Abstract
Free full text

MicroRNAs in NF-κB signaling
Abstract
Nuclear factor κB (NF-κB) is a transcriptional factor that regulates a battery of genes that are critical to innate and adaptive immunity, cell proliferation, inflammation, and tumor development. MicroRNAs (miRNAs) are short RNA molecules of 20–25 nucleotides in length that negatively regulate gene expression in animals and plants primarily by targeting 3′ untranslated regions of mRNAs. In this work, we review the convergence of miRNAs and NF-κB signaling and dysregulation of miRNAs and NF-κB activation in human diseases, particularly in cancer. The function of miR-146, miR-155, miR-181b, miR-21, and miR-301a in NF-κB activation and their impact on tumorigenesis are discussed. Given that over 1000 human miRNAs have been identified, rendering miRNAs one of the most abundant classes of regulatory molecules, deciphering their biological function and pathological contribution in NF-κB dysregulation is essential to appreciate the complexity of immune systems and to develop therapeutics against cancer.
Introduction
Nuclear factor κB (NF-κB) is a transcriptional regulator consisting of reticuloendotheliosis (Rel) protein dimers that bind a DNA sequence motif known as κB site. The Rel protein family is classified into two groups: one that requires proteolytic processing and the other that does not. The first includes NF-κB1 (also known as p105) and NF-κB2 (p100), which are processed to produce the mature p50 and p52 proteins, respectively. The second includes RelA (also known as p65), RelB, and c-Rel. All Rel proteins can form homodimers or heterodimers, except for RelB, which can only form heterodimers (Ryseck et al., 1995), while p50-RelA heterodimer is the most abundant form of NF-κB in most, if not all, unstimulated cells. We refer to the p50-RelA dimer as NF-κB in this work unless indicated otherwise. Transient NF-κB activation is regulated by two major pathways (Karin et al., 2002; Karin, 2006). The first canonical NF-κB pathway applies to dimers that are composed of RelA, c-Rel, or p50, which are retained in the cytoplasm by inhibitors of κB proteins (IκBα, IκBβ, IκBγ, IκBε, IκBζ, IκBNS, and Bcl-3). This classical pathway is normally triggered in response to microbial and viral infections and exposure to proinflammatory cytokines, such as tumor necrosis factor (TNF)-α, all of which activate the β-subunit of IκB kinase (IKKβ) complex through the toll-like receptor (TLR). IKK phosphorylates NF-κB-bound IκBs, leading to ubiquitin-dependent degradation of IκBs and translocation of NF-κB dimers to the nucleus. The second pathway is triggered by certain members of the TNF cytokine family (such as LTβ) that selectively activate the α-subunit of IKK (IKKα) through the TNF receptor, along with another protein kinase called NIK, causing phosphorylation of p100. This phosphorylation leads to polyubiquitination-dependent degradation of the C-terminal half of p100 to generate p52, allowing the formation of p52-RelB heterodimers, which then translocate to the nucleus and activate target genes.
During the past decade, much accumulated evidence has supported the role of NF-κB in linking inflammation and tumorigenesis (Karin, 2006; Inoue et al., 2007). Several pro-inflammatory cytokines and chemokines, such as TNF-α, IL-1, IL-6, and IL-8, produced upon NF-κB activation, are associated with tumor development and progression (Karin and Greten, 2005). Furthermore, NF-κB can also be activated by many oncogenes and chemopreventive chemicals to play a crucial role in tumorigenesis and tumor progression (Bharti and Aggarwal, 2002). Recently, somatic mutation has been implicated to be causative in NF-κB activation in cancers as a large number of genetic abnormalities are found in genes involved in the canonical or alternative NF-κB pathway. The CARD11 gene encoding a cytoplasmic scaffolding protein is found to be mutated in activated B-cell (ABC)-like diffuse large B-cell lymphoma (DLBCL) and such mutations result in constitutive NF-κB activation and enhanced NF-κB activity upon antigen/receptor stimulation in lymphoma cell lines (Lenz et al., 2008). Over a dozen NF-κB-relevant genes in multiple myeloma are mutated, amplified, truncated, or deleted with a few genetic alterations confirmed to activate NF-κB (Annunziata et al., 2007; Keats et al., 2007). NF-κB activation mechanisms in solid tumors, however, have not been well understood. A recent comprehensive exome analysis of 24 pancreatic tumors, a cancer with constitutively activated NF-κB (Sarkar et al., 2007), revealed that there are plenty of mutations in genes in 12 cellular signaling pathways and processes, but few in genes within the NF-κB network (Jones et al., 2008; Yachida et al., 2010).
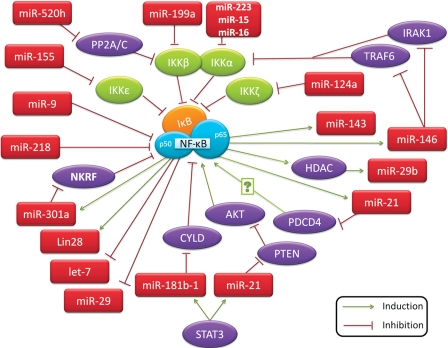
MicroRNAs (miRNAs) are a class of short (20–23 nucleotides in length), endogenous, single-stranded RNAs that regulate gene expression. miRNAs are initially transcribed by either RNA polymerase II or RNA polymerase III, as a long primary miRNA transcript (pri-miRNA). It is then cleaved in the nucleus by the microprocessor complex, Drosha-DGCR8, resulting in a precursor hairpin (pre-miRNA) ranging in length from 60 to 110 nucleotides. The pre-miRNA is exported from the nucleus to the cytoplasm by exportin-5-Ran-GTP. In the cytoplasm, Dicer, a member of the RNase III family, in complex with TRBP, cleaves the pre-miRNA hairpin to a ~22 bp miRNA duplex. The mature miRNA is incorporated with argonaute (Ago2) proteins into the RNA-induced silencing complex (RISC), where miRNA guides the complex to partial complementary binding sites located in the 3′ untranslated region (UTR) of target mRNAs to suppress gene expression. A recent report suggests that miRNA-binding sites also occur within the 5′ UTR and in the coding region (Hafner et al., 2010). It was long believed that the partial complementarity favors inhibition of translation initiation; however, recent studies suggest that miRNAs predominantly act to decrease target mRNA levels, at least in mammals (Baek et al., 2008; Guo et al., 2010). Numerous investigations have supported the role of miRNAs in the initiation and progression of human cancer, as well as in physiological function such as immune responses, cell proliferation, cell death, and inflammation, which are also known to be regulated by NF-κB (Baud and Karin, 2009). Naturally, this has led many researchers to look into the convergence of miRNAs and their target genes with NF-κB signaling cascades that are critical to tumor development and malignant progression. Here we discuss some key miRNAs intertwined with NF-κB signaling (Figure 1) and their roles in cancer.
miR-146
miR-146 was first identified as an immune system regulator in a systematic effort to find miRNAs that influence the mammalian response to microbial infection. Exposure of human monocytic THP-1 cells to lipopolysaccharides (LPS) results in rapid induction of the expression of both miR-146a and miR-146b (Taganov et al., 2006). Further characterization of miR-146a/b revealed that it is induced through TLR and the induction is NF-κB dependent. Importantly, two key adapter molecules in the TLR/NF-κB pathway, TRAF6 and IRAK1, are identified as direct targets of miR-146. This suggests a negative regulatory loop, in which NF-κB activation upregulates miR-146 gene that, upon processing and maturation, down-regulates IRAK1 and TRAF6 to reduce the activity of NF-κB. Subsequent studies have shown the pathological relevance of NF-κB/miR-146 in human breast cancer, pancreatic cancer, anaplastic thyroid carcinomas, brain tumors, and mesenchymal stem cells (Bhaumik et al., 2008; Lukiw et al., 2008; Hurst et al., 2009; Li et al., 2010b; Pacifico et al., 2010; Suzuki et al., 2010). In an experiment that directly demonstrates miR-146 as an NF-κB negative regulator, the phosphorylation of IκBα on serine 32, which is essential for its degradation, is reduced to ~40% or 20% of control levels in cells expressing miR-146a or miR-146b (Bhaumik et al., 2008). Moreover, miR-146a and miR-146b have also shown to inhibit migration and invasion in breast cancer cells (Bhaumik et al., 2008; Hurst et al., 2009).
In human lung alveolar epithelial tumor A549 cells, IL-1β induces miR-146a and negatively regulates the release of IL-8 and RANTES (regulated upon activation, normal T-cell expressed, and secreted; also known as CCL5) (Perry et al., 2008). IL-8 and RANTES are regulated by NF-κB activation, providing additional support for the negative feedback regulation of inflammation following activation of the innate immune response. However, this is only observed at high IL-1β concentrations, indicating that it may be an important feedback mechanism during severe inflammation. miR-146 is also reported to be a contributor in pancreatic β-cell function (Lovis et al., 2008). Increased levels of miR-146 are observed in pancreatic β-cells incubated with free fatty acids (FFAs) and are also found in pancreatic islets from diabetic db/db obese mice. Moreover, induction of miR-146 promotes β-cell apoptosis, while miR-146 inhibition reduces β-cell death elicited by FFAs. Activation of NF-κB is proposed to be a key event in the progressive loss of β-cells in diabetes, as inhibition of this process protects β-cells against cytokine-induced apoptosis (Roggli et al., 2010). Treating cells with oligonucleotides blocking miR-146 partially protects them against palmitate-induced apoptosis, suggesting that miR-146 contributes to the detrimental effects of palmitate on β-cells (Roggli et al., 2010). Overall, miR-146 is an NF-κB transactivational target and negatively regulates IRAK1 and TRAF6, constituting a negative feedback loop. Upregulation of miR-146a is reported in papillary thyroid carcinoma, cervical cancer, breast cancer, and pancreatic cancer, whereas reduced miR-146a expression is associated with prostate cancer (Williams et al., 2008). Thus, it is unknown whether miR-146a dysregulation is causal to cancer.
miR-155
miR-155 is processed from a non-protein-coding primary transcript, called ‘BIC’. BIC/miR-155 has been shown to be highly expressed in a variety of human B cell lymphomas, including Hodgkin lymphoma, primary mediastinal B-cell lymphoma, and DLBCL, suggesting that this miRNA may contribute to the etiology of lymphomas (Kluiver et al., 2005). In monocytes, macrophages, and myeloid dendritic cells, miR-155 is induced substantially after exposure to a variety of inflammatory cytokines such as IFN-β and IFN-γ (O'Connell et al., 2007). Yet there are some discrepancies regarding whether the induction of BIC and miR-155 is NF-κB dependent. Overexpression of an IκBα dominant negative protein does not block antigen-B cell receptor (BCR)-mediated induction of BIC in Ramos cells, a Burkitt lymphoma cell line (van den Berg et al., 2003). However, inhibition of IKK leads to a reduction in BIC and other NF-κB-regulated transcripts in ABC-like DLBCL cells with constitutively active NF-κB (Lam et al., 2008). A third report supports BCR signaling regulation of pri-miR-155 expression via activation of the PKC-NF-κB pathway in Ramos cells because blocking either PKC signaling or NF-κB activation abrogates the induction of BIC expression (Kluiver et al., 2007). Induced or ectopic expression of BIC in Ramos, HEK293, normal tonsillar B cells, and other Burkitt lymphoma cell lines results in miR-155 overexpression in all cell lines except Ramos cells (Kluiver et al., 2007), suggesting an unknown mechanism of differential miR-155 and BIC regulation in Ramos cells.
Most recent publications on miR-155 support a positive correlation between miR-155 upregulation and NF-κB activation. First, when the expression pattern of miR-155 in twenty DLBCL cell lines is examined, it is found that this miRNA is expressed at a higher level in ABC-like DLBCL cells (Rai et al., 2008). Second, both miR-155 expression and NF-κB activation are significantly elevated at early stages of choline-deficient and amino-acid-defined (CDAA) diet-induced hepatocarcinogenesis mouse model (Wang et al., 2009). Third, miR-155 is induced during Helicobacter pylori infection, which stimulates NF-κB (Xiao et al., 2009). Fourth, in human mesangial cells, IFN-γ and TNF-α induce miR-155 expression and regulate inflammatory and immune responses, which are dependent on transforming growth factor-β-activated kinase-1 (TAK1)-binding protein 2 (TAB2) and NF-κB (Imaizumi et al., 2010). Finally, in a mouse model of alcoholic liver disease, chronic alcohol consumption increases miR-155 in macrophages via NF-κB, and the increased miR-155 levels contribute to alcohol-induced elevation in TNF-α production (Bala et al., 2011).
A few target genes (FADD, IKKε, Ripk1, and PU.1) of miR-155 have been identified (Tili et al., 2007; Vigorito et al., 2007; Thompson et al., 2011). In addition, the splenocytes of Eμ-miR-155 transgenic mice, which specifically overexpress miR-155 in B cells, displayed lower levels of IKKβ transcripts than their wild type counterparts (Costinean et al., 2006). Thus, miR-155 may control the expression of both IKKβ and IKKε, which leads to repression of, or at least limitation of NF-κB activation, constituting a negative feedback loop. Taken together, these results indicate that miR-155 is an NF-κB transactivational target and is involved in a negative feedback loop through down-regulation of IKKs and other genes. miR-155 is upregulated in B-cell lymphomas and chronic lymphocytic leukemia (Eis et al., 2005), as well as in solid tumors of lung (Yanaihara et al., 2006), breast (Iorio et al., 2005), colon, pancreas (Gironella et al., 2007; Greither et al., 2010), and thyroid (Nikiforova et al., 2008), indicating its oncogenic role.
miR-181b
miR-181b-1 has recently been identified as a key player in a positive feedback loop linking inflammation to an epigenetic switch that controls cellular transformation in human mammary epithelial MCF-10A cells (Iliopoulos et al., 2010). Inhibition of miR-181b-1 in colon, prostate, and hepatocellular cancer cell lines reduced colony formation. Signal transducer and activator of transcription 3 (STAT3), a transcription factor upregulated during transformation, and miR-181b-1 expression levels are positively correlated in colon adenocarcinomas as well as in MCF-10A cells during transformation. Furthermore, miR-181b-1 and CYLD are inversely correlated in these tumors and in MCF-10A cells. CYLD is a tumor suppressor and deubiquitinating enzyme known to negatively regulate NF-κB (Trompouki et al., 2003). miR-181b-1 is found to be transactivated by STAT3, resulting in a positive feedback loop: STAT3 binds promoter regions in the miR-181b-1 gene to increase its transcription, which then inhibits CYLD production, which in turn causes increased NF-κB activation. NF-κB works to complete this feedback loop by increasing IL-6 production, leading to STAT3 phosphorylation and activation. However, miR-181b-1 is not simply a downstream effector of this signaling cascade. Transient transfection of MCF-10A cells with miR-181b-1 caused stable transformation of these cells, allowing them to be passaged for at least 30 days while retaining the ability to form colonies in soft agar, suggesting the involvement of an epigenetic switch. Therefore, miR-181b is indirectly regulated by NF-κB in a positive feedback loop (NF-κB → IL-6 → STAT3 → miR-181b → CYLD → NF-κB) and participates in an exclusive epigenetic circuit to promote cell transformation. Overexpression of miR-181b is associated with the progression of leukoplakia to oral carcinoma (Cervigne et al., 2009), as well as poor prognosis and therapeutic outcome in colon cancer (Schetter et al., 2008). Yet down-regulation of miR-181b-1 is observed in human glioma cells (Shi et al., 2008) and astrocytic tumors (Conti et al., 2009), suggesting that miR-181b may have a tumor-type-specific role.
miR-21
Unlike miR-181b-1, the function of miR-21 has been elucidated to a greater extent, its pervasive overexpression patterns in cancer have been fleshed out, and many of its predicted targets have been confirmed (Liu et al., 2010a). Instead of providing the scientific community with more answers, however, this plethora of information only serves to raise more questions. One of which is the mechanism behind miR-21's complex relationship with NF-κB (Young et al., 2010). In MCF-10A cells, miR-21 is characterized as part of the positive feedback loop linking inflammation to cellular transformation involved in STAT3 (Iliopoulos et al., 2010). Inhibition of STAT3 resulted in lower expression levels of miR-21. PTEN, a target of miR-21, is a known inhibitor of AKT phosphorylation that promotes NF-κB activation and enhances tumorigenesis. Thus, miR-21 works within the inflammation-transformation positive feedback loop by down-regulating PTEN expression to increase NF-κB activity.
Opposing this feedback loop in which miR-21 serves to enhance NF-κB activation is the recent study reporting that miR-21 is induced by LPS to attenuate pro-inflammatory effects of TLR4 signaling by negatively regulating NF-κB activity (Sheedy et al., 2010). Mice deficient in PDCD4, a confirmed miR-21 target (Lu et al., 2008), exhibit lower LPS-induced mortality rates, lower IL-6 production, and increased IL-10 protein levels. As depletion of the NF-κB subunit p65 abolished LPS-induced miR-21 expression, the authors then show that miR-21 is an NF-κB transactivational gene, which is supported in another report (Shin et al., 2011). Inhibition of miR-21 blocks PDCD4 down-regulation induced by LPS and increases NF-κB activation as well as the pro-inflammatory cytokine IL-6, yet decreases the levels of the anti-inflammatory cytokine IL-10. Thus, miR-21 acts as an anti-inflammatory agent within a negative regulatory loop: NF-κB activity is necessary for miR-21 induction, but by targeting PDCD4, miR-21 works to inhibit NF-κB and its pro-inflammatory transcriptional targets. Cell-type specificity may cause the irreconcilable difference of miR-21 in NF-κB activity: in epithelial cells, miR-21 acts to down-regulate PTEN, activate AKT, and increase NF-κB activation; in LPS-stimulated macrophages, miR-21 negatively regulates PDCD4, which activates NF-κB through an unknown mechanism. It is notable that during earlier stages of liver regeneration, miR-21 is upregulated, leading to down-regulation of pellino (Marquez et al., 2010), an activator of NF-κB, supporting that miR-21 is an NF-κB inhibitor. Work from animal models provides no insight in this regard: the expression of miR-21 target genes is not widely upregulated in tissues of miR-21 knockout mice as one would expect (Hatley et al., 2010; Patrick et al., 2010). There is an omnipresent overexpression of miR-21 in all types of human carcinomas (Liu et al., 2010a), as well as chronic lymphocytic leukemia (Fulci et al., 2007), diffuse large-B-cell lymphoma (Lawrie et al., 2007), acute myeloid leukemia (Jongen-Lavrencic et al., 2008), and Hodgkin lymphoma (Navarro et al., 2008). Interestingly, NF-κB activation has been reported in all of these cancers (Baud and Karin, 2009), underscoring the interplay of miR-21 overexpression and NF-κB activation in cancer. Further evidence is needed to dissect the role of miR-21 in NF-κB signaling and inflammation.
miR-301a
miR-301a is a newly identified miRNA that activates NF-κB. Using an NF-κB-dependent reporter screening, miR-301a stands out as the most potent NF-κB activator from hundreds of human miRNA minigenes (Lu et al., 2011). It was then demonstrated that miR-301a down-regulates NF-κB repressing factor (NKRF) and elevates NF-κB activation. When NKRF was first discovered, it was deemed to interact with specific negative regulatory elements (NREs) to mediate NF-κB's transcriptional activity, which regulates the expression of three NF-κB-responsive genes including IL-8, IFN-β, and NOS2A (Nourbakhsh and Hauser, 1999; Nourbakhsh et al., 2001; Feng et al., 2002). Yet much evidence is provided to suggest that NKRF, like IκBα, broadly inhibits the expression of NF-κB transactivational targets such as MMP2 and COX2 without NREs. As the promoter to miR-301a also contains a bona fide κB site (Lu et al., 2011), these results support a positive feedback loop as a mechanism for persistent NF-κB activation in which miR-301a represses NKRF to elevate NF-κB activity, which in turn, promotes miR-301a transcription.
Interestingly, miR-301a was first reported to be only specifically overexpressed (34.2-fold with a P-value of 1.11E−05) in pancreatic adenocarcinoma tumors and tumor cell lines compared with normal pancreas and pancreatitis, but not in other types of cancers (Lee et al., 2007). Yet, later, it was found to be upregulated (2.19-fold with a P-value of 0.0213) in hepatocellular carcinoma at a much lower level and less significantly (Jiang et al., 2008). Examination of NKRF expression reveals that it is down-regulated in human pancreatic adenocarcinoma tissues. Moreover, miR-301a inhibition or NKRF upregulation in pancreatic cancer cells led to reduced NF-κB target gene expression and attenuated xenograft tumor growth (Lu et al., 2011), indicating that miR-301a overexpression contributes to NF-κB activation. Revealing this novel mechanism of NF-κB activation by a miRNA offers new avenues for therapeutic interventions against pancreatic cancer. There are still some unanswered questions about miR-301a and NF-κB. First, what drives miR-301a overexpression in pancreatic cancer? Second, is miR-301a overexpression a cause or a consequence of pancreatic cancer pathogenesis? Third, why do mice without the NKRF allele have no obvious phenotype (Froese et al., 2006)? Answers to these questions will allow us to gain a comprehensive view of miR-301a in NF-κB signaling and pancreatic tumor development.
Other miRNAs in the NF-κB network
There are other miRNAs that suppress genes coding for NF-κB, IκB, and IKK proteins. miR-9 is another miRNA induced by LPS via MyD88 and NF-κB (Bazzoni et al., 2009). Induction of miR-9 is also mediated by the proinflamatory cytokines IL-1β and TNF-α, but not by IFN-γ. As NF-κB1 encoding p105 (processed into p50) is experimentally confirmed as a miR-9 target, a new model is proposed with the induction of miR-9 acting as a fine-tuning mechanism to prevent negative regulation by p50 homodimers in monocytes in anti-inflammatory response. Moreover, miR-9 inhibits ovarian and gastric cancer cell growth through modulation of the NF-κB pathway (Guo et al., 2009; Wan et al., 2010; Wang et al., 2010). miR-199a negatively regulates IKKβ expression to reduce NF-κB activity in ovarian cancer cells (Chen et al., 2008). MyD88-positive epithelial ovarian cancer cells have high levels of IKKβ due to lower miR-199a expression, and vice versa. Twist1 is found to regulate the expression of both miR-199a and miR-214 within the same primary transcript to modulate IKKβ/NF-κB and PTEN/AKT pathways and subsequently impact the differentiation of epithelial ovarian cancer stem cells (Yin et al., 2010). miR-124a is the first reported miRNA targeting a member of the IκB family, IκBζ, though the biological importance of this regulation remains elusive (Lindenblatt et al., 2009). miR-143 is also an NF-κB transactivational target, and it promotes live tumor cell invasion and metastasis with FNDC3B as a direct and functional target (Zhang et al., 2009). Overexpression of miR-143 decreases cell viability and increases cell death of colon cancer cells upon exposure to 5-fluorouracil with an impact on NF-κB p65 expression (Borralho et al., 2009). IKKα is targeted by miR-223, miR-15 and miR-16 during monocyte–macrophage differentiation, and decreased expression of these miRNAs may prevent macrophage hyperactivation (Li et al., 2010a).
miR-15a and miR-16-1 are encoded by the miR-15/16 cluster, which resides at chromosome 13q14.3, a genomic region frequently deleted in chronic lymphocytic leukemia (CLL) and other malignancies, such as multiple myeloma, mantle cell lymphoma, and prostate carcinoma (Aqeilan et al., 2010). miR-15a and miR-16-1 are reported to inhibit cell proliferation, induce cellular apoptosis, and suppress tumorigenesis by targeting multiple oncogenes, including Bcl-2, Mcl1, CcnD1, and Wnt3A (Bonci et al., 2008). A recent report on miR-15a, miR-16, and miR-223 suggests that these miRNAs play a role in noncanonical NF-κB pathway (Li et al., 2010a). During human monocyte–macrophage differentiation, these three miRNAs were down-regulated by 75%–90%. Li et al. (2010a) further identified that IKKα, but not IKKβ or IKKγ, is a target gene of miR-15, miR-16, and miR-223. Additionally, the expression of TRAF2, NIK, and p52, which are important components in the noncanonical NF-κB pathway, were also affected with modulation of these miRNAs in macrophage cells and/or HeLa cells. An ensuing study of miR-16 in gastric cancer cells further supports the regulation of IKKα by miR-16 and provides evidence that miR-16 is an NF-κB transactivational target (Shin et al., 2011), indicating that miR-16 modulates the noncanonical NF-κB pathway through a feedback loop.
During the past two years, more miRNAs have been found to be involved in NF-κB signaling by targeting NF-κB regulators and effectors. One high-profile paper reports that transient Src activation triggers an inflammatory response mediated by NF-κB that directly activates Lin28 transcription and rapidly reduces let-7 miRNA levels. As let-7 directly inhibits IL-6 expression, reduced let-7 expression results in high levels of IL-6 to activate STAT3 and transform epithelial cells. Src-mediated inflammation activates this positive feedback loop that maintains the epigenetic transformed state for many generations in the absence of the inducing signal. This provides direct evidence linking inflammation to cellular transformation (Iliopoulos et al., 2009). miR-520h is involved in adenovirus type 5 E1A-mediated tumor suppression through the signal cascade, E1A → miR-520h → PP2A/C → IKK → NF-κB → Twist (Su et al., 2010). miR-29b is identified as a miRNA that is repressed by NF-κB. miR-29b is found in a signal cascade, KIT (a tyrosine kinase receptor) → Sp1 → NF-κB → HDAC → miR-29b, in which miR-29b represses Sp1 expression to promote KIT-driven leukemia (Liu et al., 2010b). By repressing the expression of another target YY1, miR-29b plays the role of a tumor suppressor regulated by the NF-κB/YY1 pathway, whose abnormal expression may contribute to myogenesis and rhabdomyosarcoma (Wang et al., 2008). NF-κB repression of miR-29b is also found in cholangiocytes and cholangiocarcinoma cells (Mott et al., 2010). Another NF-κB-regulated miRNA, miR-10a, is found to be a novel regulator in smooth muscle cell differentiation from embryonic stem cells (Huang et al., 2010). Other NF-κB-regulated miRNAs include miR-125b-1, miR-30b, miR-130a, the clusters miR-17-92 and miR-23b-27b-24-1, all of which may regulate epithelial anti-microbial defenses (Zhou et al., 2009, 2010).
Conclusion
miRNAs target scores of genes encoding NF-κB, IκB, IKK, regulators, and effectors in the NF-κB signaling network with the vast majority of them participating in positive or negative feedback loops. It remains to be ascertained whether dysregulation of miRNAs (rather than accompanied NF-κB activation) is causal to tumor development and progression. Nonetheless, miRNAs seem to be potent targets as many tumor cells are sensitive to up- or down-regulation of miRNAs. The seminal work from Slack and colleagues has demonstrated that miR-21 is such an oncomiR in mouse pre-B-cell lymphoma (Medina et al., 2010). Development of miRNA replacement therapy, therefore, may potentiate our current findings into future clinical applications against cancer and other diseases that have a component of constitutive NF-κB activation. The major challenges for targeting miRNAs directly using modified DNA or RNA oligonucleotides are similar to that of clinical delivery of RNA interference (RNAi), which include (but are not limited to) biological barriers, toxicities, and tissue specificity (Pecot et al., 2011). On the other hand, targeting miRNAs that are effectors of NF-κB using small chemical inhibitors of IKK/NF-κB may show disproportionate side effects with excessive and prolonged NF-κB inhibition, given the importance of NF-κB in immunity (Baud and Karin, 2009). We are cautiously optimistic that the development of new miRNA inhibition tools such as cholesterol conjugates (Krutzfeldt et al., 2005) and locked nucleic acids (LNAs) (Elmen et al., 2008) and the discovery of dysregulation and/or mutation of specific components of NF-κB signaling in tumor initiation, progression, and maintenance will pave the way towards translating the immense popularity of miRNA-based therapeutic strategies into effective and marketable clinical solutions.
Funding
Studies on miRNAs and NF-κB in the Li laboratory are supported by an Innovative Award from University of Louisville Clinical & Translational Science Pilot Grant Program, the Diabetes and Obesity Center funded by NCRR/NIH (P20 RR024489) and the Center for Environmental Genomics and Integrated Biology funded by NIEHS/NIH (P30 ES014443). Y.L. is also supported by the Scientist Development Grant from American Heart Association (0830288N) and a R01 Grant from National Institute of Health (CA138688).
Conflict of interest: none declared.
References
- Annunziata C.M., Davis R.E., Demchenko Y., et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. 10.1016/j.ccr.2007.07.004. [Europe PMC free article] [Abstract] [Google Scholar]
- Aqeilan R.I., Calin G.A., Croce C.M. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. 10.1038/cdd.2009.69. [Abstract] [Google Scholar]
- Baek D., Villen J., Shin C., et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. 10.1038/nature07242. [Europe PMC free article] [Abstract] [Google Scholar]
- Bala S., Marcos M., Kodys K., et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 2011;286:1436–1444. 10.1074/jbc.M110.145870. [Europe PMC free article] [Abstract] [Google Scholar]
- Baud V., Karin M. Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009;8:33–40. 10.1038/nrd2781. [Europe PMC free article] [Abstract] [Google Scholar]
- Bazzoni F., Rossato M., Fabbri M., et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA. 2009;106:5282–5287. 10.1073/pnas.0810909106. [Europe PMC free article] [Abstract] [Google Scholar]
- Bharti A.C., Aggarwal B.B. Chemopreventive agents induce suppression of nuclear factor-κB leading to chemosensitization. Ann. NY Acad. Sci. 2002;973:392–395. 10.1111/j.1749-6632.2002.tb04671.x. [Abstract] [Google Scholar]
- Bhaumik D., Scott G.K., Schokrpur S., et al. Expression of microRNA-146 suppresses NF-κB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. 10.1038/onc.2008.171. [Europe PMC free article] [Abstract] [Google Scholar]
- Bonci D., Coppola V., Musumeci M., et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. 10.1038/nm.1880. [Abstract] [Google Scholar]
- Borralho P.M., Kren B.T., Castro R.E., et al. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–6700. 10.1111/j.1742-4658.2009.07383.x. [Abstract] [Google Scholar]
- Cervigne N.K., Reis P.P., Machado J., et al. Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum. Mol. Genet. 2009;18:4818–4829. 10.1093/hmg/ddp446. [Abstract] [Google Scholar]
- Chen R., Alvero A.B., Silasi D.A., et al. Regulation of IKKβ by miR-199a affects NF-κB activity in ovarian cancer cells. Oncogene. 2008;27:4712–4723. 10.1038/onc.2008.112. [Europe PMC free article] [Abstract] [Google Scholar]
- Conti A., Aguennouz M.H., La Torre D., et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J. Neuro-Oncol. 2009;93:325–332. 10.1007/s11060-009-9797-4. [Abstract] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA. 2006;103:7024–7029. 10.1073/pnas.0602266103. [Europe PMC free article] [Abstract] [Google Scholar]
- Eis P.S., Tam W., Sun L., et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 2005;102:3627–3632. 10.1073/pnas.0500613102. [Europe PMC free article] [Abstract] [Google Scholar]
- Elmen J., Lindow M., Schutz S., et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. 10.1038/nature06783. [Abstract] [Google Scholar]
- Feng X., Guo Z., Nourbakhsh M., et al. Identification of a negative response element in the human inducible nitric-oxide synthase (hiNOS) promoter: the role of NF-κB-repressing factor (NRF) in basal repression of the hiNOS gene. Proc. Natl. Acad. Sci. USA. 2002;99:14212–14217. 10.1073/pnas.212306199. [Europe PMC free article] [Abstract] [Google Scholar]
- Froese N., Schwarzer M., Niedick I., et al. Innate immune responses in NF-κB-repressing factor-deficient mice. Mol. Cell. Biol. 2006;26:293–302. 10.1128/MCB.26.1.293-302.2006. [Europe PMC free article] [Abstract] [Google Scholar]
- Fulci V., Chiaretti S., Goldoni M., et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. 10.1182/blood-2006-12-062398. [Abstract] [Google Scholar]
- Gironella M., Seux M., Xie M.-J., et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA. 2007;104:16170–16175. 10.1073/pnas.0703942104. [Europe PMC free article] [Abstract] [Google Scholar]
- Greither T., Grochola L.F., Udelnow A., et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Intl. J. Cancer. 2010;126:73–80. 10.1002/ijc.24687. [Abstract] [Google Scholar]
- Guo L.M., Pu Y., Han Z., et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-κB1. FEBS J. 2009;276:5537–5546. 10.1111/j.1742-4658.2009.07237.x. [Abstract] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. 10.1038/nature09267. [Europe PMC free article] [Abstract] [Google Scholar]
- Hafner M., Landthaler M., Burger L., et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. 10.1016/j.cell.2010.03.009. [Europe PMC free article] [Abstract] [Google Scholar]
- Hatley M.E., Patrick D.M., Garcia M.R., et al. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell. 2010;18:282–293. 10.1016/j.ccr.2010.08.013. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang H., Xie C., Sun X., et al. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J. Biol. Chem. 2010;285:9383–9389. 10.1074/jbc.M109.095612. [Europe PMC free article] [Abstract] [Google Scholar]
- Hurst D.R., Edmonds M.D., Scott G.K., et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. 10.1158/0008-5472.CAN-08-3559. [Europe PMC free article] [Abstract] [Google Scholar]
- Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-κB, Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. 10.1016/j.cell.2009.10.014. [Europe PMC free article] [Abstract] [Google Scholar]
- Iliopoulos D., Jaeger S.A., Hirsch H.A., et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39:493–506. 10.1016/j.molcel.2010.07.023. [Europe PMC free article] [Abstract] [Google Scholar]
- Imaizumi T., Tanaka H., Tajima A., et al. IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am. J. Nephrol. 2010;32:462–468. 10.1159/000321365. [Abstract] [Google Scholar]
- Inoue J., Gohda J., Akiyama T., et al. NF-κB activation in development and progression of cancer. Cancer Sci. 2007;98:268–274. 10.1111/j.1349-7006.2007.00389.x. [Abstract] [Google Scholar]
- Iorio M.V., Ferracin M., Liu C.-G., et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. 10.1158/0008-5472.CAN-05-1783. [Abstract] [Google Scholar]
- Jiang J., Gusev Y., Aderca I., et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 2008;14:419–427. 10.1158/1078-0432.CCR-07-0523. [Europe PMC free article] [Abstract] [Google Scholar]
- Jones S., Zhang X., Parsons D.W., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. 10.1126/science.1164368. [Europe PMC free article] [Abstract] [Google Scholar]
- Jongen-Lavrencic M., Sun S.M., Dijkstra M.K., et al. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. 10.1182/blood-2008-01-133355. [Abstract] [Google Scholar]
- Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. 10.1038/nature04870. [Abstract] [Google Scholar]
- Karin M., Greten F.R. NF-κB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. 10.1038/nri1703. [Abstract] [Google Scholar]
- Karin M., Cao Y., Greten F.R., et al. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. 10.1038/nrc780. [Abstract] [Google Scholar]
- Keats J.J., Fonseca R., Chesi M., et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. 10.1016/j.ccr.2007.07.003. [Europe PMC free article] [Abstract] [Google Scholar]
- Kluiver J., Poppema S., de Jong D., et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207:243–249. 10.1002/path.1825. [Abstract] [Google Scholar]
- Kluiver J., van den Berg A., de Jong D., et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. 10.1038/sj.onc.1210147. [Abstract] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., et al. Silencing of microRNAs in vivo with ‘antagomirs. Nature. 2005;438:685–689. 10.1038/nature04303. [Abstract] [Google Scholar]
- Lam L.T., Davis R.E., Ngo V.N., et al. Compensatory IKKα activation of classical NF-κB signaling during IKKβ inhibition identified by an RNA interference sensitization screen. Proc. Natl. Acad. Sci. USA. 2008;105:20798–20803. 10.1073/pnas.0806491106. [Europe PMC free article] [Abstract] [Google Scholar]
- Lawrie C.H., Soneji S., Marafioti T., et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer. 2007;121:1156–1161. 10.1002/ijc.22800. [Abstract] [Google Scholar]
- Lee E.J., Gusev Y., Jiang J., et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. 2007;120:1046–1054. 10.1002/ijc.22394. [Europe PMC free article] [Abstract] [Google Scholar]
- Lenz G., Davis R.E., Ngo V.N., et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. 10.1126/science.1153629. [Abstract] [Google Scholar]
- Li T., Morgan M.J., Choksi S., et al. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 2010a;11:799–805. 10.1038/ni.1918. [Europe PMC free article] [Abstract] [Google Scholar]
- Li Y., Vandenboom T.G., 2nd, Wang Z., et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010b;70:1486–1495. 10.1158/0008-5472.CAN-09-2792. [Europe PMC free article] [Abstract] [Google Scholar]
- Lindenblatt C., Schulze-Osthoff K., Totzke G. IκBζ expression is regulated by miR-124a. Cell Cycle. 2009;8:2019–2023. 10.4161/cc.8.13.8816. [Abstract] [Google Scholar]
- Liu M.F., Jiang S., Lu Z., et al. McQueen C.A. Comprehensive Toxicology. 2nd edn. London: Elsevier Science; 2010a. Physiological and pathological functions of mammalian microRNAs; pp. 427–446. [Google Scholar]
- Liu S., Wu L.C., Pang J., et al. Sp1/NFκB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010b;17:333–347. 10.1016/j.ccr.2010.03.008. [Europe PMC free article] [Abstract] [Google Scholar]
- Lovis P., Roggli E., Laybutt D.R., et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic β-cell dysfunction. Diabetes. 2008;57:2728–2736. 10.2337/db07-1252. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu Z., Liu M., Stribinskis V., et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. 10.1038/onc.2008.72. [Abstract] [Google Scholar]
- Lu Z., Li Y., Takwi A., et al. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. 10.1038/emboj.2010.296. [Europe PMC free article] [Abstract] [Google Scholar]
- Lukiw W.J., Zhao Y., Cui J.G. An NF-κB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008;283:31315–31322. 10.1074/jbc.M805371200. [Europe PMC free article] [Abstract] [Google Scholar]
- Marquez R.T., Wendlandt E., Galle C.S., et al. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets pellino-1, and inhibits NF-κB signaling. Am. J. Physiol. Gastroint. Liver Physiol. 2010;298:G535–G541. 10.1152/ajpgi.00338.2009. [Europe PMC free article] [Abstract] [Google Scholar]
- Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. 10.1038/nature09284. [Abstract] [Google Scholar]
- Mott J.L., Kurita S., Cazanave S.C., et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-κB. J. Cell Biochem. 2010;110:1155–1164. 10.1002/jcb.22630. [Europe PMC free article] [Abstract] [Google Scholar]
- Navarro A., Gaya A., Martinez A., et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. 10.1182/blood-2007-06-096784. [Abstract] [Google Scholar]
- Nikiforova M.N., Tseng G.C., Steward D., et al. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008;93:1600–1608. 10.1210/jc.2007-2696. [Europe PMC free article] [Abstract] [Google Scholar]
- Nourbakhsh M., Hauser H. Constitutive silencing of IFN-β promoter is mediated by NRF (NF-κB-repressing factor), a nuclear inhibitor of NF-κB. EMBO J. 1999;18:6415–6425. 10.1093/emboj/18.22.6415. [Europe PMC free article] [Abstract] [Google Scholar]
- Nourbakhsh M., Kalble S., Dorrie A., et al. The NF-κB repressing factor is involved in basal repression and interleukin (IL)-1-induced activation of IL-8 transcription by binding to a conserved NF-κB-flanking sequence element. J. Biol. Chem. 2001;276:4501–4508. 10.1074/jbc.M007532200. [Abstract] [Google Scholar]
- O'Connell R.M., Taganov K.D., Boldin M.P., et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 2007;104:1604–1609. 10.1073/pnas.0610731104. [Europe PMC free article] [Abstract] [Google Scholar]
- Pacifico F., Crescenzi E., Mellone S., et al. Nuclear factor-κB contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J. Clin. Endocrinol. Metab. 2010;95:1421–1430. 10.1210/jc.2009-1128. [Abstract] [Google Scholar]
- Patrick D.M., Montgomery R.L., Qi X., et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Inves. 2010;120:3912–3916. 10.1172/JCI43604. [Europe PMC free article] [Abstract] [Google Scholar]
- Pecot C.V., Calin G.A., Coleman R.L., et al. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11:59–67. 10.1038/nrc2966. [Europe PMC free article] [Abstract] [Google Scholar]
- Perry M.M., Moschos S.A., Williams A.E., et al. Rapid changes in microRNA-146a expression negatively regulate the IL-1β-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 2008;180:5689–5698. [Europe PMC free article] [Abstract] [Google Scholar]
- Rai D., Karanti S., Jung I., et al. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet. Cytogenet. 2008;181:8–15. 10.1016/j.cancergencyto.2007.10.008. [Europe PMC free article] [Abstract] [Google Scholar]
- Roggli E., Britan A., Gattesco S., et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic β-cells. Diabetes. 2010;59:978–986. 10.2337/db09-0881. [Europe PMC free article] [Abstract] [Google Scholar]
- Ryseck R.P., Novotny J., Bravo R. Characterization of elements determining the dimerization properties of RelB and p50. Mol. Cell. Biol. 1995;15:3100–3109. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarkar F.H., Banerjee S., Li Y. Pancreatic cancer: pathogenesis, prevention and treatment. Toxicol. Appl. Pharmacol. 2007;224:326–336. 10.1016/j.taap.2006.11.007. [Europe PMC free article] [Abstract] [Google Scholar]
- Schetter A.J., Leung S.Y., Sohn J.J., et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. J. Am. Med. Assoc. 2008;299:425–436. 10.1001/jama.299.4.425. [Europe PMC free article] [Abstract] [Google Scholar]
- Sheedy F.J., Palsson-McDermott E., Hennessy E.J., et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. 10.1038/ni.1828. [Abstract] [Google Scholar]
- Shi L., Cheng Z., Zhang J., et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. 10.1016/j.brainres.2008.07.085. [Abstract] [Google Scholar]
- Shin V.Y., Jin H., Ng E.K., et al. NF-κB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011;32:240–245. 10.1093/carcin/bgq240. [Abstract] [Google Scholar]
- Su J.L., Chen P.B., Chen Y.H., et al. Downregulation of microRNA miR-520h by E1A contributes to anticancer activity. Cancer Res. 2010;70:5096–5108. 10.1158/0008-5472.CAN-09-4148. [Europe PMC free article] [Abstract] [Google Scholar]
- Suzuki Y., Kim H.W., Ashraf M., et al. Diazoxide potentiates mesenchymal stem cell survival via NF-κB-dependent miR-146a expression by targeting Fas. Am. J. Physiol. Heart Circ. Physiol. 2010;299:H1077–H1082. 10.1152/ajpheart.00212.2010. [Europe PMC free article] [Abstract] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.-J., et al. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. 10.1073/pnas.0605298103. [Europe PMC free article] [Abstract] [Google Scholar]
- Thompson R.C., Herscovitch M., Zhao I., et al. NF-κB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J. Biol. Chem. 2011;286:1675–1682. 10.1074/jbc.M110.177063. [Europe PMC free article] [Abstract] [Google Scholar]
- Tili E., Michaille J.J., Cimino A., et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. [Abstract] [Google Scholar]
- Trompouki E., Hatzivassiliou E., Tsichritzis T., et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 2003;424:793–796. 10.1038/nature01803. [Abstract] [Google Scholar]
- van den Berg A., Kroesen B.J., Kooistra K., et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–28. 10.1002/gcc.10186. [Abstract] [Google Scholar]
- Vigorito E., Perks K.L., Abreu-Goodger C., et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. 10.1016/j.immuni.2007.10.009. [Abstract] [Google Scholar]
- Wan H.Y., Guo L.M., Liu T., et al. Regulation of the transcription factor NF-κB1 by microRNA-9 in human gastric adenocarcinoma. Mol. Cancer. 2010;9:16–20. 10.1186/1476-4598-9-16. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang H., Garzon R., Sun H., et al. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. 10.1016/j.ccr.2008.10.006. [Abstract] [Google Scholar]
- Wang B., Majumder S., Nuovo G., et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. 10.1002/hep.23100. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Q., Sun Z.X., Allgayer H., et al. Downregulation of E-cadherin is an essential event in activating beta-catenin/Tcf-dependent transcription and expression of its target genes in Pdcd4 knockdown cells. Oncogene. 2010;29:128–138. 10.1038/onc.2009.302. [Europe PMC free article] [Abstract] [Google Scholar]
- Williams A.E., Perry M.M., Moschos S.A., et al. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem. Soc. Trans. 2008;36:1211–1215. 10.1042/BST0361211. [Abstract] [Google Scholar]
- Xiao B., Liu Z., Li B.S., et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 2009;200:916–925. 10.1086/605443. [Abstract] [Google Scholar]
- Yachida S., Jones S., Bozic I., et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. 10.1038/nature09515. [Europe PMC free article] [Abstract] [Google Scholar]
- Yanaihara N., Caplen N., Bowman E., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. 10.1016/j.ccr.2006.01.025. [Abstract] [Google Scholar]
- Yin G., Chen R., Alvero A.B., et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through miR199A2/214. Oncogene. 2010;29:3545–3553. 10.1038/onc.2010.111. [Europe PMC free article] [Abstract] [Google Scholar]
- Young M.R., Santhanam A.N., Yoshikawa N., et al. Have tumor suppressor PDCD4 and its counteragent oncogenic miR-21 gone rogue? Mol. Interv. 2010;10:76–79. 10.1124/mi.10.2.5. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang X., Liu S., Hu T., et al. Up-regulated microRNA-143 transcribed by nuclear factor κB enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. 10.1002/hep.23008. [Abstract] [Google Scholar]
- Zhou R., Hu G., Liu J., et al. NF-κB p65-dependent transactivation of miRNA genes following cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:100–681. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou R., Hu G., Gong A.Y., et al. Binding of NF-κB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. 10.1093/nar/gkq056. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Journal of Molecular Cell Biology are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/jmcb/mjr007
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3104013?pdf=render
Citations & impact
Impact metrics
Article citations
The Effect of Szigetvár Medicinal Water on HaCaT Cells Exposed to Dithranol.
Life (Basel), 14(10):1318, 17 Oct 2024
Cited by: 0 articles | PMID: 39459618 | PMCID: PMC11509105
miRNAs as Epigenetic Biomarkers in the Study of the Bidirectional Relationship between Type 2 Diabetes Mellitus and Periodontitis: A Systematic Review.
Int J Mol Sci, 25(19):10723, 05 Oct 2024
Cited by: 0 articles | PMID: 39409052 | PMCID: PMC11477124
Review Free full text in Europe PMC
Expression of miRNAs (146a and 155) in human peri-implant tissue affected by peri-implantitis: a case control study.
BMC Oral Health, 24(1):856, 28 Jul 2024
Cited by: 0 articles | PMID: 39068455 | PMCID: PMC11283691
Octadecaneuropeptide, ODN, Promotes Cell Survival against 6-OHDA-Induced Oxidative Stress and Apoptosis by Modulating the Expression of miR-34b, miR-29a, and miR-21in Cultured Astrocytes.
Cells, 13(14):1188, 12 Jul 2024
Cited by: 0 articles | PMID: 39056770 | PMCID: PMC11487398
The Multifaceted Role of miR-21 in Pancreatic Cancers.
Cells, 13(11):948, 30 May 2024
Cited by: 1 article | PMID: 38891080 | PMCID: PMC11172074
Review Free full text in Europe PMC
Go to all (390) article citations
Other citations
Wikipedia
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The functional analysis of MicroRNAs involved in NF-κB signaling.
Eur Rev Med Pharmacol Sci, 20(9):1764-1774, 01 May 2016
Cited by: 33 articles | PMID: 27212168
Differential expression of miRNAs regulating NF-κB and STAT3 crosstalk during colitis-associated tumorigenesis.
Mol Cell Probes, 47:101442, 31 Aug 2019
Cited by: 16 articles | PMID: 31479716
Regulation of the MIR155 host gene in physiological and pathological processes.
Gene, 532(1):1-12, 14 Dec 2012
Cited by: 281 articles | PMID: 23246696
Review
MicroRNA-181b inhibits thrombin-mediated endothelial activation and arterial thrombosis by targeting caspase recruitment domain family member 10.
FASEB J, 30(9):3216-3226, 13 Jun 2016
Cited by: 31 articles | PMID: 27297585 | PMCID: PMC5001512
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: CA138688
Grant ID: R01 CA138688
NCRR NIH HHS (1)
Grant ID: P20 RR024489
NIEHS NIH HHS (1)
Grant ID: P30 ES014443