Abstract
Free full text

An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition
Abstract
 Epithelial-mesenchymal transition (EMT) is a form of cellular plasticity that is critical for embryonic development and tumor metastasis. A double-negative feedback loop involving the miR-200 family and ZEB (zinc finger E-box-binding homeobox) transcription factors has been postulated to control the balance between epithelial and mesenchymal states. Here we demonstrate using the epithelial Madin Darby canine kidney cell line model that, although manipulation of the ZEB/miR-200 balance is able to repeatedly switch cells between epithelial and mesenchymal states, the induction and maintenance of a stable mesenchymal phenotype requires the establishment of autocrine transforming growth factor-β (TGF-β) signaling to drive sustained ZEB expression. Furthermore, we show that prolonged autocrine TGF-β signaling induced reversible DNA methylation of the miR-200 loci with corresponding changes in miR-200 levels. Collectively, these findings demonstrate the existence of an autocrine TGF-β/ZEB/miR-200 signaling network that regulates plasticity between epithelial and mesenchymal states. We find a strong correlation between ZEBs and TGF-β and negative correlations between miR-200 and TGF-β and between miR-200 and ZEBs, in invasive ductal carcinomas, consistent with an autocrine TGF-β/ZEB/miR-200 signaling network being active in breast cancers.
Epithelial-mesenchymal transition (EMT) is a form of cellular plasticity that is critical for embryonic development and tumor metastasis. A double-negative feedback loop involving the miR-200 family and ZEB (zinc finger E-box-binding homeobox) transcription factors has been postulated to control the balance between epithelial and mesenchymal states. Here we demonstrate using the epithelial Madin Darby canine kidney cell line model that, although manipulation of the ZEB/miR-200 balance is able to repeatedly switch cells between epithelial and mesenchymal states, the induction and maintenance of a stable mesenchymal phenotype requires the establishment of autocrine transforming growth factor-β (TGF-β) signaling to drive sustained ZEB expression. Furthermore, we show that prolonged autocrine TGF-β signaling induced reversible DNA methylation of the miR-200 loci with corresponding changes in miR-200 levels. Collectively, these findings demonstrate the existence of an autocrine TGF-β/ZEB/miR-200 signaling network that regulates plasticity between epithelial and mesenchymal states. We find a strong correlation between ZEBs and TGF-β and negative correlations between miR-200 and TGF-β and between miR-200 and ZEBs, in invasive ductal carcinomas, consistent with an autocrine TGF-β/ZEB/miR-200 signaling network being active in breast cancers.
INTRODUCTION
In contrast to the traditional view of cellular differentiation as being a unidirectional and largely irreversible process, it is now recognized that many differentiated cells can retain a considerable degree of plasticity. This is exemplified by recent remarkable demonstrations of pluripotent stem cell (iPSC) generation from ostensibly fully differentiated cells through manipulation of key regulatory genes. It is also becoming apparent that formation of iPSCs from differentiated precursors has much in common with mesenchymal-epithelial transition (MET) (Li et al., 2010; Samavarchi-Tehrani et al., 2010). MET and its reverse, epithelial-mesenchymal transition (EMT), are prime examples of reversible differentiation processes that occur during normal development. Indeed, some tissues in the developing embryo are produced as the result of up to 3 successive cycles of EMT and MET (Thiery et al., 2009). Such developmental plasticity requires that cells be capable of maintaining a stable differentiated phenotype yet retain the capacity to switch to an alternative phenotype. EMT has also been extensively demonstrated to enhance the invasive properties of epithelial tumor cells and promote tumor metastasis (Thiery, 2002; Berx et al., 2007). Recent studies have shown that this process is also associated with the acquisition of tumor initiating and self-renewal properties (Mani et al., 2008) with interconversions between epithelial and mesenchymal states being postulated to influence tumor malignancy (Gupta et al., 2009; Polyak and Weinberg, 2009). Therefore, deciphering the pathways controlling epithelial cell plasticity has broad implications for our understanding of developmental processes and may represent important therapeutic targets for cellular reprogramming and tumorigenesis.
Among several growth factors that can act as inducers of EMT, transforming growth factor-β (TGF-β) has been found to play an important role in particular stages of development and in disease processes, such as fibrosis and cancer metastasis (Zeisberg and Kalluri, 2004; Derynck and Akhurst, 2007; Pardali and Moustakas, 2007). TGF-β induces the transcription factors Snail, Slug, zinc finger E-box-binding homeobox (ZEB)1, and ZEB2 (also known as SIP1), which are each implicated in mediating the effects of TGF-β at least in part through repression of E-cadherin and initiation of EMT (Peinado et al., 2007). The Madin Darby canine kidney (MDCK) cell line has been used extensively as an in vitro model of EMT. In culture, these cells have all the hallmarks of epithelial cells, but convert to mesenchymal cells in response to TGF-β. We previously used these cells to identify microRNAs that are involved in specifying epithelial phenotype and to identify a double-negative regulatory feedback loop involving microRNAs of the miR-200 family and the transcription repressors ZEB1 and ZEB2 (collectively ZEB) (Bracken et al., 2008; Gregory et al., 2008a). In this feedback loop, the miR-200 family repress ZEBs, and ZEBs, which are expressed in mesenchymal cells, repress the transcription of the two gene loci (miR-200b~200a~429 and miR-200c~141) encoding the five members of the miR-200 family (Bracken et al., 2008; Burk et al., 2008). Both miR-200 and ZEBs have independently been found to participate in the control of EMT in many different cell types and can mediate the effects of TGF-β signaling (Comijn et al., 2001; Postigo et al., 2003; Eger et al., 2005; Hurteau et al., 2007; Shirakihara et al., 2007; Burk et al., 2008; Korpal et al., 2008; Park et al., 2008).
The ZEB/miR-200 double-negative feedback loop has been postulated to explain both the stability and interchangeability of the epithelial versus mesenchymal phenotypes (Gregory et al., 2008b; Brabletz and Brabletz, 2010), but to date this has not been tested within a single cell system. In this proposed model, the miR-200 family predominates in epithelial cells and prevents expression of ZEB1 and ZEB2, allowing E-cadherin and other epithelial genes to be fully expressed, whereas in mesenchymal cells the ZEB factors prevent expression of miR-200, E-cadherin, and other epithelial genes. The ZEB/miR-200 feedback loop predicts that a perturbing influence such as TGF-β would need to change the balance between miR-200 and ZEB factors beyond a threshold, which would then be self-reinforcing and reset the state of the cells in the alternative phenotype. This new cell state would be metastable but able to be reversed if the balance of miR-200 and ZEB was altered by upstream changes in cell signaling.
In a number of EMT model systems it has been shown that autocrine TGF-β signaling contributes to the stability of the mesenchymal state in conjunction with oncogenic Ras, Raf, or Fos overexpression (Oft et al., 1996; Lehmann et al., 2000; Janda et al., 2002; Eger et al., 2004). Recently, one of the TGF-β family members, TGF-β2, was shown to be targeted by miR-141/200a in cancer cells (Burk et al., 2008), leading to the hypothesis that repression of miR-200a during EMT might facilitate induction of autocrine TGF-β signaling (Burk et al., 2008; Gregory et al., 2008b; Ocana and Nieto, 2008). The importance of these interactions in the establishment and maintenance of EMT, however, has not yet been demonstrated. Using the MDCK cell model, we show that the ZEB/miR-200 double-negative feedback loop plays a central role in controlling cell plasticity and specifying cell phenotype. By manipulating the ZEB/miR-200 ratio, the cell phenotype can be repeatedly switched between stable epithelial and mesenchymal states without the requirement for the continued influence of exogenous factors, verifying the hypothesized function of the double-negative feedback model. Furthermore, we demonstrate an essential requirement for autocrine TGF-β signaling in both the establishment and maintenance of EMT through up-regulation of ZEB1 and ZEB2. Collectively, these findings demonstrate that epithelial cell plasticity is controlled by an autocrine TGF-β/ZEB/miR-200 signaling network. Prolonged activation of this signaling network was shown to affect dynamic and reversible DNA methylation of the miR-200 family loci which may contribute to stability of the mesenchymal state.
RESULTS
Prolonged TGF-β treatment establishes a mesenchymal state that is stabilized by the ZEB/miR-200 regulatory loop
On the basis of the double-negative feedback loop model, we predicted that a critical threshold in the balance between miR-200 and ZEB levels determines whether cells stably reside in a self-reinforcing epithelial or mesenchymal state. As a consequence, cells that are induced to undergo a change in state (e.g., induced by TGF-β to undergo EMT) should remain in that state indefinitely, without the need for ongoing stimulation with the inducer. To test this proposition we induced EMT in MDCK cells with TGF-β1 and then removed it at varying time points while monitoring cell morphology, miR-200, and ZEB1 and ZEB2 expression. After 2 d of TGF-β1 treatment, the epithelial cells began to adopt a mesenchymal morphology, but at this stage only the miR-200b~200a~429 cluster (represented by miR-200b) was repressed, and ZEB1 and ZEB2 proteins were not yet detectable (Figure 1, A–C). After 5 d of treatment, both miR-200 clusters were repressed (miR-200c~141 by ~50%; represented by miR-200c), and this result was coincident with greatly elevated levels of ZEB1 mRNA and protein. When TGF-β1 was removed at this time point, the cells reverted back to an epithelial morphology and expression profile (Figure 1, A–C), suggesting that the changes in miR-200 and ZEB levels had not reached a critical threshold to maintain the mesenchymal state. By 8 d of TGF-β1 treatment, the microRNAs from both miR-200 clusters were strongly repressed, coincident with a large up-regulation of both ZEB1 and ZEB2 proteins. Of note, the change in ZEB1 and ZEB2 mRNA between days 5 and 8 was relatively small in comparison to protein level changes, suggesting that the protein elevation was caused by a loss of miR-200–mediated translational repression. Removal of TGF-β1 after 8 d of treatment resulted in the cells maintaining a mesenchymal morphology and expression profile which was stable for several months in culture (hereafter called MDCK-TGF cells) (Figure 1, A–C). In these stable mesenchymal cells, ZEB2 expression continued to increase whereas ZEB1 expression decreased from its day 8 levels, suggesting that ZEB2 function may be more important in this context. These results are consistent with the prediction that a crucial threshold in the ZEB/miR-200 balance sets the cell phenotype.
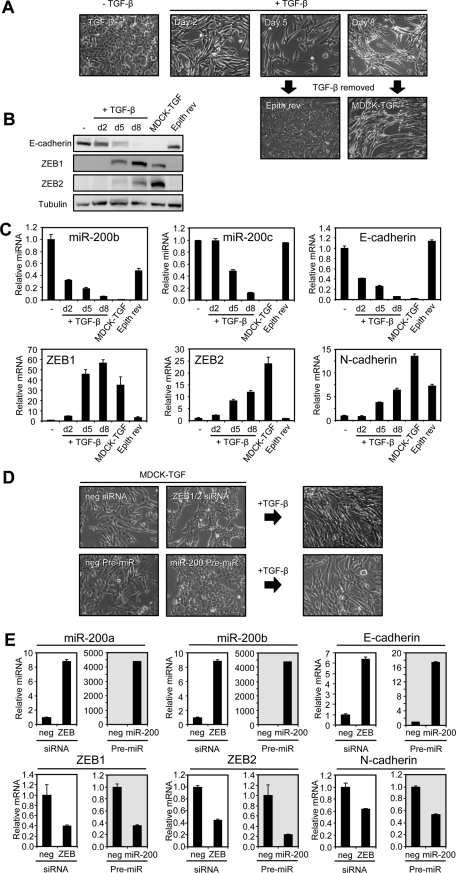
Prolonged TGF-β treatment establishes a mesenchymal state that is stabilized by the ZEB/miR-200 regulatory loop. (A) Cell morphology of MDCK cells treated with TGF-β1 over an 8-d time course as indicated, followed by its removal. MDCK-TGF designates MDCK cells that remain mesenchymal 35 d after cessation of the TGF-β1 treatment. “Epith rev” designates MDCK cells which had been treated with TGF-β1 for 5 d followed by TGF-β1 withdrawal for 12 d, resulting in reversion back to an epithelial phenotype. (B) Western blot and (C) real-time PCR of EMT markers and miR-200 family members over the TGF-β1 time course. (D) Cell morphology of MDCK-TGF transfected with ZEB1 and ZEB2 siRNAs or miR-200a and miR-200b pre-miRs (representing both “seed” sequence groupings, miR-200b/200c/429 and miR-141/200a, within the miR-200 family) or their negative controls over a 6-d period followed by treatment with TGF-β1 for 6 d. (E) Real-time PCR of EMT markers after miR-200 transfection or ZEB knockdown as shown in (D). Data are representative of triplicate experiments. Each value shown is the mean ± SD of three replicate measurements.
To determine whether the stable mesenchymal state of MDCK-TGF cells retained plasticity, we directly manipulated the ZEB/miR-200 balance by transfection of ZEB1 and ZEB2 siRNAs or miR-200a and miR-200b pre-miRs into these cells. Increasing the miR-200 levels or reducing ZEB levels restored these mesenchymal cells back to an epithelial morphology and expression profile (Figure 1, D and E). Furthermore, when we added TGF-β1 back to the epithelially reverted cells they were able to undergo yet another EMT (Figure 1D). These results show that the epithelial and mesenchymal phenotypes can be consecutively switched in either direction in response to manipulation of the ZEB/miR-200 balance.
Autocrine TGF-β signaling is required for the maintenance of the mesenchymal state of MDCK-TGF cells
In the stable mesenchymal state, decreased miR-200 levels would allow for uninhibited production of ZEB proteins, but we reasoned that sustained ZEB transcription would also be needed to maintain high levels of ZEB mRNAs. Because it has been well documented in studies with MDCK and other EMT cell culture models that TGF-β can cooperate with other signaling pathways initiated by factors including Ras, Raf, and Fos to establish autocrine TGF-β signaling (Oft et al., 1996; Lehmann et al., 2000; Janda et al., 2002; Eger et al., 2004; Medici et al., 2008), we considered that prolonged TGF-β treatment might initiate autocrine TGF-β signaling to promote the ZEB transcription that enforces the stable mesenchymal state of the MDCK-TGF cells. To test this possibility, we first measured the expression of endogenous TGF-β1, TGF-β2, and TGF-β3 mRNAs and found that they were progressively induced by TGF-β1 treatment and that TGF-β1 and TGF-β2 proteins were secreted by MDCK-TGF cells (an assay for TGF-β3 protein was not available) (Figure 2, A and B). To determine whether response to this endogenously synthesized TGF-β is important for mesenchymal stability, we treated MDCK-TGF cells with an inhibitor of TGF-β receptor 1 (RI) activity, SB-505124. Addition of this inhibitor led to a time-dependent decrease in ZEB mRNA (Figure 2C), consistent with autocrine TGF-β produced by MDCK-TGF cells being required for ZEB transcription in these cells. Concomitant with the loss of ZEB was an increase in miR-200 expression (see Figure 6C later in the paper and Supplemental Figure 3C), accompanied by hallmark epithelial features, such as expression of E-cadherin and ZO-1 on the plasma membrane, and a rearrangement of F-actin in a cortical pattern (Figure 2D). Similar effects were also observed with a different TGF-β RI inhibitor, SB-431542, confirming that the epithelial reversion was caused by TGF-β pathway inhibition (Supplemental Figure 1A).
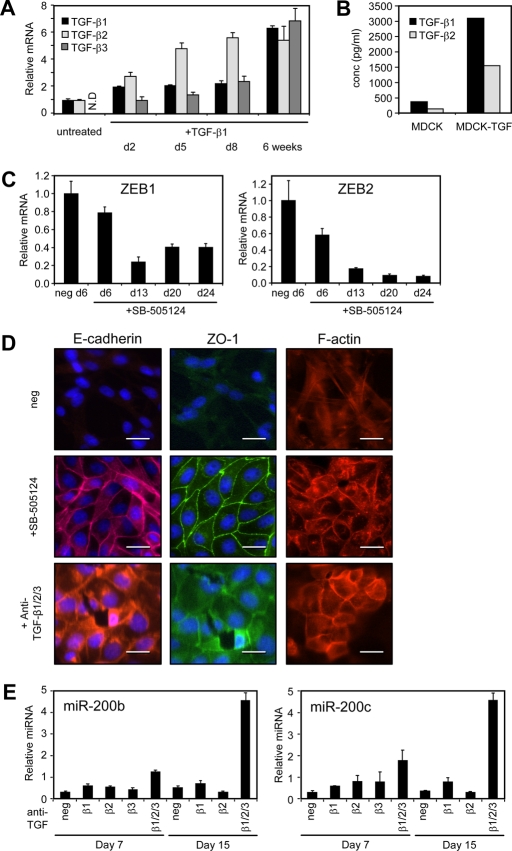
Autocrine TGF-β signaling is required for the maintenance of a mesenchymal state. (A) Real-time PCR measurements of TGF-β family members after exogenous TGF-β1 treatment of MDCK cells. TGF-β3 levels were not detected (N.D.) in untreated MDCK cells. Data are normalized to untreated levels for TGF-β1 and TGF-β2 or Day 2 levels for TGF-β3. (B) TGF-β1 and TGF-β2 secreted by MDCK and MDCK-TGF cells into the culture medium was measured by ELISA. A TGF-β3 ELISA was not available for measurement of secreted TGF-β3. (C) Real-time PCR measurement of ZEBs in MDCK-TGF cells after inhibition of TGF-β signaling using the TGF-βRI inhibitor (1 μM SB-505124; 13 d) (D) Immunofluorescence staining of E-cadherin (red) and ZO-1 (green), and rhodamine phalloidin staining of F-actin in MDCK-TGF cells either untreated or treated with a TGF-βRI inhibitor [as in (C)] or a TGF-β1/2/3 neutralizing antibody (100 μg/ml; 15 d). Cells are counterstained with DAPI to visualize nuclei. Scale bars = 50 μm. (E) Quantitation of miR-200 levels in MDCK-TGF cells treated with individual TGF-β1, -β2, -β3 and -β1/2/3 neutralizing antibodies (100 μg/ml) after 7 or 15 d of treatment. Values are the mean ± SD of three replicate measurements.
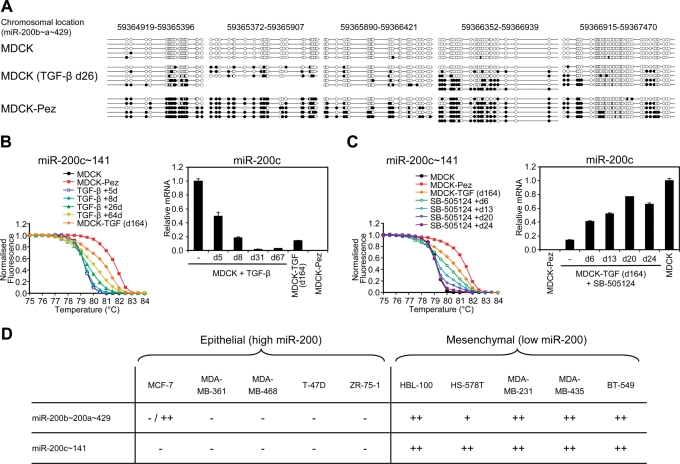
Prolonged autocrine TGF-β signaling promotes reversible DNA methylation of the miR-200 family promoters. (A) Mapping of methylated CpGs across a ~2.5 kb region spanning the miR-200b~200a~429 cluster transcription start site. Several clones were sequenced from bisulfite-modified genomic DNA derived from MDCK, MDCK cells treated with TGF-β1 for 26 d, and MDCK cells stably expressing the tyrosine phosphatase Pez (possessing autocrine TGF-β signaling). Black and white circles indicate methylated and unmethylated CpGs, respectively. (B) PCR melt curve analysis of the CpG-rich region encompassing the miR-200c~141 transcription start site (see Supplemental Figure 3A). MDCK cells were treated with TGF-β1 for the indicated time periods followed by melt curve analysis with MDCK and MDCK-Pez samples representing unmethylated and methylated baseline profiles. MDCK-TGF (d164) are cells treated with TGF-β1 for 8 d followed by its removal for 156 d. (C) TGF-β signaling was inhibited in methylated stable mesenchymal cells (MDCK-TGF d164) using the 1 μM SB-505124 over a 24-d time course and methylation of the miR-200c~141 promoter assessed by melt curve analysis. For (C) and (D), miR-200c levels were measured in these samples, or samples at similar time points, by real-time PCR. Data are representative of duplicate experiments. Each value shown is the mean ± SD of three replicate measurements. (D) Methylation of the miR-200b~200a~429 and miR-200c~141 proximal promoters in epithelial (high miR-200) or mesenchymal (low miR-200) human breast cancer cell lines (characterized for miR-200 expression in Gregory et al., 2008a). The degree of methylation was assessed by melt curve analysis and scored as follows: A melt curve that approached the unmethylated control was scored –, a melt curve that approached the methylated control was scored as ++, and a plot that was between the unmethylated and methylated controls was scored as +. MCF-7 cells were shown to have both unmethylated and methylated alleles by cloning of PCR products, which could reflect allelic specific methylation.
To confirm whether secreted TGF-β mediates autocrine TGF-β signaling in MDCK-TGF cells, we added anti-TGF-β antibodies to the culture medium. Addition of a pan TGF-β1/2/3 antibody to the culture medium caused a time-dependent increase in miR-200 levels and drove the cells toward an epithelial phenotype (Figure 2, D and E). These changes were not observed with the individual TGF-β1, -β2, or -β3 neutralizing antibodies, suggesting that there is redundancy in the function of these ligands in this cell system (Figure 2E). The redundant function of these ligands is further supported by the ability of TGF-β2 and TGF-β3 to each induce EMT in MDCK cells (Supplemental Figure 1B). Collectively, these data demonstrate that autocrine TGF-β signaling, involving induction and secretion of TGF-β1, -β2, and -β3, is required for stabilization of the mesenchymal phenotype of MDCK-TGF cells and that this is not dependent on the presence of other exogenous factors.
Autocrine TGF-β signaling maintains the stable mesenchymal state through up-regulation of ZEB1 and ZEB2
The findings reported in the preceding section suggest that autocrine TGF-β signaling maintains the stable mesenchymal state of MDCK-TGF cells through up-regulation of ZEB1 and ZEB2. To further test this possibility, we assessed whether ZEB expression can obviate the requirement for autocrine TGF-β signaling in maintaining the mesenchymal state by inhibiting TGF-β signaling in cells where ZEB1 or ZEB2 expression has been stably enforced (Bracken et al., 2008). At the same time, we tested whether the EMT-inducing transcription factor Snail could perform a similar function to ZEB by generating MDCK cell lines with constitutive Snail expression. MDCK-TGF cells were used as a control for this experiment. Individual clones from the MDCK-ZEB1, ZEB2, and Snail cell lines displayed a mesenchymal phenotype as expected, accompanied by an increase in TGF-β1, -β2, and -β3 levels relative to empty vector (EV) clones (Figure 3A). Addition of the TGF-β RI inhibitor, SB-505124, caused MDCK-TGF and MDCK-Snail cells to revert to an epithelial phenotype over time with increased levels of miR-200 expression (Figure 3, B and C). In contrast, the MDCK-ZEB1 and MDCK-ZEB2 cells were resistant to the effects of TGF-β inhibition, with miR-200 levels remaining repressed and the cells maintaining a mesenchymal morphology after 25 d of treatment (Figure 3, B and C). Removal of the TGF-β inhibitor after this time point allowed MDCK-Snail cells to transition back to a mesenchymal morphology with decreased miR-200 levels after 13 d, whereas the MDCK-TGF cells remained stably epithelial and maintained miR-200 expression for several months in culture (44-d time point shown; Figure 3, B and C). These data demonstrate that enforced ZEB1 or ZEB2, but not Snail, expression is sufficient to prevent the mesenchymal cells from increasing miR-200 expression and undergoing MET in response to TGF-β pathway inhibition. Although enforced Snail expression cannot counteract the effects of TGF-β pathway inhibition, it is able to drive cells back into EMT when this inhibition is removed, suggesting that Snail expression is able to influence the ZEB/miR-200 balance. Collectively, these data support the notion that autocrine TGF-β signaling acts to maintain the mesenchymal state through up-regulation of ZEB1 and ZEB2 and repression of miR-200.
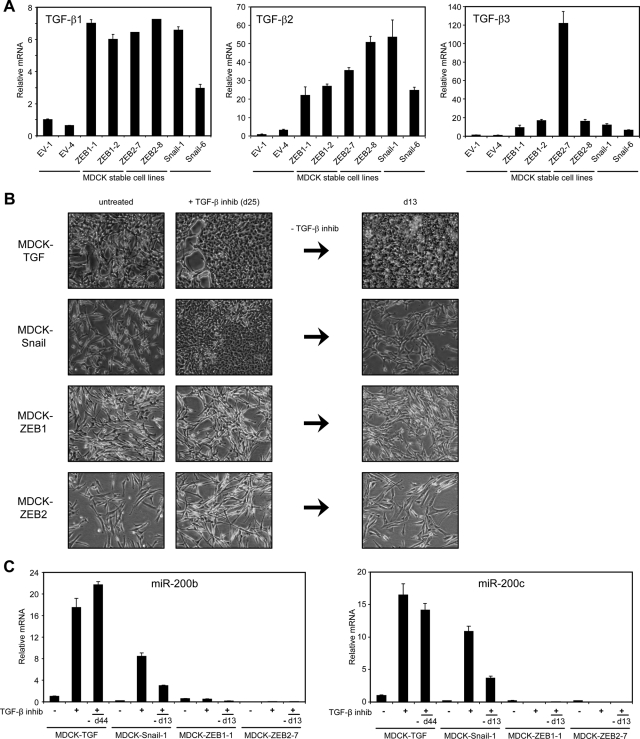
Ectopic expression of ZEB1 or ZEB2 abrogates MET following inhibition of autocrine TGF-β signaling. (A) Real-time PCR measurements of TGF-β family members in two individual EV, MDCK-ZEB1, MDCK-ZEB2, and MDCK-Snail clones. (B) Phase contrast micrographs of MDCK-stable cells before and after treatment with 1 μM SB-505124 (25 d) and 13 d after removal of the inhibitor. (C) Real-time PCR measurements of miR-200b and miR-200c in MDCK-stable cells treated as in (B). The measurement for the MDCK-TGF cells was taken at day 44 after removal of the TGF-βRI inhibitor showing the stability of the reverted epithelial state.
Manipulation of miR-200 and ZEB levels influences TGF-β production
It has recently been shown that TGF-β2 is directly targeted by miR-141/200a in breast and colon cancer cell lines (Burk et al., 2008), suggesting that the loss of miR-200 family members during EMT might enhance autocrine TGF-β signaling through removal of this repression of TGF-β2. TGF-β1 and TGF-β3 are not predicted to be direct targets of miR-200 but may be influenced indirectly by the ZEB/miR-200 loop. We therefore investigated the extent to which the ZEB/miR-200 feedback loop impacts TGF-β production in epithelial and mesenchymal cells by directly manipulating their levels. First, we measured TGF-β mRNA levels in MDCK-TGF cells following ectopic expression of miR-200a and miR-200b or knockdown of ZEB1 and ZEB2 as shown in Figure 1. Both of these treatments decreased each of the TGF-β mRNAs, with the strongest effect being on TGF-β3 (Figure 4A). Next, we inhibited endogenous miR-200 expression in MDCK cells using a locked nucleic acid (LNA) anti-miR designed to bind all members of the miR-200 family. Knockdown of miR-200 family caused an increase in each of the TGF-β mRNAs, with the strongest effect being on TGF-β3 (Figure 4B). These changes occur concomitantly with increases in ZEB mRNA levels but before alterations in cell morphology and E-cadherin expression (Supplemental Figure 2), suggesting that autocrine TGF-β induction by miR-200 repression precedes acquisition of a mesenchymal phenotype. Taken together, these data indicate that manipulation of miR-200 and ZEB levels influences the expression of all three TGF-β isoforms, most likely by direct and indirect mechanisms given the lack of putative miR-200 target sites in TGF-β1 and TGF-β3.
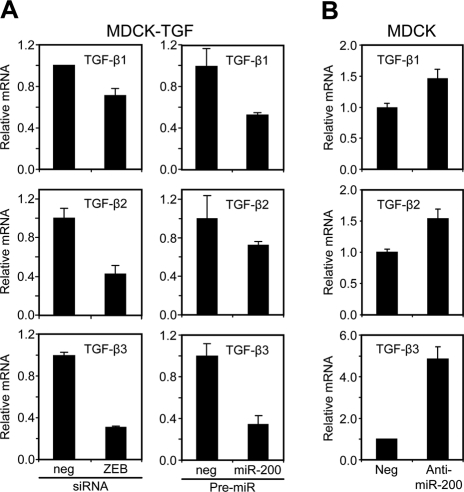
The ZEB/miR-200 regulatory loop regulates TGF-β production. (A) Real-time PCR measurement of TGF-β isoform levels following transfection of MDCK-TGF cells with ZEB1 and ZEB2 siRNAs or miR-200a and miR-200b pre-miRs relative to their negative controls for 6 d. (B) Measurement of TGF-β isoforms after transfection of MDCK cells with a control or LNA miR-200 family Anti-miR (10 d). Data are representative of duplicate experiments. Each value shown is the mean ± SD of three replicate measurements.
Autocrine TGF-β signaling is required for ZEB up-regulation during EMT induction
As shown earlier in the text and in previous studies, inhibition of miR-200 is able to initiate an EMT of MDCK cells, and this initiation of EMT is dependent on up-regulation of ZEB (Gregory et al., 2008a). In the studies described here, we have found that autocrine TGF-β signaling is required to maintain the mesenchymal state through up-regulating ZEB levels. To determine whether autocrine TGF-β signaling is also required for ZEB up-regulation during the induction of EMT, we treated MDCK cells with the SB-505124 TGF-β RI inhibitor while repressing the miR-200 family. Blockade of TGF-β signaling largely prevented the ability of the miR-200 anti-miR to up-regulate ZEB mRNA (Figure 5A) and to transition MDCK cells toward a mesenchymal phenotype, as shown by maintenance of E-cadherin and ZO-1 expression on the plasma membrane (Figure 5B). To verify that miR-200 was effectively inhibited by the anti-miR in the cells treated with SB-505124, we checked whether derepression of an unrelated target, CFL2 (Burk et al., 2008), occurred in the presence of SB-505124. Unlike ZEB, CFL2 was derepressed by the miR-200 anti-miR equally well in the presence or absence of the TGF-β signaling inhibitor (Figure 5A), demonstrating that autocrine TGF-β signaling is specifically required for ZEB up-regulation, even in the absence of miR-200 functional activity. Furthermore, these data show that, in the absence of TGF-β signaling, cells can remain in an epithelial state despite the lack of miR-200 activity.
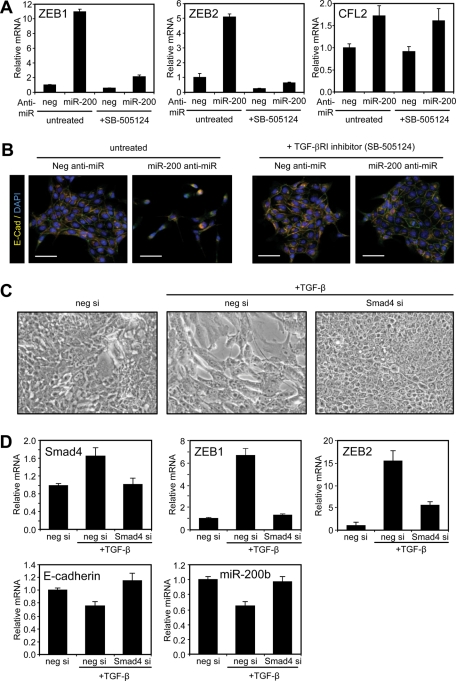
Autocrine TGF-β signaling is required for EMT induction by the ZEB/miR-200 regulatory loop. (A) Quantitation of ZEB1, ZEB2, and CFL2 levels in MDCK cells transfected with a control (neg) or LNA miR-200 family Anti-miR in the absence or presence of a TGF-β RI inhibitor (1 μM SB-505124; 14 d). Data are representative of duplicate experiments. Each value shown is the mean ± SD of three replicate measurements. (B) Merged images showing E-cadherin (red), ZO-1 (green), and nuclei (blue) staining of the cells. Scale bars = 100 μm. (C) Phase contrast micrographs of MDCK cells after transfection with control (neg) or Smad4 siRNAs followed by TGF-β1 treatment for 3 d. (D) Real-time PCR of Smad4 and EMT markers in cells described in (C). Data are representative of duplicate experiments. Each value shown is the mean ± SD of three replicate measurements.
TGF-β is known to signal through phosphorylation-mediated activation of Sma and Mad related family (Smad) transcription factors and in some cases by activation of the phosphoinositide 3-kinase and extracellular-signal regulated kinase/mitogen-activated protein kinase (ERK/MAPK) pathways to induce EMT (Miyazono, 2009). Smads have been previously shown to interact with the ZEB2 promoter and activate its transcription in MCF10A cells (Lu et al., 2009), suggesting that Smad signaling might be important for ZEB up-regulation during EMT. To investigate this possibility, we treated MDCK cells with TGF-β1 in the presence of an siRNA toward the Smad2/3 binding partner Smad4. Smad4 knockdown almost completely suppressed up-regulation of ZEB1 and ZEB2 mRNA and prevented induction of EMT (Figure 5, C and D). These data indicate that autocrine TGF-β signaling through the Smad pathway is required for ZEB up-regulation during the induction of EMT. Collectively, our findings demonstrate that the induction and maintenance of EMT is integrally controlled by a tripartite autocrine TGF-β/ZEB/miR-200 signaling network, with the balance of each factor determining the outcome of epithelial or mesenchymal cell phenotype.
Prolonged autocrine TGF-β signaling promotes reversible DNA methylation of the miR-200 family promoters
Although we have shown an important role for the autocrine TGF-β/ZEB/miR-200 network in maintaining the mesenchymal stability of MDCK-TGF cells, it is possible that epigenetic changes may be reinforcing this state. Recent reports have implicated a role for sustained TGF-β signaling in the DNA hypermethylation of E-cadherin and other genes silenced in basal-like breast cancers (Dumont et al., 2008; Papageorgis et al., 2010). In independent studies, the miR-200 loci have been shown to be subject to epigenetic repression through hypermethylation in gastric and breast cancer cell lines (Neves et al., 2010; Vrba et al., 2010; Wiklund et al., 2011). We hypothesized that prolonged exposure to TGF-β may lead to DNA hypermethylation of the miR-200 promoters and long-term suppression of its expression. To test this hypothesis, we first examined CpG methylation of the miR-200b~200a~429 proximal promoter in cells treated with TGF-β1 for 26 d and in MDCK-Pez cells which have been stably mesenchymal for 12 mo as a result of autocrine TGF-β production due to overexpression of the tyrosine phosphatase Pez (Wyatt et al., 2007). Sequencing of bisulfite-modified DNA showed that TGF-β induced de novo CpG methylation of several promoter regions that were unmethylated in parental MDCK cells (Figure 6A). DNA methylation across these regions was more pronounced in MDCK-Pez cells, suggesting that prolonged TGF-β exposure may enhance this process. We next examined CpG methylation of the miR-200b and miR-200c promoters over an extended TGF-β1 time course using PCR melt curve analysis (Supplemental Figure 3A). The DNA methylation of both miR-200 loci progressively increased with the duration of TGF-β exposure (by either exogenous or autocrine TGF-β); this increase was accompanied by a progressive decrease in miR-200 expression, consistent with a role for de novo DNA methylation in repressing miR-200 expression (Figure 6B and Supplemental Figure 3B).
To determine whether sustained TGF-β signaling was required for maintenance of miR-200 promoter methylation, we measured DNA methylation in MDCK-TGF cells treated with the TGF-β RI inhibitor SB-505124. In accordance with the progressive increase in miR-200 levels, the DNA methylation across both miR-200 promoters progressively decreased to a level at which little or none was detected at 24 d (Figure 6C and Supplemental Figure 3C). Collectively, these data demonstrate that prolonged autocrine TGF-β signaling promotes de novo CpG methylation of the miR-200 loci which is reversible upon inhibition of TGF-β signaling. In accordance with previous reports (Neves et al., 2010; Vrba et al., 2010) we also observed DNA hypermethylation of both miR-200 promoters in mesenchymal breast cancer cell lines in which miR-200 is repressed, but not in epithelial breast cancer cell lines with high miR-200 levels (Figure 6D) (Gregory et al., 2008a). This finding suggests that DNA hypermethylation of the miR-200 promoters may be an important mechanism for maintaining prolonged miR-200 repression during breast cancer progression.
Invasive ductal breast carcinomas display evidence of an operative autocrine TGF-β/ZEB/miR-200 signaling network
The TGF-β pathway plays a complex role in tumor progression, acting as a tumor suppressor in early-stage carcinoma but stimulating tumor cell migration and EMT in advanced cancer (Bierie and Moses, 2006). Recent gene-profiling studies have identified TGF-β–responsive signatures that correlate with breast cancer metastasis, reinforcing the role of this pathway as a potent driver of breast cancer progression (Padua et al., 2008). Considering the interconnection between TGF-β signaling and the ZEB/miR-200 regulatory loop, we examined invasive ductal carcinomas (IDCs) for evidence of this signaling network in invasive breast cancers. Real-time PCR was performed using RNA obtained from regions of 27 high grade IDCs that were histologically defined to contain primarily tumor cells. Comparing miR-200c~141 cluster expression with TGF-β1, TGF-β2, ZEB1, and ZEB2, we found highly significant inverse correlations (p < 0.01) for each pairwise comparison, the only exception being that miR-141 and TGF-β1 levels were not significantly correlated (Figure 7 and Supplemental Table 1). Strong direct correlations were also observed among all three TGF-β isoforms and ZEB1 and ZEB2 (p < 0.002), consistent with a role for autocrine TGF-β signaling in activating ZEB transcription (Figure 7 and Supplemental Table 1). Interestingly, we did not find significant correlations involving the miR-200b~200a~429 cluster and the TGF-βs or ZEB, or with any of the miR-200 family and TGF-β3 (Supplemental Table 1). Collectively, these data support a potential role for an autocrine TGF-β/ZEB/miR-200 signaling network in invasive breast cancers and indicate that there may be some specificity of interaction among miR-200, ZEB, and TGF-β family members in breast cancer cells.
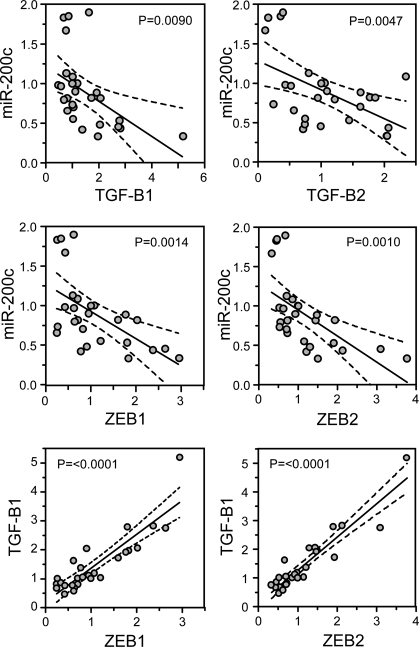
Invasive ductal breast carcinomas display evidence of an operative autocrine TGF-β/ZEB/miR-200 signaling network. Real-time PCR was performed using total RNA isolated from 27 invasive ductal breast cancer samples and the expression of miR-200c, ZEB1, ZEB2, TGF-β1, and TGF-β2 was compared in a pairwise manner. The line of best fit was obtained by linear regression analysis with 95% confidence intervals shown. p values were calculated using Pearson correlation coefficients.
DISCUSSION
In this study, we demonstrate that epithelial cell plasticity is regulated by a tripartite autocrine TGF-β/ZEB/miR-200 signaling network which provides a mechanistic explanation for the stable and yet reversible nature of EMT observed in many developmental and pathological scenarios. In response to TGF-β stimulation, MDCK cells transition toward a mesenchymal state which is stabilized only after 5–8 d of exogenous TGF-β1 exposure. This finding indicates that threshold changes in the level of ZEB, miR-200, and TGF-β are important in determining the final outcome of cell state. These findings are consistent with the proposed function of the ZEB/miR-200 double-negative feedback loop model in which self-reinforcing, opposing expression of miR-200 and ZEB develops over time and eventually leads to a stable change in cell state (Gregory et al., 2008b; Brabletz and Brabletz, 2010). This model also predicts that the endpoint state would remain stable and be buffered against subthreshold changes in miR-200 and ZEB. In support of this concept, we observed that short-term TGF-β1 treatment (<5–8 d) induces only a transient EMT which was reversible upon factor withdrawal. These data are also consistent with the hypothesis that the epithelial phenotype is the default state in the absence of factors that induce transition toward a mesenchymal state (Frisch, 1997; Polyak and Weinberg, 2009). To confirm the importance of the ZEB/miR-200 feedback loop in determining cell state, we altered the balance of these factors either directly or indirectly (with TGF-β) and showed that we could repeatedly switch cells between epithelial and mesenchymal states. Integral to this process, however, was the influence of these factors on autocrine TGF-β signaling. Autocrine TGF-β signaling was initiated and regulated by the ZEB/miR-200 loop and was necessary for the induction and maintenance of ZEB expression in the mesenchymal state. These findings demonstrate that a tripartite autocrine TGF-β/ZEB/miR-200 signaling network controls both the establishment and maintenance of EMT.
The mechanisms through which the ZEB/miR-200 feedback loop regulates and is controlled by autocrine TGF-β is not yet fully elucidated but is likely to involve both direct and indirect interactions. In accordance with previous observations that Smads interact with the ZEB2 promoter (Lu et al., 2009), we observed that knockdown of Smad4 prevented up-regulation of ZEB mRNAs and induction of EMT. Autocrine TGF-β was also shown to be required for the maintenance of the mesenchymal state of MDCK-TGF cells as inhibition of this signaling pathway resulted in cells reverting to an epithelial phenotype. By ectopically expressing either ZEB or Snail in MDCK cells, we provide evidence that autocrine TGF-β signaling acts through up-regulation of ZEB1 and ZEB2, but not Snail, to repress miR-200 and enforce the mesenchymal phenotype. These observations indicate that a specific interaction of autocrine TGF-β signaling with ZEB is needed for stability of the mesenchymal state. The fact that ectopically expressed Snail did not repress miR-200 expression when TGF-β signaling was blocked indicates that Snail does not directly repress miR-200, but acts indirectly through stimulating autocrine TGF-β. Snail has been shown to be essential for the initial induction of ZEB1 in NMuMG cells (Dave et al., 2011), suggesting that Snail is an essential early mediator of activation of the TGF-β/ZEB/miR-200 pathway. Conversely, we also demonstrated that direct manipulation of miR-200 or ZEB levels could influence expression of TGF-β1, TGF-β2, and TGF-β3. Previous studies have shown that miR-141/200a can directly target TGF-β2 (Burk et al., 2008; Wang et al., 2011), leading to the proposal that low miR-200 levels may promote autocrine TGF-β signaling (Burk et al., 2008; Gregory et al., 2008a). We observed, however, that TGF-β3 experienced the largest change in its levels after miR-200 manipulation. Considering that TGF-β1 and TGF-β3 are not predicted to be direct targets of the miR-200 family, it is likely that changes in TGF-β expression by miR-200 in MDCK cells are caused by a combination of direct and indirect effects. Although individual TGF-β isoforms are known to have context-specific functions (Romano and Runyan, 2000; Nawshad et al., 2007), we observed a redundancy in the function of the TGF-β isoforms in maintaining the mesenchymal state of MDCK-TGF cells. Given that TGF-β isoforms are also known to regulate the expression of one another in MDCK cells (Medici et al., 2008), it is likely that the interactions between the ZEB/miR-200 loop and autocrine TGF-β signaling are complex. Interestingly, several other components within the TGF-β signaling pathway (including Smad2 and TGF-βRI) have recently been shown to be targeted by the miR-200 family in anaplastic thyroid carcinomas (Braun et al., 2010), and these interactions may also be relevant in promoting autocrine TGF-β signaling and epithelial cell plasticity in this and other contexts.
Although we have shown that the autocrine TGF-β/ZEB/miR-200 signaling network is central to the initiation and maintenance of EMT in MDCK cells, a number of other EMT-inducing transcription factors may also have functions within this context. This is particularly evident at the early stages of TGF-β1–induced EMT in MDCK cells where the transcription factors Snail and Slug have been shown to be rapidly induced within 24 h of treatment (Peinado et al., 2003; Medici et al., 2006; Morita et al., 2007). We found that ZEB1 and ZEB2 mRNAs are induced within 2 d of TGF-β treatment but that their protein levels remain undetectable for several more days. This finding is consistent with the high levels of miR-200 acting to repress translation of these mRNAs and suggests that factors other than the ZEB/miR-200 feedback loop are likely to be driving the initial changes in marker expression and cell morphology. Interestingly, the down-regulation of the miR-200b~200a~429 but not miR-200c~141 cluster appeared to precede detectability of ZEB1 and ZEB2 proteins, suggesting that other factors may be responsible for the initial repression of the miR-200b~200a~429 cluster. These factors might facilitate activation of the ZEB/miR-200 feedback loop, which would otherwise be inhibited by high miR-200 levels squelching ZEB translation. The induction of Snail by TGF-β in MDCK cells has been studied in most detail and shown to involve both Smad- and MAPK-dependent pathways (Peinado et al., 2003; Medici et al., 2006). Snail and Slug in turn have been shown to up-regulate TGF-β3 by a TCF4-β-catenin–dependent mechanism (Medici et al., 2008). Our findings are consistent with this model in that Snail can induce autocrine TGF-β, but we find that Snail remains upstream in this pathway and is not sufficient to maintain the mesenchymal state, which requires ongoing ZEB expression.
Our findings in this study with MDCK cells share similarities and differences with other EMT cell culture models. In the normal mouse mammary epithelial cell NMuMG cell model, prolonged TGF-β1 stimulation (>2 wk) also induces a full EMT, but, unlike MDCK cells, they do not sustain the mesenchymal state long term after TGF-β withdrawal (Gal et al., 2008). TGF-β1 treatment has been shown to reduce the expression of the miR-200 family in NMuMG cells (Korpal et al., 2008); however, we have found that these cells express much lower levels of miR-200 than do MDCK cells (unpublished data). Therefore, it is feasible that the ZEB/miR-200 feedback loop may not play a dominant role in NMuMG cells. In support of this observation, ZEB1 and ZEB2 induction have been shown to be required for TGF-β–mediated repression of E-cadherin but not for induction of mesenchymal markers in NMuMG cells (Shirakihara et al., 2007). In contrast, we have previously shown that enforced expression of miR-200 in MDCK cells prevents up-regulation of ZEB1 and ZEB2 as well as changes in epithelial and mesenchymal markers in response to TGF-β (Gregory et al., 2008a), confirming that changes in miR-200 are needed for a complete EMT. Participation of autocrine TGF-β signaling in the maintenance of the mesenchymal state has been previously observed in MDCK cells where it was found that activation of the Ras/Erk/MAPK pathway by stable expression of Raf caused cells to undergo a stable EMT associated with the induction of autocrine TGF-β signaling (Lehmann et al., 2000). Constitutive activation of the MAPK pathway by Ras was also required for TGF-β to induce a stable mesenchymal state in mouse mammary EpH4 cells (Oft et al., 1996; Janda et al., 2002). We did observe an increase in MAPK activity in MDCK-TGF cells; however, inhibition of this pathway was not sufficient to revert these cells to an epithelial phenotype (unpublished data). It will be interesting to further examine whether the TGF-β and MAPK pathways converge to regulate expression of the ZEB/miR-200 feedback loop.
A major finding of this study is that TGF-β signaling induces reversible DNA methylation of the miR-200 promoters. Although it is well established that DNA hypermethylation of specific genes occurs in advanced cancers, links between EMT and de novo DNA methylation have only recently been described (Dumont et al., 2008; Papageorgis et al., 2010). Here we have shown that the miR-200b and miR-200c loci are subject to de novo DNA methylation upon prolonged TGF-β signaling and that this was reversible upon inhibition of TGF-β signaling. Changes in the degree of miR-200 promoter methylation closely correlated with miR-200 expression, implicating a role for this process in miR-200 repression. The mechanism through which TGF-β signaling controls DNA methylation of miR-200 is not clear at this stage, but may involve active DNA methyltransferases (DNMTs). DNMT activity has been linked with the LSD1 histone demethylase complex (Wang et al., 2009), of which ZEB1 is a component that facilitates complex recruitment to ZEB binding sites (Wang et al., 2007), providing a potential connection between ZEB and miR-200 gene methylation. In addition to the MDCK EMT model, we found that invasive mesenchymal breast cancer cell lines also exhibit methylated miR-200 promoters, in contrast to epithelial cells in which the miR-200 promoters were unmethylated. Similar findings have recently been observed in primary mesenchymal breast cells and invasive bladder tumors (Vrba et al., 2010; Wiklund et al., 2011). Taken together, these data suggest that DNA methylation of the miR-200 promoters contributes to their silencing during EMT and cancer progression.
TGF-β expression is often increased in tumor cells and can act in an autocrine and paracrine manner within the tumor microenvironment to enhance cancer progression (Bierie and Moses, 2006). Our data suggest that the autocrine TGF-β/ZEB/miR-200 signaling axis may be involved in mediating progression of breast cancer. This finding is supported by a recent study in which a TGF-β–responsive signature, including elevated ZEB1 levels, was found to be an independent predictor of breast cancer metastasis to the lung (Padua et al., 2008). Knockdown of ZEB1 in cancer cell lines has been shown to reduce both tumor size and metastases in xenograft mouse models, verifying its ability to enhance tumor progression (Spaderna et al., 2008; Wellner et al., 2009). Several reports have shown that enforced miR-200 expression correlates with reduced ZEB expression and invasive potential in a range of cancer cell lines (Burk et al., 2008; Gibbons et al., 2009; Kong et al., 2009). We found that increased expression of TGF-β1 and TGF-β2 correlated with low miR-200c and high ZEB expression in invasive ductal breast cancer samples. Interestingly, these correlations were not observed generally with all TGF-β isoforms and miR-200 family members, although strong correlations were observed with all TGF-β isoforms and ZEBs. These data are consistent with a role for autocrine TGF-β signaling in up-regulating ZEB in breast cancer cells, but suggest that there may be differential regulation of the miR-200 family members in this context.
In summary, we have identified a central role for an autocrine TGF-β/ZEB/miR-200 signaling network in controlling the transition between epithelial and mesenchymal states. Prolonged activation of this pathway leads to dynamic epigenetic changes in miR-200 and may contribute to invasive breast cancer progression. In light of these findings, a remarkable connection between EMT and breast cancer stem cells was recently demonstrated where TGF-β treatment was shown to initiate EMT and concomitant acquisition of tumor-initiating and self-renewal properties (Mani et al., 2008). Independently of these studies, the miR-200 family and ZEB1 have been shown to be key regulators of these stem-like properties (Shimono et al., 2009; Wellner et al., 2009; Iliopoulos et al., 2010). These observations provide an intriguing link between the autocrine TGF-β/ZEB/miR-200 signaling network and the plasticity of EMT and the stem-like properties of cells during cancer progression and metastasis (Gupta et al., 2009). Similar links among the TGF-β–related factors, the bone morphogenetic proteins, and the miR-200 family have recently been described in somatic cell reprogramming (Samavarchi-Tehrani et al., 2010). It will be of significant interest to examine the importance of the autocrine TGF-β/ZEB/miR-200 signaling network in governing cell plasticity and stemness in developmental and pathological scenarios.
MATERIALS AND METHODS
Cell line maintenance and treatments
MDCK cells and their derivatives and human breast cancer cell lines were cultured as previously described (Bracken et al., 2008). The generation of MDCK-EV, MDCK-ZEB1, MDCK-ZEB2, and MDCK-Pez stable cell lines has been previously described (Wyatt et al., 2007; Bracken et al., 2008). MDCK-Snail cells were made by transfection of pcDNA3-mSnail (a gift from Antonio Garcia de Herreros, IMIM-Hospital del Mar, Barcelona, Spain) and selection of single clones as previously described for the ZEB1 and ZEB2 stable cell lines (Bracken et al., 2008). TGF-β1, -β2, and -β3 ligands, anti–TGF-β1, -β2, -β3, and pan anti–TGF-β1/β2/β3 (R&D Systems, Minneapolis, MN) were used at 1 ng/ml or 100 μg/ml, respectively. The TGF-β R1 inhibitors SB-505124 and SB-431542 (Calbiochem, San Diego, CA) were used at a 1 μM final concentration. Treatments of MDCK and derivative cell lines were commenced 1 d after plating and were readministered at time of passage or transfection (~3–4 d).
Isolation of RNA and real-time PCR
Total RNA was isolated from cell lines, and real-time PCR was performed by using primers as previously described (Gregory et al., 2008a). Primers for canine TGF-β isoforms and CFL2 are as follows: caTGF-β1 (5′-TGGACACGCAGTACAGCAA-3′; 5′-TAGTACACGATGGGCAGTGG-3′), caTGF-β2 (5′-CGTTTACAGAACCCGAAAGC-3′; 5′-TCGTCTTGACGACTTTGCTG-3′), caTGF-β3 (5′-CTTGCACCACCTTGGACTTT-3′; 5′-CTGTTGTAAAGGGCCAGGAC-3′), and caCFL2 (5′-ATGGCTTCTGGAGTTACAGTGA-3′; 5’-GCATATCGGCAATCATTCAGAGG-3′). MicroRNA PCRs were performed using TaqMan microRNA assays (Applied Biosystems, Foster City, CA). Real-time PCR data for mRNA and microRNA are expressed relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6, respectively.
Transfection of microRNAs and siRNAs
MDCK and its derivatives were transfected at low density using HiPerFect transfection reagent (Qiagen, Chatsworth, CA). For concurrent knockdown of ZEB1 and ZEB2, 10 nM of each siRNA (Invitrogen; 40 nM total) or a control siRNA was transfected every 3 d for a total of 6 d as previously described (Gregory et al., 2008a). Ectopic expression of miR-200a and miR-200b (20 nM; Shanghai GenePharma, Shanghai, China) was performed in a similar manner. For Smad4 knockdown, 200 nM of siRNA (100 nM each Smad siRNA) or control (Shanghai GenePharma) was transfected before the addition of TGF-β. The sequences for the canine Smad4 siRNAs are 5′-AGACAGAGCAUCAAAGAAATT-3′ and 5′-UCAGGUGGCUGGUCGGAAATT-3′. Inhibition of all members of the miR-200 family was carried out by transfection of a modified LNA Anti-miR with the sequence 5′-mU*+C*mG*mU+CmUmU+TmAmC+CmAmG+GmCmA+GmUmA*+T*mU*mA-3′, where mN is a 2′O-methyl base, + is a LNA base, and * is a phosphorothioate bond. Anti-miR transfections were performed as mentioned earlier in the text for 10–14 d using a 300 nM final concentration.
Western blotting and immunofluorescence
Western blotting was performed as previously described (Gregory et al., 2008a). The following primary antibodies were used: ZEB1 (1:2000 vol:vol, A301–922A, Bethyl Laboratories, Montgomery, TX), ZEB2 (1:5000 vol:vol; Christoffersen et al., 2007), E-cadherin (1:1000 vol:vol, 610182; BD Transduction Laboratories), and tubulin (1:5000 vol:vol, ab7291; Abcam, Cambridge, MA). Membranes were exposed using enhanced chemiluminescence (GE Healthcare, Buckinghamshire, UK) and imaged using the LAS4000 Luminescent Image Analyzer (Fujifilm, Tokyo, Japan). For immunofluorescence, cells were plated on fibronectin-coated chamber slides (Nunc, Rochester, NY) and stained using anti-E-cadherin (1:500 vol:vol, as mentioned earlier in the text), ZO-1 (1:500 vol:vol; Zymed, South San Francisco, CA), or F-actin as previously described (Gregory et al., 2008a). Nuclei were visualized by costaining with DAPI. Cells were visualized on an Olympus IX81 microscope, and pictures were taken using a Hamamatsu Orca camera. Images were analyzed with Olympus Cell software.
Enzyme-linked immunosorbent assay (ELISA) of TGF-β isoforms
Secreted TGF-β1 and TGF-β2 levels were determined using the Quantikine human TGF-β1 and -β2 kits (R&D Systems) as per the manufacturer's instruction. Prior to assay, MDCK cells were treated with TGF-β1 for 12 d and then grown for a further 6 d without exogenous TGF-β1. A stable mesenchymal phenotype was confirmed by cell morphology and real-time PCR analysis of epithelial and mesenchymal gene expression. Equal numbers of MDCK and MDCK-TGF cells were then plated in 12-well trays, grown for 1 d, then grown overnight in serum-free medium that was collected for analysis.
Analysis of primary human breast cancer samples
Formalin-fixed, paraffin-embedded (FFPE) deidentified sections from invasive ductal breast cancer samples were derived from the Breast Biomarker Project at Royal Melbourne Hospital and from the Department of Tissue Pathology at SA Pathology. Access to patient tumor samples was approved by the appropriate institutional human ethics review boards. Sections were hematoxylin and eosin stained, and regions that contained primarily tumor cells were marked for further analysis. Using duplicate unstained sections, marked areas of the tumor were scraped into tubes where total RNA was isolated using an miRNeasy FFPE kit (Qiagen). cDNA was specifically primed, then real-time PCR analysis for mRNA was performed using TaqMan assays (Applied Biosystems) and expressed relative to GAPDH. Samples were obtained from one to two distinct regions from each patient specimen. Each was separately assayed, and the triplicate values were averaged and then treated as individual data points. Primers for the TaqMan Gene Expression assays (with appropriate Applied Biosystems Assay ID) were as follows: hGAPDH (Hs99999905_m1), hTGF-β1 (Hs00171257_m1), hTGF-β2 (Hs01548875_m1), hTGF-β3 (Hs00234245_m1), hZEB1 (Hs00232783_m1), hZEB2 (Hs00207691_m1). Multiplex miRNA cDNA was prepared, then miRNA PCRs were performed using TaqMan microRNA assays (Applied Biosystems). Real-time PCR data for microRNA are expressed relative to five control miRNAs (U6, miR-16, miR-24, miR-93, and let-7a). Significance of correlation between normalized mRNA and miRNA data was calculated using the Pearson correlation.
DNA methylation analysis of the miR-200 loci
Genomic DNA was isolated from cells using TRIzol (Invitrogen). The DNA was quantitated on a NanoDrop-1000 (Thermo Fisher Scientific, Scoresby, VIC, Australia), and 500 ng was bisulfite modified with the EZ DNA Methylation-Gold Kit according to the manufacturer's protocol (Zymo Research, Irvine, CA). For bisulfite sequencing, bisulfite-modified DNA was PCR amplified using the bisulfite sequencing primers specified in Supplementary Table 2. The size of the PCR products was confirmed by electrophoresis on 2% agarose gels (Bio-Rad, Hercules, CA) stained with ethidium bromide. The PCR products were purified from the agarose gels using the QIAquick Gel Extraction Kit (Qiagen). The PCR products were then cloned into pGEM-T Easy Vector (Promega, Madison, WI) and selected after transformation into JM109 competent cells (Promega) and plating onto LB Agar plates containing 100 μg/ml ampicillin, 0.5 mM isopropyl-β-d-thiogalactopyranoside, and 80 μg/ml X-Gal. White (positive) colonies were selected and cultured overnight, and plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen). On purification, three to six cloned fragments were sequenced using a pUC/M13 Reverse (22 mer) Sequencing Primer (Promega) and BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) to detect methylated and unmethylated cytosine residues.
For melt curve analysis, bisulfite-modified DNA was PCR amplified and melted as described previously (Smith et al., 2009). The PCR primer sets and conditions used did not discriminate between methylated and unmethylated DNA and did not amplify unmodified DNA (Supplemental Table 1). For melt curve analysis of the canine miR-200 loci, bisulfite-modified DNA from MDCK (unmethylated reference), MDCK-Pez (methylated reference), and unmodified DNA from MDCK (negative control) was included in each PCR. For melt curve analysis of the human miR-200 loci, bisulfite-modified MDA-MB-361 (unmethylated reference), HBL-100 (methylated reference), and unmodified human donor lymphocyte DNA (negative control) was included in each PCR. The PCR was performed using a Rotor-Gene 3000 (Corbett Life Science, Sydney, NSW, Australia) with a 95°C activation step for 15 min; 95°C for 30 s, 55°C for 60 s for 45 cycles; and a final extension step of 72°C for 4 min. The melt of the PCR product was performed from 60 to 90°C, rising in 0.5°C increments, waiting for 30 s at the first step and for 5 s at each step thereafter, and acquiring fluorescence at each temperature increment. The raw melt data were normalized as described previously (Smith et al., 2009).
Acknowledgments
We thank Narrelle Mancini for excellent technical assistance and members of the Goodall and Khew-Goodall labs for helpful discussions. This work was supported by fellowships from the National Breast Cancer Foundation Australia (to P.A.G. and C.P.B.) and by grants from the National Health and Medical Research Council (to G.J.G and Y.K-G) and the Cancer Council South Australia (to G.J.G., Y.K.-G., and P.A.D.). The Breast Biomarker Project, which contributed breast cancer samples, was supported by The Victorian Cancer Agency.
Abbreviations used:
| DNMT | DNA methyltransferase |
| EMT | epithelial-mesenchymal transition |
| ERK/MAPK | extracellular-signal regulated kinase/mitogen-activated protein kinase |
| EV | empty vector |
| FFPE | formalin-fixed, paraffin-embedded |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| IDC | invasive ductal carcinoma |
| iPSC | induced pluripotent stem cell |
| LNA | locked nucleic acid |
| MDCK | Madin Darby canine kidney |
| MET | mesenchymal-epithelial transition |
| RI | receptor-1 |
| Smad | Sma and Mad related family |
| TGF-β | transforming growth factor-β |
| ZEB | zinc finger E-box-binding homeobox |
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0103) on March 16, 2011.
REFERENCES
- Berx G, Raspe E, Christofori G, Thiery JP, Sleeman JP. Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clin Exp Metastasis. 2007;24:587–597. [Abstract] [Google Scholar]
- Bierie B, Moses HL. Tumor microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. [Abstract] [Google Scholar]
- Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer. EMBO Rep. 2010;11:670–677. [Europe PMC free article] [Abstract] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. [Abstract] [Google Scholar]
- Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. [Abstract] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. [Europe PMC free article] [Abstract] [Google Scholar]
- Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–1178. [Europe PMC free article] [Abstract] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. [Abstract] [Google Scholar]
- Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, Garcia de Herreros A. Functional cooperation between Snail1 and Twist in the regulation of Zeb1 expression during epithelial-to-mesenchymal transition. J Biol Chem. 2011;2011 Feb 12. [Epub ahead of print] [Europe PMC free article] [Abstract] [Google Scholar]
- Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat Cell Biol. 2007;9:1000–1004. [Abstract] [Google Scholar]
- Dumont N, Wilson MB, Crawford YG, Reynolds PA, Sigaroudinia M, Tlsty TD. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci USA. 2008;105:14867–14872. [Europe PMC free article] [Abstract] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. [Abstract] [Google Scholar]
- Eger A, Stockinger A, Park J, Langkopf E, Mikula M, Gotzmann J, Mikulits W, Beug H, Foisner R. beta-Catenin and TGFbeta signaling cooperate to maintain a mesenchymal phenotype after FosER-induced epithelial to mesenchymal transition. Oncogene. 2004;23:2672–2680. [Abstract] [Google Scholar]
- Frisch SM. The epithelial cell default-phenotype hypothesis and its implications for cancer. Bioessays. 1997;19:705–709. [Abstract] [Google Scholar]
- Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A. Sustained TGF beta exposure suppresses Smad and non-Smad signaling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene. 2008;27:1218–1230. [Abstract] [Google Scholar]
- Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. [Europe PMC free article] [Abstract] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008a;10:593–601. [Abstract] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008b;7:3112–3118. [Abstract] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–1012. [Abstract] [Google Scholar]
- Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. [Abstract] [Google Scholar]
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. [Europe PMC free article] [Abstract] [Google Scholar]
- Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156:299–313. [Europe PMC free article] [Abstract] [Google Scholar]
- Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. [Europe PMC free article] [Abstract] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. [Europe PMC free article] [Abstract] [Google Scholar]
- Lehmann K, Janda E, Pierreux CE, Rytomaa M, Schulze A, McMahon M, Hill CS, Beug H, Downward J. Raf induces TGFbeta production while blocking its apoptotic but not invasive responses: a mechanism leading to increased malignancy in epithelial cells. Genes Dev. 2000;14:2610–2622. [Europe PMC free article] [Abstract] [Google Scholar]
- Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. [Abstract] [Google Scholar]
- Lu J, et al. 14–3–3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell. 2009;16:195–207. [Europe PMC free article] [Abstract] [Google Scholar]
- Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. [Europe PMC free article] [Abstract] [Google Scholar]
- Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17:1871–1879. [Europe PMC free article] [Abstract] [Google Scholar]
- Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. [Europe PMC free article] [Abstract] [Google Scholar]
- Miyazono K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:314–323. [Europe PMC free article] [Abstract] [Google Scholar]
- Morita T, Mayanagi T, Sobue K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J Cell Biol. 2007;179:1027–1042. [Europe PMC free article] [Abstract] [Google Scholar]
- Nawshad A, Medici D, Liu CC, Hay ED. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J Cell Sci. 2007;120:1646–1653. [Europe PMC free article] [Abstract] [Google Scholar]
- Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S, Uhrberg M. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. [Europe PMC free article] [Abstract] [Google Scholar]
- Ocana OH, Nieto MA. A new regulatory loop in cancer-cell invasion. EMBO Rep. 2008;9:521–522. [Europe PMC free article] [Abstract] [Google Scholar]
- Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. [Abstract] [Google Scholar]
- Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. [Europe PMC free article] [Abstract] [Google Scholar]
- Papageorgis P, et al. Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res. 2010;70:968–978. [Europe PMC free article] [Abstract] [Google Scholar]
- Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta 1775. 2007:21–62. [Abstract] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. [Europe PMC free article] [Abstract] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumor progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. [Abstract] [Google Scholar]
- Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. [Abstract] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. [Abstract] [Google Scholar]
- Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. [Europe PMC free article] [Abstract] [Google Scholar]
- Romano LA, Runyan RB. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev Biol. 2000;223:91–102. [Abstract] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. [Abstract] [Google Scholar]
- Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. [Europe PMC free article] [Abstract] [Google Scholar]
- Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith E, Jones ME, Drew PA. Quantitation of DNA methylation by melt curve analysis. BMC Cancer. 2009;9:123. [Europe PMC free article] [Abstract] [Google Scholar]
- Spaderna S, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. [Abstract] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumor progression. Nat Rev Cancer. 2002;2:442–454. [Abstract] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. [Abstract] [Google Scholar]
- Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang B, et al. miR-200a prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes 60, 280–287. 2011 [Europe PMC free article] [Abstract] [Google Scholar]
- Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. [Abstract] [Google Scholar]
- Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. [Abstract] [Google Scholar]
- Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. [Abstract] [Google Scholar]
- Wiklund ED, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer 128, 1327–1334. 2011 [Abstract] [Google Scholar]
- Wyatt L, Wadham C, Crocker LA, Lardelli M, Khew-Goodall Y. The protein tyrosine phosphatase Pez regulates TGFbeta, epithelial-mesenchymal transition, and organ development. J Cell Biol. 2007;178:1223–1235. [Europe PMC free article] [Abstract] [Google Scholar]
- Zeisberg M, Kalluri R. The role of epithelial-to-mesenchymal transition in renal fibrosis. J Mol Med. 2004;82:175–181. [Abstract] [Google Scholar]
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Relationship of Signaling Pathways between RKIP Expression and the Inhibition of EMT-Inducing Transcription Factors SNAIL1/2, TWIST1/2 and ZEB1/2.
Cancers (Basel), 16(18):3180, 17 Sep 2024
Cited by: 0 articles | PMID: 39335152 | PMCID: PMC11430682
Review Free full text in Europe PMC
TGF-β Modulated Pathways in Colorectal Cancer: New Potential Therapeutic Opportunities.
Int J Mol Sci, 25(13):7400, 05 Jul 2024
Cited by: 1 article | PMID: 39000507
Review
Cell-state transitions and density-dependent interactions together explain the dynamics of spontaneous epithelial-mesenchymal heterogeneity.
iScience, 27(7):110310, 19 Jun 2024
Cited by: 0 articles | PMID: 39055927 | PMCID: PMC11269952
MLK3 promotes prooncogenic signaling in hepatocellular carcinoma via TGFβ pathway.
Oncogene, 43(30):2307-2324, 10 Jun 2024
Cited by: 1 article | PMID: 38858590
Comprehensive molecular interaction map of TGFβ induced epithelial to mesenchymal transition in breast cancer.
NPJ Syst Biol Appl, 10(1):53, 17 May 2024
Cited by: 1 article | PMID: 38760412
Go to all (385) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mesenchymal stem cells play a potential role in regulating the establishment and maintenance of epithelial-mesenchymal transition in MCF7 human breast cancer cells by paracrine and induced autocrine TGF-β.
Int J Oncol, 41(3):959-968, 03 Jul 2012
Cited by: 50 articles | PMID: 22766682
A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition.
PLoS One, 7(4):e35440, 13 Apr 2012
Cited by: 107 articles | PMID: 22514743 | PMCID: PMC3325969
Specificity protein 1 (Sp1) maintains basal epithelial expression of the miR-200 family: implications for epithelial-mesenchymal transition.
J Biol Chem, 289(16):11194-11205, 13 Mar 2014
Cited by: 41 articles | PMID: 24627491 | PMCID: PMC4036258
ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer.
Int J Cancer, 132(4):745-754, 21 Jul 2012
Cited by: 173 articles | PMID: 22753312
Review




