Abstract
Free full text

Differential Roles of Hypoxia-Inducible Factor 1α (HIF-1α) and HIF-2α in Hypoxic Gene Regulation
Abstract
Transcriptional responses to hypoxia are primarily mediated by hypoxia-inducible factor (HIF), a heterodimer of HIF-α and the aryl hydrocarbon receptor nuclear translocator subunits. The HIF-1α and HIF-2α subunits are structurally similar in their DNA binding and dimerization domains but differ in their transactivation domains, implying they may have unique target genes. Previous studies using Hif-1α−/− embryonic stem and mouse embryonic fibroblast cells show that loss of HIF-1α eliminates all oxygen-regulated transcriptional responses analyzed, suggesting that HIF-2α is dispensable for hypoxic gene regulation. In contrast, HIF-2α has been shown to regulate some hypoxia-inducible genes in transient transfection assays and during embryonic development in the lung and other tissues. To address this discrepancy, and to identify specific HIF-2α target genes, we used DNA microarray analysis to evaluate hypoxic gene induction in cells expressing HIF-2α but not HIF-1α. In addition, we engineered HEK293 cells to express stabilized forms of HIF-1α or HIF-2α via a tetracycline-regulated promoter. In this first comparative study of HIF-1α and HIF-2α target genes, we demonstrate that HIF-2α does regulate a variety of broadly expressed hypoxia-inducible genes, suggesting that its function is not restricted, as initially thought, to endothelial cell-specific gene expression. Importantly, HIF-1α (and not HIF-2α) stimulates glycolytic gene expression in both types of cells, clearly showing for the first time that HIF-1α and HIF-2α have unique targets.
Oxygen (O2), the final electron acceptor during oxidative phosphorylation, is absolutely required for invertebrate and vertebrate life. The immediate response to O2 deprivation (hypoxia) is a defense phase, which suppresses ATP consumption by arresting protein translation and ion channel activity, two major ATP sinks during normoxia. During a rescue phase, in spite of a general reduction in RNA synthesis, transcription of some genes increases dramatically under low O2 (21, 34). These hypoxia-responsive genes are involved in glucose transport, glycolysis, erythropoiesis, angiogenesis, vasodilation, and respiratory rate, and together they function to minimize the effects caused by low O2 at cellular, tissue and systemic levels (93, 106).
The activation of many O2-regulated genes is mediated by hypoxia-inducible factor (HIF), a heterodimer consisting of HIF-1α and HIF-1β (also called the aryl hydrocarbon receptor nuclear translocator [ARNT]) in most cells (52, 104, 105). Both HIF-1α and ARNT belong to the basic helix-loop-helix (bHLH)-Per-Arnt-Sim (PAS) family of transcription factors, which share several conserved structural domains, including a bHLH region for DNA binding and two PAS domains for target gene specificity and dimerization (102). Although ARNT is absolutely required for HIF activity (63, 110), HIF function is primarily regulated by HIF-1α protein stability (37, 46, 84). Under normoxia, HIF-1α is ubiquitinated through interaction with the von Hippel-Lindau tumor suppressor protein (pVHL) and subsequently degraded by the 26S proteasome (7, 18, 50, 70, 99, 111). pVHL, the recognition component of an E3 ubiquition-ligase complex, binds HIF-1α when it is hydroxylated at proline residues 402 and 577 (41, 43, 112). Proline hydroxylation of HIF-1α is catalyzed by prolyl hydroxylase domain-containing (PHD) proteins, members of the 2-oxoglutarate-dependent dioxygenase superfamily whose activity requires O2 as a cofactor, implying that these HIF-PHDs may directly sense low O2 (10, 27). However, using mitochondrial DNA-depleted cells and a number of mitochondrial electron transport inhibitors, we and others demonstrated that mitochondria act as the proximal O2 sensor during hypoxia via increased production of reactive O2 species to stabilize HIF-α proteins (14, 15, 89). Therefore, it remains a formal possibility that reactive O2 species act upstream of HIF-PHD function. Under hypoxia, the activity of HIF-PHDs is suppressed, resulting in reduced HIF-1α proline hydroxylation and association with pVHL. The stabilized HIF-1α subunit translocates to the nucleus, dimerizes with ARNT, and transactivates hypoxia-responsive genes through binding to hypoxia response elements (HREs) located in the promoter or enhancer regions of hypoxia-inducible genes (45, 94, 103).
In addition to HIF-1α, another mammalian bHLH-PAS protein, HIF-2α (also called EPAS1/HRF/HLF/MOP2), has been implicated in executing the hypoxia response (26, 30, 35, 101). Structurally, HIF-2α and HIF-1α are closely related, sharing 48% overall amino acid identity. In particular, there is 83% identity in their bHLH domains and approximately 70% homology in their PAS regions; these findings are in agreement with the data from in vitro functional studies indicating that HIF-2α also binds ARNT and transactivates HRE-containing target genes such as those for erythropoietin and vascular endothelial growth factor (VEGF) (26, 30, 35, 101). The two HIF-α subunits also exhibit a high degree of homology in their oxygen-dependent-degradation domains, including the two critical proline residues. Thus, both proteins appear to be subject to identical protein stability regulation by O2 as described above (73, 80, 97, 108). Interestingly, the 50 amino acids located at the C-termini of HIF-1α and HIF-2α are also highly conserved and appear to be important for O2-regulated interaction with the transcriptional coactivator p300 (13, 22, 25, 33, 57, 62, 85). Besides the described similarities, HIF-2α gene targeting experiments also demonstrate that HIF-2α regulates the expression of several hypoxia-inducible genes, including those for tyrosine hydroxylase in the organ of Zuckerkandl (78, 100) and VEGF in type II pneumocytes in vivo (19). Taken together, these studies demonstrate that HIF-2α, like HIF-1α, is involved in hypoxic responses.
While HIF-1α is ubiquitously expressed, HIF-2α is detected most prominently in vascular endothelial cells during embryonic development (26, 30, 44, 101). Recently, HIF-2α protein expression has been observed postnatally in a number of cell populations in different tissues of mice treated with hypoxia or hypoxia-mimetic agents (82, 107). In addition to being present in endothelial cells, HIF-2α mRNA has also been detected in kidney fibroblasts, liver hepatocytes, epithelial cells of the intestinal lumen, pancreatic interstitial cells, heart myocytes and interstitial cells, and lung type II pneumocytes (82, 107). In contrast to in vivo restricted expression patterns, almost all transformed cell lines exhibit HIF-2α expression (98, 108). Moreover, HIF-2α has been shown to be expressed in tumor vascular cells, parenchymal cells, and infiltrating macrophages (58, 72). These data indicate that HIF-2α might play an important role in a broad range of cells in addition to endothelial cells as well as in tumorigenesis.
There has been a long-term interest in distinguishing the roles of HIF-1α and HIF-2α. Hif-1α−/− mice exhibit mid-gestation lethality and severe blood vessel defects (42, 83). Interestingly, Hif-2α−/− mice also exhibit embryonic lethality, abnormal lung maturation, and blood vessel defects (19, 78, 100), indicating nonredundant roles of HIF-α subunits during development. Alternatively, their nonoverlapping expression patterns may contribute to mutant lethality even if they regulate similar genes, revealing the limitation of gene targeting in analysis of HIF-α function. Importantly, multiple studies of Hif-1α−/− embryonic stem (ES) cells indicate a critical role for HIF-1α in general hypoxia responses (12, 42, 83). In addition, HIF-1α has been shown to be necessary for the Pasteur effect (decreased oxidative phosphorylation and increased anaerobic fermentation under normal O2 tension) of mouse embryonic fibroblasts (MEFs) (91). However, these experiments did not address the function of other hypoxia-inducible factors, notably HIF-2α. It is important to determine if HIF-2α is expressed in these cells and whether HIF-2α is functional, if present. In the absence of HIF-2α (or in the presence of inactive HIF-2α), these hypoxia-stimulated genes identified in Hif-1α−/− ES cells or MEFs can only be interpreted as HIF-1α-inducible genes, not uniquely regulated by HIF-1α. We have determined that HIF-2α is nonfunctional in murine ES cells (C.-J. Hu et al., unpublished data). In addition, while the report of the present study was being prepared, HIF-2α was shown to be transcriptionally inactive in MEFs (75). Thus, studies using ES cells and MEFs did not reveal any function for HIF-2α. Recently, HIF-2α (and not HIF-1α) has been shown to promote tumor growth in a renal carcinoma xenograft model (53, 64), suggesting an important and unique role for HIF-2α. Thus, it is important to analyze the target genes of HIF-1α and HIF-2α and understand their individual functions.
Using DNA microarray analysis, we studied HIF-2α-inducible genes in renal cell carcinoma 786-O WT-8 cells that exclusively express HIF-2α and not HIF-1α (66). We determined that HIF-2α stimulates a number of hypoxia-inducible genes in renal cells, including some novel hypoxia-responsive genes, such as that for adipose differentiation-related protein (ADRP), which may be important for the clear cell phenotype of renal carcinomas. Importantly, in the presence of functional HIF-2α, glycolytic genes are not induced. However, exogenously introduced HIF-1α restores the hypoxia response of glycolytic genes of 786-O cells. To definitively show that hypoxia-inducible genes identified in 786-O WT-8 cells are regulated by HIF-2α and extend the observation that HIF-1α exclusively regulates glycolytic gene expression to another cell type, HEK293 tetracycline (TET)-on cells (cells to which TET was added to activate gene expression) expressing stabilized HIF-1α or HIF-2α proteins under control of doxycycline were constructed. In the TET-on system, we found that HIF-2α is functional and stimulates transcription of hypoxia-inducible genes identified in 786-O WT-8 cells but not glycolytic genes. Therefore, by analyzing endogenous gene expression, we show for the first time that HIF-2α regulates hypoxia-inducible genes involved in diverse functions, suggesting that HIF-2α function is not restricted to vascular endothelial cell-specific gene expression as previously thought. Importantly, in two HIF-2α functional cells, the classic hypoxia-inducible genes encoding glycolytic enzymes are determined to be exclusive targets of HIF-1α, providing a molecular explanation for why HIF-1α is necessary for the Pasteur effect (91).
MATERIALS AND METHODS
Cell culture.
786-O renal carcinoma cells stably transfected with either pRc/CMV empty vector (PRC3) or pRc/CMV-HA-VHL (WT-8) were gifts from William G. Kaelin (Boston, Mass.) (39). WT-8 cells stably transfected with pcDNA3-hygromycin vector (WT-8V) or pcDNA3-HIF-1α-hygromycin plasmid (WT-8H and WT-8L) were selected in the presence of hygromycin. RCC-4 renal carcinoma cells stably transfected with pcDNA3-HA-VHL (RCC-4T 3-14) were selected in the presence of 1 mg of G418/ml. These and HEK293, HepG2, and Hep3B cells were cultured in Dulbecco's modified Eagle’s medium containing 4.5 g of glucose/ml, 1% fetal calf serum, 25 mM HEPES, under 1.5% O2 for hypoxia or 21% O2 for normoxia experiments. Human umbilical vein endothelial cells (HUVEC) and human microvascular endothelial cells-lung (HMVEC-L) were purchased from Clonetics (Walkersville, Md.) and grown in EGM-2MV Bulletkit as suggested by the supplier.
RNA preparation and Northern blot analysis.
Total RNA was isolated from cells with TRIzol reagent according to the manufacture's protocol. Northern blot analysis was performed according to standard protocols (36). Human DNA fragments composed of the indicated nucleotides (nt) (with section numbers of the genes from GenBank) were generated through reverse transcription-PCR and cloned into pCRII-topo vector to serve as templates for Northern probes: ADRP, nt 992 to 1764 (BC005127); N-myc downstream regulated 1 (NDRG-1), nt 2078 to 2919 (D87953); adrenomedullin (ADM), nt 515 to 1381 (D14874); VEGF, nt 1756 to 2712 (AF022375); growth arrest- and DNA damage-induced gene (GADD45A), nt 197 to 804 (GI11423262); CHOP, nt 87 to 806 (BC003637); interleukin-6 (IL-6), nt 530 to 1058 (X04430); CA-12, nt 1412 to 2539 (AF037335); FLG, nt 2182 to 2848 (M60502); CCND-1, nt 2699 to 3927 (X59798); PI3K, nt 2074 to 3267 (Z29090); HIF-1α, nt 2247 to 2975 (U22431); HIF-2α, nt 1649 to 2695 (GI4503576); GLUT-1, nt 1697 to 2692 (K03195); GLUT-3, nt 1719 to 3678 (GI5902089); HK-1, nt 2495 to 3547 (BC008730); HK-2, nt 3458 to 4410 (GI4504392); GPI, nt 437 to 1637 (BC004982); PFKL, nt 1311 to 2303 (BC008964); ALDA, nt 1241 to 2257 (BC004333); ALDC, nt 577 to 1435 (BC003613); TPI, nt 143 to 1534 (M10036); glyceraldehyde-3-phosphate dehydrogenase, nt 377 to 1239 (AF261085); phosphoglycerate kinase 1 (PGK-1), nt 252 to 1678 (XM 010102); phosphoglucomutase 1 (PGM-1), nt 1340 to 2243 (gi13632895); ENO-1, nt 526 to 1702 (BC004458); pyruvate kinase M (PKM), nt 1388 to 2183 (BC007952); and lactate dehydrogenase A (LDHA), nt 517 to 1450, (X02152).
cRNA preparation and DNA microarray.
For DNA microarray analyses, triplicate sets of RNA samples were prepared from 786-O WT-8 cells grown under 1.5 or 21% O2 for 16 h. cDNA was generated from total RNA with the Superscript Choice system (Gibco BRL Life Technologies) and T7-(dT)24 primers. Subsequently, biotin-labeled ribonucleotides were synthesized with a BioArray high-yield RNA transcript labeling kit (Enzo Diagnostic, Inc.). Fragmented cRNA was tested for quality first by a test array and then subjected to hybridization with human genome U95A arrays containing approximately 12,000 known mRNA transcripts (Affymetrix). Data analysis was performed by Genechip expression analysis software. First, individual samples were analyzed for absolute call and average differentiation. Then gene expression levels from two samples (normoxia and hypoxia) were subjected to comparative analysis for all three sets of data.
Nuclear extract preparation and Western blot analysis of HIF-1α, HIF-2α, and ARNT.
Nuclear extracts were prepared in the presence of proteinase inhibitors as well as 200 μM deferoxamine (DFX) as described previously (36). Comparable amounts of protein as determined by Bradford protein assay (Bio-Rad) were resolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After blockage in Tris-buffered saline containing 5% nonfat milk, membranes were probed with anti-HIF-1α monoclonal antibody (NB 100-105; Novus Biological, Inc), anti-HIF-2α polyclonal antibody (NB 100-122; Novus), or anti-ARNT monoclonal antibody (NB 100-124; Novus) in Tris-buffered saline with 3% nonfat milk. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G for polyclonal primary antibody or horseradish peroxidase-linked anti-mouse immunoglobulin G for monoclonal primary antibody in combination with SuperSignal Chemiluminescent Substrate were used for signal detection.
Plasmid construction.
Full-length murine HIF-1α or HIF-2α cDNAs were generated by reverse transcription-PCR and inserted into a pcDNA.3 expression vector by using primers incorporated with restriction sites. For HIF-1α, the primer pairs are 5′-ATAggatcc(BamHI)ATGGAGGGCGCCGGCGGCGAGAAC-3′ (sense) and 5′-CGCctcgag(XhoI)TCAGTTAACTTGATCCAAAGCTCTG-3′ (antisense). For HIF-2α, the primer pairs are 5′-CATggtacc(KpnI)CGGCGACAATGACAGCTGACAAGGAG-3′ (sense) and 5′-CATtctaga(XbaI)TCAGGTGGCCTGGTCCAGAGCTCTGAG-3′ (antisense). Underlined letters indicate the start codon (ATG) and stop codon (TCA) of HIF-1α and HIF-2α. Lowercase letters indicate restriction sites.
A 10-amino-acid c-myc tag was further inserted at the C terminus of both HIF-α cDNAs by using a Pfu-PCR-based protocol. For HIF-α proline mutants, the same Pfu-PCR protocol was used to change proline residues to alanines by amplifying the full-length plasmid with primers incorporated with mutation (indicated in lowercase) as follows: for HIF-1α P577A, 5′-gccTATCCCAATGGATGATGATTTCCAG-3′ and 5′-AGCCAGCATCTCCAAATCTAAATCAG-3′; for HIF-1α P402A, 5′-GCTGCCGGCGACACCATCATCTCTC-3′ and 5′-ggcAGCCAGCAGAGTGAGAGCATCAGGC-; for HIF-2α P530A, 5′-gcc-TACATCCCTATGGACGGCGAGG-3′ and 5′-TGCCAAGGTCTCCAAATCCAGTTC-3′; and for HIF-2α P405A, 5′-ACCCCAGGAGATGCCATTATTTCTC-3′ and 5′-ggcGGCCAACTGGGCCAGCTCCTCGGGCTC-3′.
Construction of HEK293 TET-on HIF-αDPA clones.
Full-length cDNA of HIF-1αmycDPA (double prolines to alanines) or HIF-2αmycDPA in pcDNA.3 was amplified and inserted into a TET response element (TRE) vector (pTRE2hyg; Clontech) by using the following primer pairs: for HIF-1α, 5′-ACCAacgcgt(MluI)CCGCCATGGAGGGCGCCGGCGGCGAGAAC-3′ and 5′-ACCAgcggccgc(NotI)TCACAGATCCTCTTCTGAGATGAG-3′; for HIF-2α, 5′-ACCAacgcgt(MluI)CCGCCATGACAGCTGACAAGGAGAAAAAAAGG-3′ and 5′-ACCAgcggccgc(NotI)TCACAGATCCTCTTCTGAGATGAG-3′.
HEK293 TET-on HIF-αDPA clones were constructed by transfection of HEK293 TET-on cells (Clontech) with pTRE2hygHIF-1αDPA or pTRE2hygHIF-2αDPA in addition to pTet-tTS vector in the presence of 50 μg of hygromycin/ml and 100 μg of G418/ml.
Transient transfection.
Transfection of HEK293 cells with reporter and HIF-α expression plasmid vectors was performed with Fugene 6 (Roche) according to manufacturer's instructions. The wild-type HRE-luciferase reporter as well as a mutant HRE-luciferase reporter were previously described (3). A total of 50 to 60% confluent HEK293 cells in 6-well plates were transfected with 200 ng of reporter, 200 ng of expression plasmid, and 100 ng of reference plasmid murine sarcoma virus β-galactosidase. After transfection for 40 h, cells were collected in 1× lysis buffer (Promega) and assayed for luciferase activity as a function of HIF-α and β-galactosidase activity to monitor the transfection efficiency.
Glucose assay and VEGF enzyme-linked immunosorbent assay.
For glucose concentration and VEGF protein secretion assays, the same number of HEK293 TET-on HIF-1αDPA or HEK293 TET-on HIF-2αDPA or HEK293 TET-on parental cells were grown in high-glucose Dulbecco's modified Eagle’s medium (4.5 g/ml) with (1 μg/ml) or without doxycycline. A total of 400 μl of the medium was collected for the glucose and VEGF assay at indicated times. The amount of glucose was quantitated with Sigma Diagnostics glucose reagents. VEGF quantitation was performed with a QuantiKine human VEGF immunoassay kit (R&D Systems).
RESULTS
Hypoxia-inducible genes in 786-O WT-8 cells.
Both HIF-1α and HIF-2α have been implicated in hypoxia responses (19, 106). However, their relative contributions to hypoxic gene induction are unknown, since most O2-regulated genes have been identified in cells that express both HIF-1α and HIF-2α. A well-studied cell line, 786-O WT-8, was chosen to evaluate HIF-2α target genes (38, 39, 66). 786-O cells were originally isolated from patients with renal clear cell carcinoma and exhibited mutated VHL. Stable transfection with either pRc/CMV empty vector or pRc/CMV-HA-tagged VHL gave rise to 786-O PRC-3 and 786-O WT-8, respectively (38). Northern blot analysis of total RNA from 786-O cells detected no HIF-1α mRNA (Fig. (Fig.1,1, lanes 1 to 4), a result that is consistent with previous reports (66). Importantly, no HIF-1α protein was detected even in the VHL-defective 786-O PRC-3 cells (Fig. (Fig.1,1, lanes 1 and 2). However, 786-O cells do express HIF-2α protein. Consistent with their VHL status, 786-O PRC-3 cells grown at both 21 or 1.5% O2 expressed similar levels of HIF-2α (Fig. (Fig.1,1, lanes 1 and 2), but 786-O WT-8 cells exhibited O2-dependent regulation of HIF-2α protein (Fig. (Fig.1,1, lanes 3 and 4).
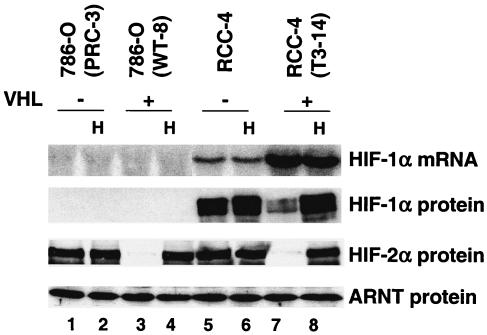
786-O cells express HIF-2α but not HIF-1α. 786-O (PRC-3) is a VHL-defective renal carcinoma cell line, whereas 786-O (WT-8) cells have been stably transfected with functional VHL (38). RCC-4 is another VHL-mutant renal carcinoma cell line, expressing both HIF-1α and HIF-2α. The RCC-4 (T3-14) cells have been stably transfected with functional VHL. Enhanced expression of HIF-1α mRNA in VHL-transfected RCC-4 cells has previously been observed (55). Hypoxia (H) treatment (1.5% O2 for 16 h) stabilized HIF-2α protein in 786-O (WT-8) cells and stabilized both HIF-1α and HIF-2α in RCC-4 (T3-14) cells as shown by HIF-1α and HIF-2α Western blots. The ARNT Western blot served as a protein loading control.
Another VHL-defective renal carcinoma cell line, RCC-4, expresses both HIF-1α and HIF-2α proteins, independent of O2 tension (Fig. (Fig.1,1, lanes 5 and 6). Stable transfection of pcDNA3-HA-tagged VHL (RCC-4T 3-14) reduced normoxic HIF-α protein levels, restoring hypoxia inducibility (Fig. (Fig.1,1, lanes 7 and 8).
To systematically study HIF-2α-inducible genes, triplicate RNA samples were prepared from 786-O WT-8 cells cultured under normoxia (21% O2) or hypoxia (1.5% O2) for 16 h. Double-stranded cDNA was generated from total RNA with the Superscript Choice System (Gibco BRL Life Technologies) and T7-(dT)24 primers. Subsequently, antisense biotin-labeled RNA was synthesized with T7 DNA polymerase. The RNA probe was fragmented and subjected to hybridization with human genome U95A arrays containing approximately 12,000 known mRNA transcripts (Affymetrix). A comparative analysis of genes expressed under hypoxia versus normoxia indicated that 1.5% O2 treatment stimulated ~30 genes in the triplicate RNA samples. However, expression of some genes is reduced by hypoxia (data not shown). The induced genes included novel hypoxia-inducible genes such as those for ADRP, chemokine growth-regulated oncogene 2 (GRO-2), baculoviral inhibitor of apoptosis (IAP) repeat-containing protein 3 (BIRC-3; also called cIAP-2), and known HIF target genes encoding VEGF and ADM in 786-O cells (Table (Table1).1). However, another known HIF target in 786-O cells, platelet-derived growth factor, B chain (PDGF-B), is not on the human genome U95A arrays. ADRP protein is frequently found at the surface of lipid droplet (the spherical organelles for neutral lipid storage) and has been shown to promote lipid accumulation and lipid droplet formation (40, 90). Its overexpression in VHL-defective renal clear cell carcinoma may explain the clear cell phenotype, a phenomenon that arises when a large amount of lipid droplets in such cells is removed during sample preparation for microscopy.
TABLE 1.
Genes induced in triplicate sets of hypoxic mRNAs as determined by DNA microarray assaya
| Gene | AC | Description | Avg fold induction | Confirmation |
|---|---|---|---|---|
| ADRP | X97324 | Adipose differentiation-related protein | 2.1 | Y |
| NDRG-1 | D87953 | N-myc downstream regulated-1 | 2.3 | Y |
| APG5L | Y11588 | APG5 (autophagy 5, S. cerevisiae)-like | 1.8 | ND |
| DMXL-1 | AJ005821 | Dmx-like 1 | 2.4 | Y |
| GRO-2 | M36820 | GRO2 oncogene | 2.3 | ND |
| CST | D88667 | Cerebroside (3′-phosphoadenylylsulfate:galactosylceramide 3′) sulfotransferase | 1.6 | ND |
| NFIL-3 | X64318 | Nuclear factor, interleukin 3 regulated | 1.5 | ND |
| AKAP-12 | U81607 | A kinase (PRKA) anchor protein (gravin) 12 | 1.6 | ND |
| POU5F-1 | Z11898 | POU domain, class 5, transcription factor 1 | 1.4 | ND |
| BIRC-3 | U45878 | Baculoviral IAP repeat-containing 3 | 1.8 | ND |
| AA142964 | ESTs, weakly similar to ORF YOR126c [S. cerevisiae] | 1.4 | ND | |
| KIAA0203 | D86958 | KIAA0203 gene product | 1.9 | ND |
| PIK3CA | Z29090 | Phosphoinositide-3-kinase, catalytic, alpha polypeptide | 1.9 | Y |
| DUSP-7 | A1655015 | Dual-specificity phosphatase 7 | 2.1 | ND |
| TPP-2 | M73047 | Tripeptidyl peptidase II | 2.1 | ND |
| SLC16A4 | U59185 | Solute carrier family 16 (monocarboxylic acid transporters), member 4 | 3.1 | ND |
| KIAA0710 | AB014610 | KIAA0710 gene product | 3.1 | ND |
| NDUFB6 | A1302176 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 6 (17 kDa, B17) | 2.4 | ND |
| GADD45A | M60974 | Growth arrest and DNA-damage-inducible, alpha | 1.5 | Y |
| GADD153 | BC003637 | Similar to DNA-damage-inducible transcript 3 | 1.7 | Y |
| IGFBP-3 | M35878 | Insulin-like growth factor binding protein 3 | 1.5 | ND |
| UNG | Y09008 | Uracil-DNA glycosylase | 1.7 | ND |
| VEGF | AF024710 | Vascular endothelial growth factor | 1.4 | Y |
| IL-6 | X04430 | Interleukin 6 (interferon, beta 2) | 2.0 | Y |
| CA-12 | AF037335 | Carbonic anhydrase XII | 2.2 | Y |
| FLG | M60502 | Filaggrin | 1.5 | Y |
| CCND-1 | X59798 | Cyclin D1 (PRAD1: parathyroid adenomatosis 1) | 1.8 | Y |
| SLC2A1 | K03195 | Solute carrier family 2, member 1 | 2.0 | Y |
| ADM | D14874 | Adrenomedullin | 2.3 | Y |
Some of these hypoxia-inducible genes identified by DNA microarray assays were further tested, and all those tested were confirmed by Northern blot analysis in triplicate RNA samples, including VEGF, which exhibited a 1.4-fold induction in the DNA microarrays (Table (Table1).1). Since the expression pattern of these genes was identical in all three sets of RNA samples, only one set of Northern blotting results is presented. Figure Figure2A2A shows the results for several genes, likely to be HIF-2α targets, including a novel hypoxia-inducible gene, the ADRP gene (induction, 2.6-fold based on phosphorimager analysis of Northern blots), and three known hypoxia-inducible genes, NDRG-1 (2.0-fold), ADM (1.8-fold), and VEGF (1.8-fold) genes (Fig. (Fig.2A,2A, lane 4 versus lane 3). To separate HIF-dependent transcription stimulation from other hypoxia effects, such as hypoxia-induced mRNA stability (59, 61, 95) or hypoxia-induced AP-1 (4, 5) and NF-κB activity (29, 88), target gene expression was also assessed in otherwise genetically identical VHL-defective 786-O PRC-3 cells. Interestingly, high levels of gene expression were observed in 786-O PRC cells, independent of O2 tension (Fig. (Fig.2A,2A, lanes 1 and 2 versus lane 3). The target gene expression (Fig. (Fig.2A)2A) clearly correlated with HIF-2α protein levels (Fig. (Fig.1),1), suggesting that HIF-2α, and not other hypoxia-stimulated transcription factors such as AP-1 and NF-κB, activates the expression of these genes. ADRP (induction, 2.5-fold), NDRG-1 (2.7-fold), ADM (2.9-fold), and VEGF (2.4-fold) were also stimulated by hypoxic treatment of RCC-4T 3-14 (with functional pVHL) (Fig. (Fig.2A,2A, lane 9 versus lane 8) and their expression levels correlated well with HIF-α protein levels (Fig. (Fig.1,1, lanes 5 to 8). However, we could not confirm which HIF-α subunit plays a role in RCC-4 gene activation, as RCC-4 cells express both HIF-1α and HIF-2α.
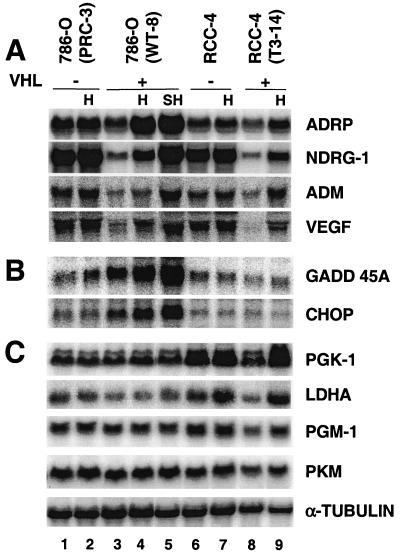
Confirmation of DNA microarray data by Northern blot analyses. (A) Hypoxia (H) (1.5% O2 for 16 h) stimulates expression of ADRP, NDRG-1, ADM, and VEGF in both 786-O (WT-8) and RCC-4 (T3-14) cells and is likely to be HIF-dependent since these genes are overexpressed in the parental 786-O (PRC-3) and RCC-4 cells independent of O2. Severe hypoxia (SH) (0.1% O2 for 16 h) further enhances hypoxic gene expression in comparison to 1.5% O2. (B) GADD45A and CHOP are hypoxia-responsive genes but are independent of HIF activity. (C) PGK-1, LDHA, PGM-1, and PKM mRNAs, which encode four glycolytic enzymes, are not stimulated by hypoxic treatment (1.5 or 0.1% O2) of 786-O cells but are induced in RCC-4 cells.
The expression of GADD45A (induction, 1.9-fold) and GADD153 (also called CHOP; induction, 1.8-fold) were also induced by low O2 (Fig. (Fig.2B,2B, lane 4 versus lane 3) as previously reported (79). However, in contrast to the expression of the above-described genes, the expression of GADD45A and CHOP appears to be independent of the HIF-α/VHL pathway, since their expression did not correlate with HIF-2α protein levels. In conclusion, analysis of HIF-2α target genes by DNA microarray and Northern blot analysis in VHL-defective cells as well as in cells with functional VHL demonstrated that HIF-2α is active in 786-O cells. Moreover, several novel functionally important genes were identified (see Discussion).
HIF-2α is not involved in regulating the glycolytic pathway.
As shown by numerous reports, genes involved in glucose metabolism are ubiquitously expressed and stimulated by hypoxia (92, 93, 106). With similar DNA microarray analyses, most glycolytic genes were induced in hypoxia-treated wild-type murine ES cells (Hu et al., unpublished data). Surprisingly, no glycolytic gene induction was observed in O2-deprived 786-O WT-8 cells, although all the glycolytic genes are present on the human genome U95A DNA microarray. The expression of 13 glycolytic genes was individually tested by Northern blot analysis, confirming the DNA microarray data indicating that these genes are not induced by hypoxia in 786-O WT-8 cells (Fig. (Fig.2C,2C, lane 4 versus lane 3; also data not shown). Figure Figure2C2C shows the results of four representative genes, including two strongly hypoxia-responsive genes, corresponding to PGK-1 and LDHA, and two moderately hypoxia-responsive genes, PGM-1 and PKM. Expression of the four genes was not responsive to hypoxia treatment in 786-O WT-8 cells (Fig. (Fig.2C,2C, lane 4 versus 3). Severe hypoxia treatment (0.1% O2) also failed to enhance the expression of these particular genes (Fig. (Fig.2C,2C, lane 5 versus lane 3). In contrast, elevated induction at 0.1% O2 was observed for HIF-2α-responsive genes, such as those for ADRP (3.5-fold), NDRG-1 (6.6-fold), ADM (3.7-fold), and VEGF (3.1-fold) (Fig. (Fig.2A,2A, lane 5 versus 3). It is noteworthy that the normoxic level of glycolytic genes in 786-O WT-8 cells was comparable to that in VHL-defective parental 786-O PRC-3 cells (Fig. (Fig.2C,2C, lane 1 versus lane 3), indicating that HIF-2α had no effect on their expression. In contrast, O2-deprived RCC-4T 3-14 cells (which express both HIF-1α and HIF-2α) exhibited hypoxic induction of PGK (2.5-fold), LDHA (3.0 old), PGM-1 (1.6-fold), and PKM (1.5-fold) gene expression in parallel experiments (Fig. (Fig.2C,2C, lane 9 versus lane 8). The glycolytic genes also appeared to be direct targets of HIF, since VHL status clearly affected the expression level of each gene (Figure C, lanes 6 to 9).
HIF-1α is associated with up-regulation of glycolytic genes in multiple cell types.
The presence of HIF-1α in RCC-4 cells and the absence of HIF-1α in 786-O cells may explain why glycolytic genes were induced by hypoxia only in RCC-4 cells. To investigate the connection between HIF-1α and hypoxic glycolytic gene induction, we surveyed HIF-1α and HIF-2α expression and glycolytic gene inducibility in multiple cell lines. HUVECs and HMVEC-Ls are two primary cells from normal donors, while the remaining three cell lines, HEK293, HepG2, and Hep3B, are transformed. All five cell lines expressed similar levels of HIF-1α mRNA, while the two endothelial cells expressed higher levels of HIF-2α mRNA (Fig. (Fig.3A),3A), a result that is consistent with a previous study showing that the levels of HIF-1α mRNA varied much less than that of HIF-2α mRNA in a number of cell types (108). The levels of HIF-α mRNA under hypoxia were slightly reduced relative to that under normoxia, indicating that transcription of HIF-1α and HIF-2α is not elevated by hypoxia. However, hypoxia-mimetic agents increased the levels of both HIF-1α and HIF-2α proteins in all five cell types (Fig. (Fig.3B).3B). In general agreement with HIF-1α mRNA levels, all the five cell lines (except HMVEC-L) exhibited similar levels of HIF-1α protein. The much lower level of HIF-1α protein in HMVEC-L cells (primary cells from lung) is consistent with a previous study with similar HMEC-1 cells (simian virus 40 large T antigen transformed from dermal vessels) (108). The two endothelial cells exhibited higher levels of HIF-2α protein than 293 and HepG2 cells expressed, in keeping with their mRNA levels. In contrast, Hep3B cells expressed the highest levels of HIF-2α protein among all five cell lines although it expresses less HIF-2α mRNA than either endothelial cell line. Interestingly, different levels of hypoxic gene induction were observed in various cell lines. While high levels of glycolytic gene induction were exhibited by HepG2 (PGK, 7.5-fold; LDHA, 4.4-fold) and Hep3B cells (PGK, 4.5-fold; LDHA, 3.5-fold), low levels of induction were exhibited by HUVEC (PGK, 1.5-fold; LDHA, 1.8-fold), HMVEC-L (PGK, 1.5-fold; LDHA, 2.4-fold), and HEK293 cells (PGK, 2.5-fold; LDHA, 2.4-fold). However, this low level of induction is not specific for glycolytic genes, since HIF-2α inducible genes (NDRG-1 and VEGF genes) also exhibited low levels of induction in HUVECs (NDRG-1, 1.4-fold; VEGF, 1.6-fold), HMVEC-Ls (NDRG-1, 2.4-fold; VEGF, 2.4-fold), and HEK293s (NDRG-1, 2.8-fold; VEGF, 2.5-fold) and higher levels of induction in HepG2s (NDRG-1, 12-fold; VEGF, 9.5-fold) and Hep3Bs (NDRG-1, 6-fold; VEGF, 3.0-fold). Cell type rather than the amount of HIF-α may determine the level of hypoxic gene induction, since low levels of induction were observed in HUVEC-L cells expressing high levels of both HIF-1α and HIF-2α. Importantly, all five HIF-1α-expressing cells exhibited glycolytic gene induction under low O2 levels, consistent with a role for HIF-1α in regulating glycolytic enzyme expression (Fig. (Fig.3C3C).
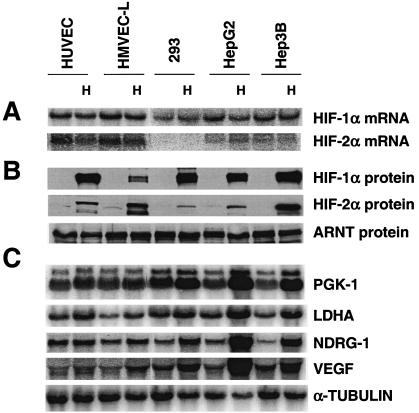
HIF-1α is associated with hypoxic induction of glycolytic genes in multiple cell types. (A) Northern blot analysis of HIF-1α and HIF-2α expression in two primary endothelial cell lines and three transformed cell lines. HIF-1α is expressed in all five lines with similar levels, whereas HIF-2α is expressed highly in two endothelial cells. (B) Western blot analysis of HIF-1α and HIF-2α protein in cells treated with hypoxia-mimetic DFX (100 μM) for 6 h. The levels of HIF-1α and HIF-2α protein expression are in general agreement with their mRNA levels, except for the levels of HIF-1α in HMVEC-L cells and HIF-2α in Hep3B cells. (C) Hypoxic treatment (1.5% O2 for 16 h) increases the expression of the glycolytic genes (PGK and LDHA) and HIF-2α inducible genes (NDRG-1 and VEGF) in all five cell lines.
Stable transfection of HIF-1α into 786-O WT-8 cells restores glycolytic gene induction.
To formally prove that a lack of HIF-1α, and not genetic defects in glycolytic genes, leads to glycolytic gene unresponsiveness to hypoxic treatment of 786-O cells, we introduced wild-type murine HIF-1α cDNA into 786-O WT-8 cells and made stable HIF-1α transfectants. As shown in Fig. Fig.4A,4A, WT-8L cells express low levels of transfected HIF-1α mRNA (Fig. (Fig.4A,4A, lanes 5 and 6), whereas WT-8H cells express high levels of HIF-1α mRNA (Fig. (Fig.4A,4A, lanes 7 and 8). The empty vector-transfected WT-8V cells (Fig. (Fig.4A,4A, lanes 3 and 4) and parental 786-O WT-8 cells (Fig. (Fig.4A,4A, lanes 1 and 2) do not express HIF-1α mRNA. Although both WT-8L and WT-8H clones express detectable HIF-1α mRNA levels, no HIF-1α protein was detected under normal O2 concentrations (Fig. (Fig.4A,4A, lanes 5 and 7). Hypoxic treatment stabilized HIF-1α protein at levels correlating with mRNA levels (Fig. (Fig.4A,4A, lanes 6 and 8) with WT-8H cells exhibiting a level of HIF-1α protein similar to that of control HEK293 cells under hypoxia (Fig. (Fig.4A,4A, lane 10). Identical amounts of HIF-2α protein were detected in hypoxia-treated WT-8 and WT-8H cells, indicating that expression of HIF-1α had no effect on the level of HIF-2α (data not shown). Importantly, glycolytic genes such as those for LDHA and PGK-1 were induced in the HIF-1α transfectants (Fig. (Fig.4B).4B). The level of induction also appeared to correlate with HIF-1α protein levels, as stronger induction was observed in the WT-8H cells (PGK, 1.7-fold; LDHA, 2.1-fold), which express higher levels of HIF-1α (Fig. (Fig.4B,4B, lane 8 versus lane 7) and lower induction in the WT-8L cells (PGK, 1.3-fold; LDHA, 1.4-fold) (Fig. (Fig.4B,4B, lane 6 versus lane 5). This level of induction in WT-8H cells is comparable to that seen in HEK293 cells (PGK, 1.9-fold; LDHA, 1.8-fold) (Fig. (Fig.4B,4B, lane 10 versus lane 9). These results demonstrated that the absence of hypoxic glycolytic gene induction in 786-O WT-8 cells is due to the lack of HIF-1α in these cells, suggesting that HIF-1α is the only hypoxia-inducible factor able to target glycolytic gene expression.
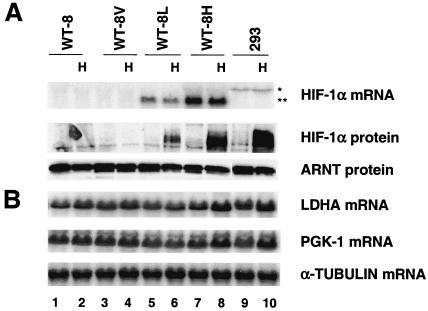
HIF-1α expression in 786-O cells restores the glycolytic gene response to hypoxia treatment. (A) Establishment and characterization of 786-O (WT-8) clones stably transfected with HIF-1α. WT-8L cells express low levels of HIF-1α, whereas WT-8H cells express higher levels of HIF-1α as demonstrated by HIF-1α Northern blot and Western blot assays. The pcDNA3 vector-transfected control WT-8V cells and parental WT-8 cells express no HIF-1α RNA or protein. HEK293 cells, whose endogenous HIF-1α mRNA (asterisk) exhibits a larger size than transfected HIF-1α mRNA (double asterisk), served as positive control for HIF-1α protein. (B) Northern blot analysis of two glycolytic genes in HIF-1α-rescued WT-8 cells, showing that LDHA and PGK-1 are induced in WT-8H cells after treatment with 1.5% O2 for 16 h. The level of induction in WT-8H is comparable to that in HEK293 cells by endogenous HIF-1α.
Generation of cell lines expressing stabilized HIF-1α and HIF-2α.
Our data raise several important questions. (i) Is the inability of HIF-2α to regulate glycolytic genes a cell-specific event? (ii) Can HIF-1α also regulate the hypoxia-responsive genes identified in 786-O WT-8 cells, or are these hypoxia-inducible genes uniquely regulated by HIF-2α? (iii) Is HIF-2α or some other factor responsible for the observed hypoxia response in 786-O cells? To address these questions, we employed a TET-on gene expression system in which the expression of HIF-1α or HIF-2α mRNA is regulated by the addition of doxycycline, allowing us to study individual functions of HIF-α subunits. Expression of wild-type HIF-α mRNA did not result in expression of the HIF-α protein under normoxia, as expected (Fig. (Fig.4A).4A). To stabilize transfected HIF-α protein under normoxia, two hydroxylated prolines in both HIF-1α and HIF-2α oxygen-dependent-degradation domains (41, 43, 65) were mutated to alanine (Fig. (Fig.5A).5A). Using transient transfection and Western blot assays, we evaluated HIF-α protein normoxic stability by comparing protein levels recovered from transfected cells with or without 100-μM DFX treatment for 6 h. Consistent with previous reports, HIF-1α P577A or HIF-2α P530A exhibited higher protein levels than wild-type HIF-α under normoxia, but DFX treatment could still enhance protein levels of these single proline mutants (41, 43). The double proline mutants exhibited even higher stability than single proline mutants, and DFX lost its stabilizing effect (Fig. (Fig.5B)5B) (65). Importantly, double proline mutants functioned like the wild-type HIF-α, as demonstrated by transactivation of the wild-type HRE-Luc reporter but not the mutant HRE-Luc reporter under normoxia (Fig. (Fig.5C).5C). Higher activation of these reporters by HIF-α proline mutants likely reflects their increased stability.
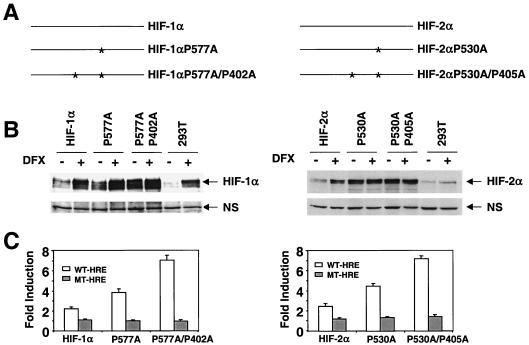
Mutagenesis of hydroxylated prolines confers HIF-1α and HIF-2α function to normoxic cells. (A) Schematic representation of HIF-1α and HIF-2α single and double proline mutants. (B) Double proline HIF-1α and HIF-2α mutants are stabilized under normal O2 tension. HEK293-T cells transfected with wild-type single-proline mutant and double-proline mutant HIF-α subunits were split into two dishes, one of which was treated with DFX for 6 h before nuclear extraction preparation. The amount of HIF-α protein was measured by Western blot analysis with specific antibodies and a nonspecific (NS) band is shown for protein loading control. (C) HIF-α double proline mutants are highly active in wild-type (WT)-HRE-Luc reporter transactivation assays in normoxic cells. The mutant (MT)-HRE-Luc reporters are not regulated by HIF-1α or HIF-2α.
HIF-1αDPA and HIF-2αDPA cDNAs under transcriptional control of the TRE were stably transfected into HEK293 TET-on cells, which express the reverse TET-controlled transactivator. To block background expression of transfected HIF-α mRNA from these constructs, TET-controlled transcription silencer (tTS) was also cotransfected. In the absence of doxycycline, tTS binds to the TET operator (TET O) DNA sequence in the TRE region, preventing HIF-α expression. Addition of doxycycline dissociates tTS from the TET O sequence and activates transcription by the TET-controlled transactivator. HEK293 TET-on HIF-1αDPA clones (Fig. (Fig.6A,6A, left) and HIF-2α DPA clones (Fig. (Fig.6A,6A, right) exhibited tight regulation of expression of transfected HIF-α mRNA. No exogenous HIF-α mRNA was detected without doxycycline. Transfected HIF-α mRNA expression was induced when cells were treated with 0.1 μg of doxycycline/ml and higher induction was obtained in a dose-dependent manner which reached a plateau at 1 μg of doxycycline/ml (Fig. (Fig.6A).6A). However, expression of endogenous HIF-1α mRNA was not changed by exogenous HIF-1α mRNA (Fig. (Fig.6A6A left), verifying our experimental design that only exogenous HIF-α function will be evaluated. The induction of HIF-α mRNA by doxycycline occurred rapidly. HIF-α mRNA was detected after cells were treated with 1 μg of doxycycline/ml for 4 h and maximal levels were reached at 20 h, sustaining the high levels of HIF-α mRNA for at least 48 h (data not shown). Importantly, the addition of doxycycline induced expression of not only HIF-α mRNA but also the HIF-α protein under normoxia, as demonstrated by Western blot analysis of HIF-α proteins in nuclear extracts of two independent HEK293 TET-on HIF-1α DPA clones (Fig. (Fig.6B,6B, left) and two independent HEK293 TET-on HIF-2α DPA clones (Fig. (Fig.6B,6B, right). The establishment of HEK293 TET-on HIF-αDPA clones allows assessment of HIF-1α and HIF-2α function in the absence of other hypoxic mechanisms.
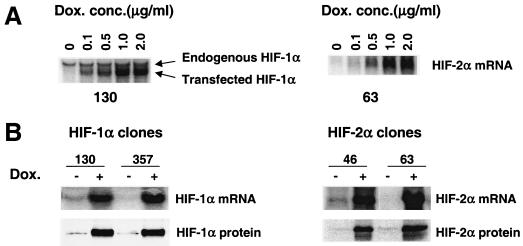
Establishment of HEK293 TET-on HIF-1αDPA and HIF-2αDPA clones. (A) HIF-α mRNA expression is tightly regulated by doxycycline. Northern blot analysis of HIF-1α and HIF-2α mRNA expression in HEK293 TET-on HIF-1αDPA clone 130 (left) and HEK293 TET-on HIF-2αDPA clone 63 (right) treated with different amounts of doxycycline for 20 h. (B) Doxycycline (1 μg/ml for 20 h) induced HIF-1α mRNA and protein expression in two independent HEK293 TET-on HIF-1αDPA clones (left) and HIF-2α mRNA and protein expression in two independent HEK293 TET-on HIF-2αDPA clones (right). Dox. conc., concentration of doxycycline.
HIF-1α is a master regulator of hypoxia responses.
Total RNA was harvested from two independent HEK293 TET-on HIF-1αDPA clones (130 and 357) and HEK293 TET-on parental cells, after treatment with (1 μg/ml) or without doxycycline for 20 h under normal O2 levels. The addition of doxycycline in HEK293 TET-on HIF-1αDPA clones 130 and 357 induced hypoxic gene expression, including PGK-1 (2.4- and 2.6-fold, respectively), LDHA (2.2- and 1.5-fold, respectively), NDRG-1 (2.6- and 3.6-fold, respectively), and VEGF (2.6 and 2.2-fold, respectively) (Fig. (Fig.7A).7A). Other hypoxia-responsive genes, such as those corresponding to PGM-1, PKM, ALDA, ADRP, ADM, and GLUT-1, were also induced by HIF-1α in HEK293 TET-on HIF-1αDPA cells (data not shown). Importantly, doxycycline had no effect on the expression of these genes in HEK293 TET-on parental cells (data not shown). These data suggest that HIF-1α is a master hypoxic regulator and can activate all HIF-dependent hypoxia-inducible genes we have identified thus far. Interestingly, expression of the GADD45A and CHOP genes was not induced by HIF-1α (data not shown), confirming our previous observation in 786-O cells that hypoxic regulation of these genes is independent of HIF activity.
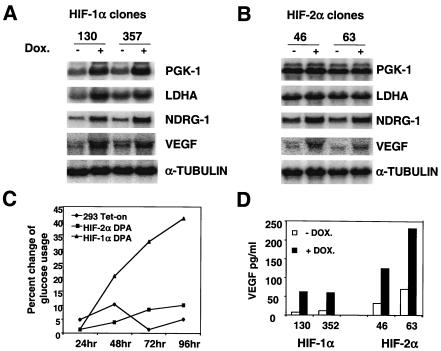
HIF-1α and not HIF-2α stimulates genes encoding glycolytic enzymes in HEK293 TET-on cells. (A) Northern blot analysis of two glycolytic genes and two HIF-2α-responsive genes in HEK293 TET-on HIF-1αDPA clones treated with 1 μg of doxycycline/ml for 20 h (+). −, no doxycycline. HIF-1α increases the transcription of all four genes. (B) Northern blot analysis of two glycolytic genes and two HIF-2α-responsive genes in HEK293 TET-on HIF-2αDPA clones treated with 1 μg of doxycycline/ml for 20 h. HIF-2α induced NDRG-1 and VEGF expression but not glycolytic gene expression. (C) Addition of doxycycline (1 μg/ml) increases the glucose usage of a HEK293 TET-on HIF-1DPA clone but not a HEK293 TET-on HIF-2DPA clone or HEK293 TET-on parental cells. The percent change of glucose usage was defined as [glucose (−Dox) − glucose (+DOX)]/glucose (−DOX). (D) Doxycycline treatment (1 μg/ml for 48 h) increases VEGF protein secretion in both HEK293 TET-on HIF-1DPA and HIF-2DPA clones.
Expression of some hypoxia-inducible genes was induced in 786-O cells (Fig. (Fig.2A).2A). We assumed that these genes are regulated by HIF-2α and not by HIF-3α (or even unidentified HIFs) in 786-O cells. The HEK293 TET-on HIF-2αDPA clones allowed us to confirm that these genes are bona fide HIF-2α targets. Expression of the HIF-2αDPA protein in clones 46 and 63 increased NDRG-1 (2.2- and 2.1-fold, respectively), VEGF (2.5- and 1.9-fold, respectively),ADRP, ADM, and GLUT-1 expression (Fig. (Fig.7B7B and data not shown), consistent with the Northern blot data in Fig. Fig.2,2, demonstrating that HIF-2α is functional and capable of regulating these hypoxia-regulated genes in both HEK293 and 786-O cells. In contrast, HIF-2α did not activate PGK-1, LDHA, PGM-1, PKM and ALDA (Fig. (Fig.7B7B and data not shown), providing independent confirmation that HIF-2α does not transactivate glycolytic genes in another cell type.
HIF-1α, but not HIF-2α, promotes cellular glucose consumption.
To further study the role of HIF-1α and HIF-2α in glucose metabolism, glucose consumption was determined in HEK293 TET-on HIF-1αDPA, HIF-2αDPA and parental HEK293 TET-on cells by assaying glucose concentration in the culture media as a function of HIF-α expression. Results are presented as percent change of glucose usage, defined as [glucose (no doxycycline) − glucose (with doxycycline)]/glucose (no doxycycline). The addition of doxycycline significantly increased glucose consumption by HEK293 TET-on HIF-1α DPA clones after 48 h (Fig. (Fig.7C7C and data not shown). Although glycolytic gene expression was induced as early as 4 h posttreatment (data not shown), no significant glucose concentration change was observed until 48 h, indicating that there is a delay between glycolytic gene expression and increased glucose catabolism. However, doxycycline treatment of HEK293 TET-on HIF-2αDPA and parental HEK293 TET-on cells had no effect on glucose consumption, which is consistent with a Northern blot analysis showing that HIF-1α (but not HIF-2α) increases glycolytic gene expression. On the other hand, doxycycline treatment enhanced VEGF protein secretion in both HEK293 TET-on HIF-1αDPA and HIF-2αDPA clones, correlating well with VEGF mRNA expression. These results indicate that both HIF-1α and HIF-2α regulate VEGF expression. Using the TET-on system in combination with normoxia functional HIF-1α and HIF-2α, we clearly demonstrate a unique role for HIF-1α in glycolytic gene regulation and a common function for HIF-1α and HIF-2α in regulating a subset of hypoxia-responsive genes.
DISCUSSION
Functional studies of HIF-1α and HIF-2α are complicated by the fact that most cell lines express both HIF-1α and HIF-2α (see Fig. Fig.3A)3A) (98, 108). Hif-1α−/− and Hif-2α−/− ES cells would provide a perfect system to study HIF-2α target genes in Hif-1α−/− ES cells or HIF-1α target genes in Hif-2α−/− ES cells. Analysis of Hif-1α−/− ES cells suggested that HIF-1α, but not HIF-2α, plays a fundamental role in hypoxia responses (12, 42, 83). In agreement with this suggestion, analysis of Hif-2α−/− ES cells indicated that HIF-2α is not involved in hypoxia in ES cells (11). In contrast to the ES cell studies, HIF-2α has been shown to be important in stimulating several hypoxia-inducible genes (including those for tyrosine hydroxylase and VEGF) during embryonic development (19, 78, 100). Different temporal and spatial expression patterns of HIF-1α and HIF-2α in the developing mouse embryo (44) could result in distinct phenotypes in mutant mice, even if the proteins regulate identical target genes. This possibility limits the usefulness of gene targeting experiments in studying HIF-1α and HIF-2α target genes. To avoid these issues, we began our studies of HIF-1α and HIF-2α in a defined cell type, 786-O cells. 786-O is one of the few cell lines in existence that is VHL-deficient with an accompanying genetic rescue of VHL (38, 39), allowing us to distinguish HIF-dependent regulation from HIF-independent hypoxic induction. In addition, 786-O cells express HIF-2α but not HIF-1α (66), enabling us to study the function of HIF-2α exclusively. Furthermore, 786-O WT-8 cells exhibit hypoxic induction of GLUT-1, VEGF, and PDGF-B, suggesting that the HIF-2α in 786-O cells is functional (39).
A genome-wide analysis of hypoxia-inducible genes with a DNA microarray and Northern blots demonstrated that a number of genes are induced by hypoxic treatment of 786-O WT-8 cells (Fig. (Fig.22 and Table Table1).1). Several novel hypoxia-inducible genes were identified, including those for ADRP, GRO-2, BIRC-3, and the Dmx-like 1 (DMXL-1) and nuclear factor IL-3-regulated (NFIL-3) genes. Interestingly, ADRP is frequently found at the surface of lipid droplets and appears to be important for lipid accumulation and lipid droplet formation (40, 90). This suggests a possible link for its function with the clear cell phenotype in VHL-defective renal carcinoma (49). Several studies indicated that GRO-1, a homologue of GRO-2, acts synergistically with PDGF-B (hypoxia-stimulated in the 786-O cells) to regulate oligodendrocyte precursor proliferation in vitro and in vivo in the rat central nervous system (69, 81). Moreover, the enhanced expression of GRO-1 by squamous cell carcinoma leads to accelerated tumor growth, angiogenesis, and metastasis in vivo (81). The genes for ADRP and GRO-2 and other genes identified in our experiments (those for growth factors and cytokines IL-6 and IL-8 and angiogenic factors VEGF and ADM) appear to be important for cell growth and may play an important role in renal cell tumorigenesis.
Analysis of the pattern of gene induction in VHL-deficient 786-O PRC-3 cells in combination with 786-O WT-8 (VHL functional) cells strongly suggests that increased expression of ADRP, NDRG-1, and VEGF is a result of HIF-2α activity. However, increased expression of GADD45A and CHOP is HIF-independent. Interestingly, glycolytic genes are not induced in 786-O cells, suggesting that HIF-2α is not able to target genes involved in glucose metabolism. In contrast, another renal clear cell carcinoma cell line expressing both HIF-1α and HIF-2α (RCC-4), exhibits HIF-α-dependent regulation of glycolytic gene expression (Fig. (Fig.2C),2C), implying that HIF-1α is critical for glycolysis during O2 deprivation. Furthermore, analysis of HIF-1α mRNA expression and glycolytic gene induction in multiple cell lines established a link between HIF-1α and glycolytic gene induction. Importantly, introduction of HIF-1α restores the glycolytic gene hypoxia response in otherwise genetically identical 786-O cells, demonstrating that HIF-1α exclusively targets glycolytic gene expression in this cell line (Fig. (Fig.4).4). Although HIF-2α protein levels correlate well with HIF target gene expression in 786-O cells, there remains a slim possibility that hypoxic gene induction is regulated by some other HIF-α subunit(s), such as HIF-3α (32) and/or unidentified HIF-α. Alternatively, HIF-2α is functional, but its ability to regulate glycolytic genes is inhibited by HIF-3α. The existence of multiple HIF-α subunits and potential unidentified HIF-α subunits compelled us to establish a system that would allow one to study the function of individual exogenous HIF-α subunits.
The TET-regulated expression system is very useful for controlled gene expression. By using such a system, the function of HIF-α subunits can be studied by directly comparing two genetically identical cells, one expressing HIF-α by the addition of doxycycline and one lacking HIF-α expression. This method of comparison avoids the clonal variability observed when the function of HIF-α is assessed by comparing parental cells with HIF-α transfectants. The TET-on system was chosen instead of the TET-off system in consideration of HIF-α protein toxicity to cells. In addition, tTS, which blocks leaky expression, is only compatible with the TET-on system. The mutation of two critical proline residues in both HIF-α subunits stabilized HIF-1α and HIF-2α under normoxia, allowing us to analyze the function of transfected HIF-α only (65). Clearly, HIF-1α activates glycolytic genes as well as other hypoxia-inducible genes identified in 786-O cells. However, as in 786-O cells, HIF-2α induced some hypoxia-inducible genes, but not the glycolytic genes, in HEK293 cells. Furthermore, glucose consumption was increased by the expression of HIF-1α and not HIF-2α. On the other hand, VEGF protein secretion was enhanced by both factors. The data from the HEK293 TET-on system independently confirmed our results from 786-O cells indicating that only HIF-1α regulates glycolytic gene expression during hypoxia. Thus, from studies of two cell lines (HEK293 and 786-O cells), we suggest that glycolytic genes are the unique targets of HIF-1α. This finding is consistent with previous data indicating that HIF-1α is necessary for glycolytic capacity in multiple cells, such as granulocytes, monocytes/macrophages (20), chondrocytes (86), and MEFs (91). Therefore, HIF-2α does not target glycolytic genes in many cell types.
Besides protein stability, HIF transcriptional activity also appears to be regulated by O2 levels (56, 57). Hydroxylation of a critical asparagine residue in the C-terminal transactivation domain is suppressed in hypoxic cells, promoting the interaction of HIF with the p300/CBP transcription coactivators and enhancing HIF transcriptional activity (2, 13, 56, 57, 67, 85). However, stabilized HIF-1α and HIF-2α appear functional in normoxic HEK293 cells as demonstrated by the transactivation of endogenous hypoxia-responsive gene expression (Fig. (Fig.7).7). No elevated induction of hypoxia-responsive gene expression was observed in the HEK293 TET-on HIF-αDPA cells treated with doxycycline in combination with 1.5% O2 (data not shown). It will be interesting to determine if the requirement for p300 is a cell type-specific event or whether other alternative factor(s) can substitute for the function of p300.
HIF-1α has been shown to be critical in regulating a variety of hypoxia-inducible genes in several systems. Hif-1α−/− ES cells lost the hypoxic response of glycolytic genes GLUT-1 and VEGF (12, 42, 83). This outcome is consistent with our results indicating that glycolytic genes are exclusively regulated by HIF-1α but inconsistent with our data stating that GLUT-1 and VEGF are also regulated by HIF-2α. However, we found that HIF-2α is not functional in ES cells, although the stability and translocation of the HIF-2α protein into the nucleus are normally regulated in ES cells (Hu et al., unpublished data). Therefore, it is understandable that the loss of HIF-1α leads to a complete loss of HIF-dependent hypoxia responses in ES cells. Consistent with our results, HIF-2α has been shown to be dispensable for hypoxia responses with Hif-2α−/− ES cells (11). The critical role of HIF-1α in glycolysis has been documented in vivo as well as in other cells. With in situ hybridization, PGK-1 expression is totally eliminated in the margins of neural folds in Hif-1α−/− embryos (83). Furthermore, no Pasteur effect is observed in Hif-1α−/− MEFs (91). However, a recent study shows that HIF-2α is not functional in MEFs, due to its inability to translocate to the nucleus (75). In contrast, our results clearly show that HIF-2α is functional in both 786-O cells and HEK293 cells but unable to regulate glycolytic gene expression.
ATP production is critical for the survival of any living organism. Shifting from oxidative phosphorylation to the O2-independent glycolytic pathway is the most ancient cellular response to hypoxia, occurring even in Caenorhabditis elegans (77). Interestingly, HIF-1α is conserved from C. elegans to humans and HIF-2α only exists in more complicated vertebrates such as chicken, quail, and mammals (24, 28, 47). Involvement of HIF-1α, and not HIF-2α, in this ancient glycolytic pathway makes sense based on the fact that HIF-1α is evolutionarily conserved and ubiquitously expressed. Although HIF-2α does not regulate glycolytic genes, it does stimulate GLUT-1 expression in 786-O and HEK293 TET-on cells. Increased glucose transport into cells may provide a survival advantage, since glucose is required for Akt-mediated inhibition of cytochrome c release and apoptosis (30, 74). Akt has been shown to be activated by hypoxia (1, 8, 16, 31, 48, 76). Besides promoting glucose uptake, expression of the GLUT-1 protein itself also has been shown to be critical for cell survival under unfavorable conditions (6, 17, 51, 60, 68). Specifically, overexpression of GLUT-1 reduces hypoxia-induced apoptosis, possibly through down-regulation of c-Jun-NH2-terminal kinase activity (60).
Loss of HIF-1α renders normal cells incapable of supplying sufficient ATP through glycolysis, which explains why extensive endothelial and mesenchymal cell death was observed in Hif-1α−/− embryos in the presence of normal amounts of VEGF (42, 54). This result demonstrates that HIF-1α-dependent regulation of glycolytic genes plays an important role in normal embryonic development. However, increased glycolysis caused by HIF-1α may not be beneficial for tumor cells which already exhibit elevated glycolysis due to other genetic mutations, since increased fermentation results in overproduction of lactic acid. Several reports indicate that p53 is only activated to promote apoptosis and cell cycle arrest in acidic environments (71, 74, 87, 109). Furthermore, the hypoxia-inducible proapoptotic gene bNIP-3 is also only active in acidic environments (9, 96). This unique function of HIF-1α in regulating the glycolytic pathway may partially explain why HIF-2α, and not HIF-1α, promotes tumorigenesis in 786-O renal clear cell carcinoma (53, 64).
We identified several HIF-2α-inducible genes; however, these genes are also regulated by HIF-1α (Table (Table2).2). A number of in vitro functional studies with reporter assays indicate that several endothelial cell-specific genes (Tie-2 and Flk-1) are exclusively regulated by HIF-2α (23, 101). Currently, a search for genes uniquely regulated by HIF-2α is under way. We demonstrated that HIF-2α does not regulate endogenous glycolytic gene induction in 786-O and 293 cells; however, HIF-2α does activate the PGK-derived HRE-luciferase reporter (three tandem copies of HRE from PGK with thymidine kinase promoter) (Fig. (Fig.5C),5C), indicating that HIF-1α and HIF-2α may recognize similar core HREs and further suggesting that the structure of the whole promoter including the number and position of HREs may play an important role. HIF-2α activates some genes but not others in the same cells (786-O and 293), showing a clear promoter selectivity. However, the cis-acting element important for the function of HIF-2α may not be the HRE. It will be interesting to see which cis-acting regulatory elements in hypoxia-inducible genes determine their distinct responsiveness to HIF-1α or HIF-2α.
TABLE 2.
Summary of HIF-1α and HIF-2α target genes analyzed in this study
| Gene type | Proteins |
|---|---|
| HIF-1α unique | Hexokinase 2, glucosephosphate isomerase, phosphofructokinase, aldoase A, aldolase C, triosephosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, PGK-1, PGM-1, enolase 1, LDHA |
| HIF-1α and HIF-2α commona | Glucose transporter 1, ADRP, NDRG-1, DMXL-1, IL-6, Carbonic anhydrase XII, filaggrin, ADM, VEGF |
Acknowledgments
We thank William G. Kaelin and Andrew Arsham for generously providing us with several reagents.
This work was supported by grant 66310 from the National Institutes of Health (M.C.S.), the Howard Hughes Medical Institute, and the Abramson Family Cancer Research Institute. M.C.S. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.23.24.9361-9374.2003
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc309606?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.23.24.9361-9374.2003
Article citations
Hypoxia-mediated rescue of retinal ganglion cells deficient in mitochondrial complex I is independent of the hypoxia-inducible factor pathway.
Sci Rep, 14(1):24114, 15 Oct 2024
Cited by: 0 articles | PMID: 39406814 | PMCID: PMC11480089
Indole Derivatives: A Versatile Scaffold in Modern Drug Discovery-An Updated Review on Their Multifaceted Therapeutic Applications (2020-2024).
Molecules, 29(19):4770, 09 Oct 2024
Cited by: 0 articles | PMID: 39407697 | PMCID: PMC11477627
Review Free full text in Europe PMC
HIF-2α-dependent induction of miR-29a restrains TH1 activity during T cell dependent colitis.
Nat Commun, 15(1):8042, 14 Sep 2024
Cited by: 0 articles | PMID: 39271652 | PMCID: PMC11399416
Hypoxic reactivation of Kaposi's sarcoma associated herpesvirus.
Cell Insight, 3(6):100200, 07 Sep 2024
Cited by: 0 articles | PMID: 39391006 | PMCID: PMC11466537
Review Free full text in Europe PMC
Mitochondrial complex I promotes kidney cancer metastasis.
Nature, 633(8031):923-931, 14 Aug 2024
Cited by: 0 articles | PMID: 39143213 | PMCID: PMC11424252
Go to all (876) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 22 of 22)
- (1 citation) ENA - M60502
- (1 citation) ENA - AF022375
- (1 citation) ENA - AF261085
- (1 citation) ENA - K03195
- (1 citation) ENA - Z29090
- (1 citation) ENA - M10036
- (1 citation) ENA - X59798
- (1 citation) ENA - BC008730
- (1 citation) ENA - D87953
- (1 citation) ENA - BC004458
- (1 citation) ENA - X04430
- (1 citation) ENA - BC003613
- (1 citation) ENA - X02152
- (1 citation) ENA - BC004982
- (1 citation) ENA - BC004333
- (1 citation) ENA - BC007952
- (1 citation) ENA - BC005127
- (1 citation) ENA - BC008964
- (1 citation) ENA - AF037335
- (1 citation) ENA - U22431
- (1 citation) ENA - D14874
- (1 citation) ENA - BC003637
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells.
Mol Cell Biol, 26(9):3514-3526, 01 May 2006
Cited by: 179 articles | PMID: 16611993 | PMCID: PMC1447431
Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha.
J Biol Chem, 279(15):14871-14878, 26 Jan 2004
Cited by: 285 articles | PMID: 14744852
Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells.
FASEB J, 18(12):1462-1464, 01 Jul 2004
Cited by: 263 articles | PMID: 15240563
The HIF pathway: implications for patterns of gene expression in cancer.
Novartis Found Symp, 240:212-25; discussion 225-31, 01 Jan 2001
Cited by: 28 articles | PMID: 11727931
Review





