant GM68804 (to Y.Z.), and support to the Division of Intramural Research Program of National Heart, Lung, and Blood Institute from the NIH (to K.Z). S.I. is a research fellow of the Japan Society for the Promotion of Science. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2036011.
Supplemental material is available for this article.
References
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [Europe PMC free article] [Abstract] [Google Scholar]
- Cedar H, Bergman Y 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304 [Abstract] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [Abstract] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [Europe PMC free article] [Abstract] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T 2010. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5: e15367 10.1371/journal.pone.0015367 [Europe PMC free article] [Abstract] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A 2010. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5: e8888 10.1317/journal.pone.0008888 [Europe PMC free article] [Abstract] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [Europe PMC free article] [Abstract] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH 2008. An integrated software system for analyzing ChIP–chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [Europe PMC free article] [Abstract] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125 10.1093/nar/gkq223 [Europe PMC free article] [Abstract] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [Europe PMC free article] [Abstract] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 10.1371/journal.pgen.1000242 [Europe PMC free article] [Abstract] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [Europe PMC free article] [Abstract] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [Europe PMC free article] [Abstract] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [Europe PMC free article] [Abstract] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [Abstract] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [Europe PMC free article] [Abstract] [Google Scholar]
- Ooi SK, O'Donnell AH, Bestor TH 2009. Mammalian cytosine methylation at a glance. J Cell Sci 122: 2787–2791 [Europe PMC free article] [Abstract] [Google Scholar]
- Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM 2008. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res 18: 1652–1659 [Europe PMC free article] [Abstract] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [Abstract] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [Europe PMC free article] [Abstract] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. 2011. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 29: 68–72 [Europe PMC free article] [Abstract] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H 2010. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 38: e181 10.1093/nar/gkq684 [Europe PMC free article] [Abstract] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [Europe PMC free article] [Abstract] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [Abstract] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [Europe PMC free article] [Abstract] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun Y, Zhang Y 2011. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 10.1038/nature09934 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.2036011
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/25/7/679.full.pdf
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/reprint/25/7/679
Free to read at www.genesdev.org
http://www.genesdev.org/cgi/content/abstract/25/7/679
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/content/full/25/7/679
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101878313
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gad.2036011
Article citations
5-Hydroxymethylcytosine: Far Beyond the Intermediate of DNA Demethylation.
Int J Mol Sci, 25(21):11780, 02 Nov 2024
Cited by: 0 articles | PMID: 39519332 | PMCID: PMC11546248
Review Free full text in Europe PMC
Cell-Free DNA Hydroxymethylation in Cancer: Current and Emerging Detection Methods and Clinical Applications.
Genes (Basel), 15(9):1160, 03 Sep 2024
Cited by: 0 articles | PMID: 39336751 | PMCID: PMC11430939
Review Free full text in Europe PMC
Non-canonical bases differentially represented in the sex chromosomes of the dioecious plant Silene latifolia.
J Exp Bot, 75(13):3849-3861, 01 Jul 2024
Cited by: 1 article | PMID: 38652039 | PMCID: PMC11233409
Epigenetic landscape of 5-hydroxymethylcytosine and associations with gene expression in placenta.
Epigenetics, 19(1):2326869, 20 Mar 2024
Cited by: 0 articles | PMID: 38507502 | PMCID: PMC10956631
One-pot trimodal mapping of unmethylated, hydroxymethylated, and open chromatin sites unveils distinctive 5hmC roles at dynamic chromatin loci.
Cell Chem Biol, 31(3):607-621.e9, 27 Dec 2023
Cited by: 0 articles | PMID: 38154461 | PMCID: PMC10962225
Go to all (383) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE26833
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells.
Cell Cycle, 10(15):2428-2436, 01 Aug 2011
Cited by: 91 articles | PMID: 21750410 | PMCID: PMC3180185
Review Free full text in Europe PMC
Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells.
Mol Cell, 42(4):451-464, 21 Apr 2011
Cited by: 408 articles | PMID: 21514197 | PMCID: PMC3099128
Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells.
Nature, 473(7347):389-393, 30 Mar 2011
Cited by: 446 articles | PMID: 21451524 | PMCID: PMC3539771
Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer.
Adv Exp Med Biol, 754:57-79, 01 Jan 2013
Cited by: 39 articles | PMID: 22956496
Review
Funding
Funders who supported this work.
Intramural NIH HHS
NIGMS NIH HHS (2)
Grant ID: GM68804
Grant ID: R01 GM068804




