Abstract
Free full text

Identification and Characterization of a Loss-of-Function Human MPYS Variant
Abstract
MPYS, also known as STING and MITA, is an IFNβ stimulator essential for host defense against RNA, DNA viruses and intracellular bacteria. MPYS also facilitates the adjuvant activity of DNA vaccines. Here we report identification of a distinct human MPYS haplotype that contains three non-synonymous SNPs, R71H-G230A-R293Q (thus named the HAQ haplotype). We estimate, in two cohorts (1074 individuals), that ~3% of Americans are homozygous for this HAQ haplotype. HAQ MPYS exhibits a >90% loss in the ability to stimulate IFNβ production. Furthermore, fibroblasts and macrophage cells expressing HAQ are defective in Listeria monocytogenes infection-induced IFNβ production. Lastly, we find that the loss of IFNβ activity is due primarily to the R71H and R293Q SNPs in HAQ. We hypothesize that individuals carrying HAQ may exhibit heightened susceptibility to viral infection and respond poorly to DNA vaccines.
Introduction
Infectious diseases kill more people worldwide than any other single cause. It has been recognized that genetic variations, e.g. single nucleotide polymorphisms or SNPs, of key molecular components in human host defense can play critical roles in determining the susceptibility of individuals to infection1. The D299G SNP found in the extracellular domain of TLR4 is associated with decreased airway response to inhaled bacterial LPS2 and enhanced susceptibility to Gram negative infection3. The S183I SNP, which occurs in the second caspase recruitment domain repeat of RIG-I confers a dominant inhibitory function4.
MPYS was originally identified as a mediator of anti-MHC II mAb-induced apoptosis in B lymphoma cells5. It was subsequently discovered that MPYS is essential for innate immune responses to RNA, DNA viruses and intracellular bacteria, coupling infection to IFNβ production6. Recently, it was shown that MPYS also facilitates the adjuvant activity of DNA vaccines7. The molecular mechanism of MPYS function is not completely understood. It is known that MPYS induction of cell death requires ERK activation5. MPYS also activates IRF38 and TBK1 is required for MPYS induced Type I IFN production6.
Human MPYS has three validated non-synonymous SNPs: R71H (rs11554776), H232R (rs1131769) and R293Q (rs7380824). The nature and location of these SNPs suggest that they may alter the activity of the molecule. Both the H232R and R293Q are in the cytoplasmic tail of MPYS adjacent to conserved Traf2 binding motifs: 226PxQxxD231 and 294TLED/E. R71H lies in an ectodomain of MPYS adjacent to a Leucine Rich Repeat (LRR) motif of unknown function. Interestingly, in most other species, Arg71 is replaced by Cys71.
Here we report that R71H and R293Q SNPs are in linkage disequilibrium and, together with a novel non-synonymous SNP G230A, form a distinct haplotype termed HAQ. More importantly, HAQ MPYS is defective in IFNβ stimulation and this defect is primarily attributable to R71H and R293Q SNPs.
Results
Identification of a R71H (rs11554776)-G230A (rs78233829)-R293Q (rs7380824) (HAQ) human MPYS haplotype
MPYS has three validated non-synonymous SNPs R71H (rs11554776), H232R (rs1131769) and R293Q (rs7380824) in dbSNPs. To identify individuals carrying these MPYS SNPs, we screened human genomic DNA samples from two cohorts of 1074 total individuals (Table 1). We found that R71H SNP is in linkage disequilibrium with R293Q SNP (H71-Q293 haplotype) (Fig. 1a, b). Sequencing the entire coding region of the MPYS gene from individuals carrying the H71-Q293 SNPs haplotype revealed a new SNP (G230A) that co-segregates with the H71-Q293 SNPs haplotype (Fig. 1a, b). Thus these three non-synonymous SNPs, R71H, G230A and R293Q, form a distinct SNP haplotype (Fig. 1c) which we refer as HAQ based on the amino acids encoded by the SNPs.
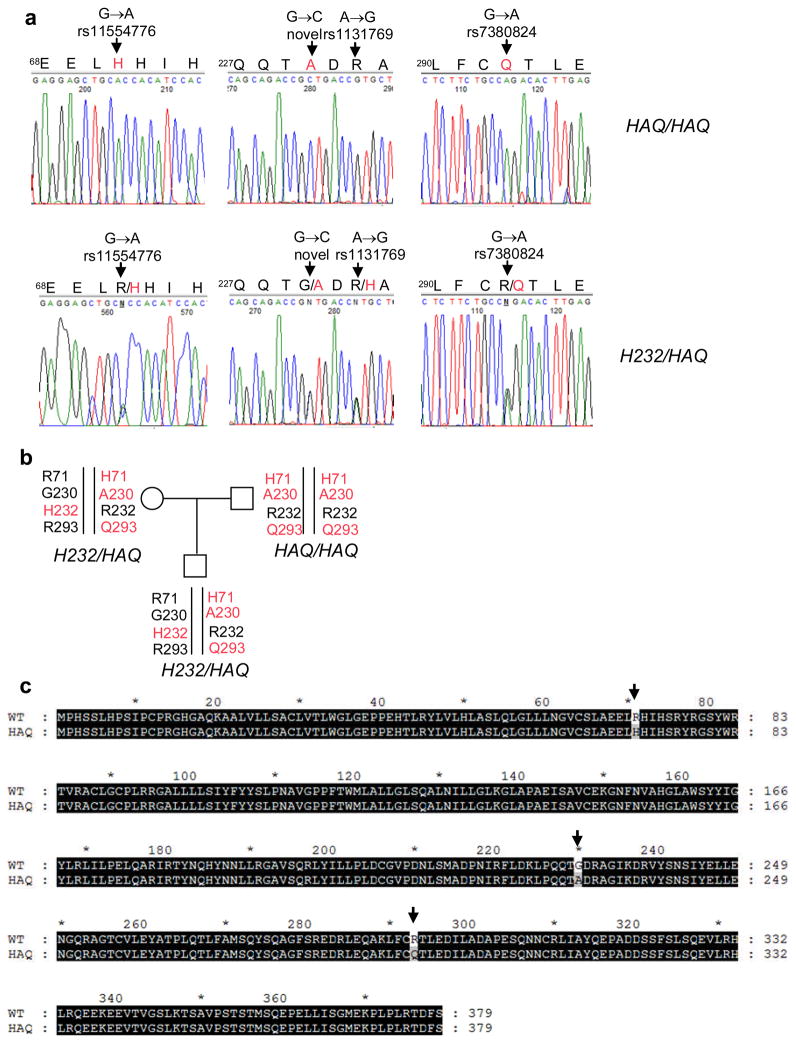
a. PCR sequencing of mpys exons from human samples that are homozygous (HAQ/HAQ) (upper three panels) and heterozygous (H232/HAQ) (lower three panels) for the HAQ haplotype. Red letters indicate the altered amino acids by SNPs. b. A family consists of homozygous (HAQ/HAQ) and heterozygous (H232/HAQ) of HAQ MPYS. c. Alignment of HAQ with WT MPYS. Altered amino acids due to SNPs are indicated by arrows.
Table 1
Two cohorts were genotyped for three validated MPYS SNPs, R71H, R232H and R293Q. The number in the table shows the individuals that carry the specific SNP alleles. Noteworthy, every individual that carries the H71 allele also has the Q293 allele (H71-Q293 haplotype). It is likely that the R71H SNP was introduced specifically to the Q293 carrier during the evolution.
| Cohort I | |||||
|---|---|---|---|---|---|
| SNPs | WT | H232 | Q293 | H71-Q293 | Total |
| WT | 248 | / | / | / | / |
| H232 | 86 | 10 | / | / | / |
| Q293 | 58 | 4 | 24 | / | / |
| H71-Q293 | 83 | 11 | 4 | 17 | 545 |
| Cohort II | ||||
|---|---|---|---|---|
| Genotype | no HQ | HQ heterozygous | HQ homozygous | Total |
| Individuals | 401 | 112 | 16 | 529 |
The HAQ haplotype also contains the reported H232R SNP (HAQ-R232 haplotype, Fig. 1a, 1b). However, in cohort I, we found that only 2% people (10 out of 545) are homozygous for the H232 allele while 45% people (248 out of 545) are homozygous for the R232 allele, which is the allele that co-segregates with HAQ (Fig. 1b). Thus, the R232 allele is prevalent and we consider it as “wildtype (WT)” (Fig. 1c). It is therefore noteworthy that the previous reported human MPYS (STING/MITA) is the rare H232 allele6, 8. Based on data from two cohorts (Table 1), we estimate that ~3% of Americans are homozygous for this HAQ haplotype.
HAQ MPYS is defective in stimulation of IFNβ production
MPYS is an IFNβ stimulator6. To examine the relative functional activity of the MPYS SNP haplotypes observed in human populations ( Table 1) we expressed each of the four haplotypes, i.e. WT, H232, Q293 and HAQ, in the 293MT cells and measured activation of IFNβ production. In addition, we measured phosphorylation of IRF3 which lies downstream from MPYS in the IFNβ stimulation pathway8. 293MT, a derivative of HEK293, was employed because it expresses no detectable endogenous MPYS (Fig. 2b). Relative to WT, HAQ MPYS displayed a 90% reduced ability to activate IFNβ promoter (Fig. 2a) and IRF3 phosphorylation (Fig. 2b). The ability of HAQ to induce IFNβ protein production detected by ELISA was similarly reduced (Fig. 2c).
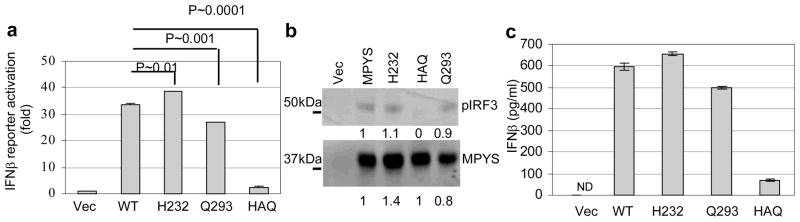
a. 293MT cells (~1×105) were transfected with indicated plasmid (100ng) along with IFNβ-luciferase reporters. After 24 hrs, the luciferase activity was measured as described in Methods. Error bars represent SD of a duplicate. P value was calculated by student T-test (one-tailed). b. 293MT cells were transfected with indicated plasmids. The blot was probed with anti-MPYS or p-IRF3 Ab (4D4G). c. 293MT cells were transfected with indicated plasmids as before. The supernatant was collected and IFNβ proteins were measured as described in Methods. Error bars represent SD of a duplicate. ND: not detected.
About 20% of subjects in our cohorts have one copy of HAQ. Most of them are WT/HAQ (~15%) (Table 1). We found that this genotype (WT/HAQ) also has decreased IFNβ responses (Fig. 3a and 3b) and IRF3 activation (Fig. 3c). Considering the fact that MPYS functions as a homodimer 5, 9, this result suggests that the HAQ-MPYS maybe a dominant allele.
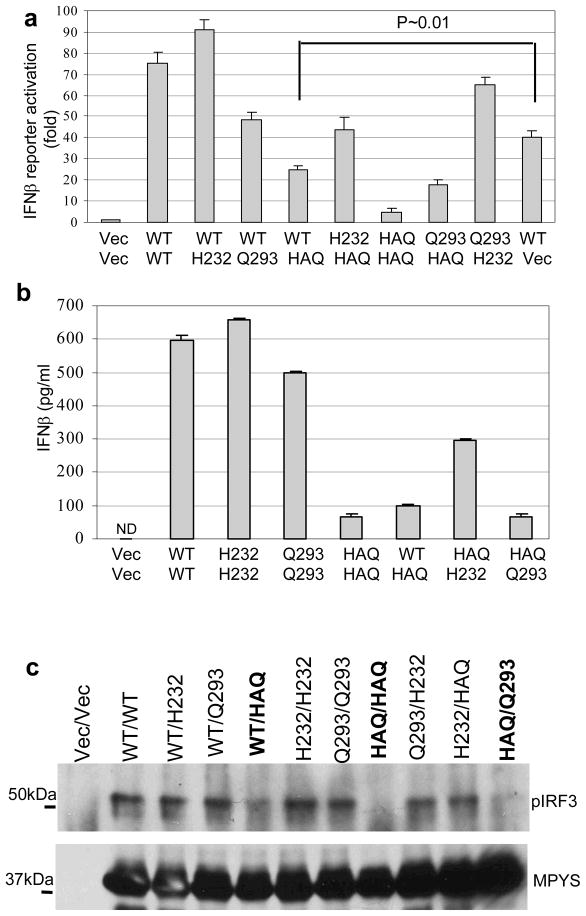
a. 293MT cells were transfected with 100ng of each indicated plasmid (e.g 100ng each of WT and HAQ for the WT/HAQ) along with IFNβ-luciferase reporters. After 24 hrs, the luciferase activity was measured as before. Error bars represent SD of a duplicate. b. 293MT cells were transfected with indicated plasmids as before. The supernatant was collected and IFNβ proteins were measured as described in Methods. Error bars represent SD of a duplicate. ND: not detected. c. 293MT cells were transfected with indicated plasmids. The blot was probed as before. Highlighted are the genotypes that are defective in IFNβ stimulation.
Listeria monocytogenes induction of IFNβ production is impaired in cells expressing HAQ
MPYS is required for Listeria monocytogenes-induced IFNβ responses in mouse embryonic fibroblasts6. We produced 293MT cells stably expressing HAQ, WT-MPYS or Vector (Fig. 4a) and examined Listeria monocytogenes infection induction of IFNβ production by ELISA. HAQ-MPYS transduced dramatically decreased IFNβ responses to Listeria monocytogenes relative to WT-MPYS (Fig. 4b). We also reconstituted bone-marrow-derived-macrophages (BMM) from STING/MPYS deficient mice with WT-MPYS, HAQ-MPYS and Vec-control (Fig 4c) and found that HAQ-BMM has dramatically decreased IFNβ production similar to the result in 293MT cells (Fig. 4d).
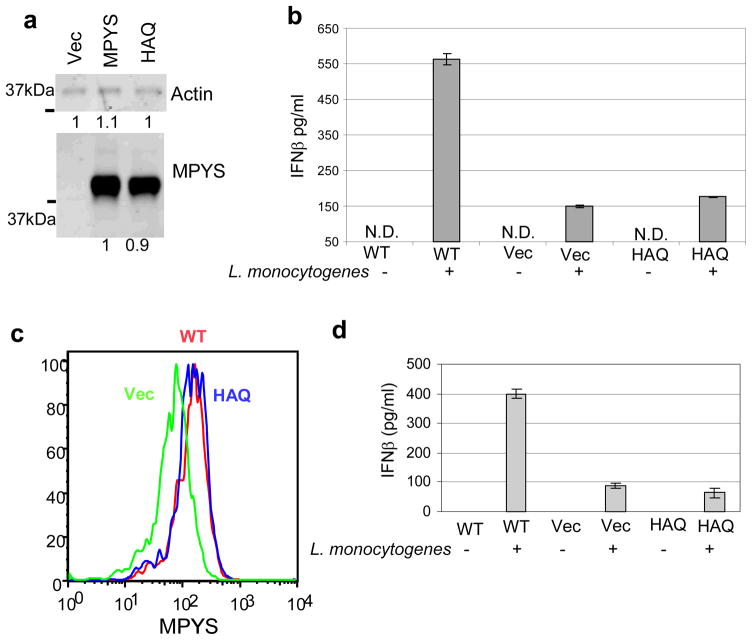
a. 293MT cells stably expressing Vec, WT or HAQ MPYS, were lysed in RIPA buffer. MPYS was detected as before. b. 293MT cells stably expressing Vec, WT or HAQ MPYS, were infected with Listeria monocytogenes as described in Methods. The IFNβ protein was measured as before. Error bars represent SD of a duplicate. ND: not detected. c. BMM cells reconstituted with MPYS, HAQ or Vec control were fixed, permeabilized and stained with α-MPYS rabbit Ab followed by anti-rabbit-Alexa647 as in Methods. d. BMM cells were infected with Listeria monocytogenes as describe in Methods. IFNβ production was measured after 20 hrs by ELISA.
The IFNβ stimulation defect of HAQ MPYS can be ascribed to R71H and R293Q SNPs
We next sought to determine which SNPs in HAQ are responsible for defective MPYS function. HAQ contains three SNPs, R71H, G230A and R293Q. Among them, amino acid 230 is not conserved among species (Fig. 5e). More importantly, HQ (H71-G230-Q293) was found to have the same IFNβ-stimulating defect as the HAQ (H71-A230-Q293) (Fig. 5a, 5b, 5c). Furthermore, the G230A SNP alone has similar IFNβ stimulation ability as the WT MPYS (data not shown). Thus, the R293Q and R71H SNPs are likely the primary determinants of the decreased IFNβ-stimulating ability of HAQ MPYS.
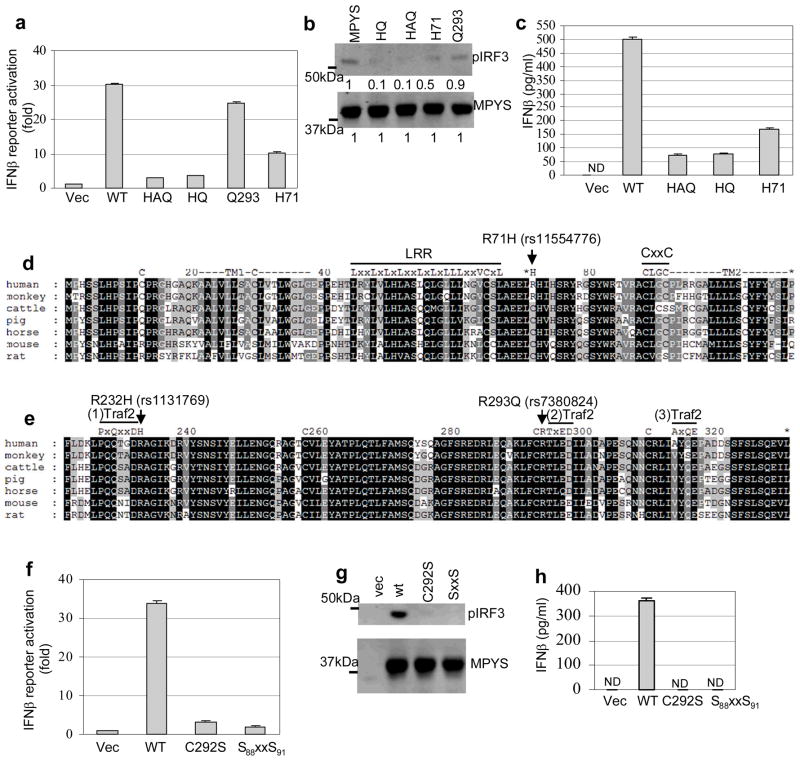
a. 293MT cells were transfected with indicated plasmid (100ng) along with IFNβ-luciferase reporters. Luciferase activity was measured after 24 hrs as before. Error bars represent SD of a duplicate. b. 293MT cells were transfected with indicated plasmids. Phosphorylated IRF3 was probed as before. c. 293MT cells were transfected with indicated plasmids as before. The supernatant was collected and IFNβ proteins were measured as before. Error bars represent SD of a duplicate. ND: not detected. d&e. Alignment of MPYS sequence from 7 different species. The locations of motifs and SNPs were indicated. f. 293MT cells were transfected with indicated plasmid (100ng) along with IFNβ-luciferase reporters. Luciferase activity was measured after 24 hrs as before. Error bars represent SD of a duplicate. g. 293MT cells were transfected with indicated plasmids. Phosphorylated IRF3 was probed as before. h. 293MT cells were transfected with indicated plasmids as before. The supernatant was collected and IFNβ proteins were measured as before. Error bars represent SD of a duplicate. ND: not detected.
The Q293 allele has decreased ability to activate IFNβ production (Fig. 2), as does WT/Q293 genotype (Fig. 3). However, the IFNβ-stimulating defect seen in cells expressing Q293 is mild compared to those expressing HAQ (Fig. 2, ,3,3, ,5).5). The H71 allele has a more severe IFNβ-stimulating defect than the Q293 allele (Fig. 5a, 5b). Yet H71 allele alone is a better IFNβ stimulator than HAQ (Fig. 5a, 5b, 5c). We concluded that both the Q293 and H71 alleles are responsible for the IFNβ-stimulating defect of HAQ.
MPYS cysteines adjacent to R293Q and R71H SNPs are critical for IFNβ stimulation
We then tried to understand how R293Q and R71H SNPs mediate their effects on MPYS ability to stimulate IFNβ production. We noted that Arg71 is replaced by a conserved Cys71 in five species including mouse (Fig. 5d). Arg293 is adjacent to a conserved Cys292 (Fig. 5e). We thus hypothesized these cysteines may be important for MPYS functions and R71H, R293Q SNPs may affect IFNβ stimulation by influencing the function of these Cysteines.
The existence of a conserved cysteine, Cys292, near the conserved Arg293 (Cys292-Arg293) is significant because the positive charge of the Arg293 residue could lower the pKa of Cys292 favoring the disulfide bond formation or other Cys-related modifications10. The charge change in R293Q may thus affect the function of Cys292. Consistent with this possibility we found that mutating Cys292 to Ser292 completely destroyed MPYS ability to induce IFNβ expression and IRF3 phosphorylation (Fig. 5f, 5g, 5h).
Extending the concept that cysteines play critical roles in MPYS signaling is the fact that Arg71, which lies in the first ectodomain loop of MPYS near a predicted LRR (Fig. 5d), is proximal to a C88xxC91 redox motif. Mutation of the C88xxC91 to S88xxS91, eliminated MPYS ability to induce IRF3 phosphorylation and IFNβ production (Fig. 5f, 5g, 5h).
We conclude that cysteines near R293Q and R71H SNPs, Cys-292 and C88xxC91 are important for IFNβ stimulation and speculate that the amino acids changes in these two SNPs may influence the functions of these critical cysteines.
Discussion
In this report, we identified a human MPYS SNP haplotype (HAQ) that encodes a R71H-G230A-R293Q amino acid sequence variant defective in stimulation of IFNβ production. In addition, we show that the IFNβ-stimulating defect of HAQ is caused by the R71H and R293Q.
The identification and study of this IFNβ-stimulating deficient SNPs haplotype (HAQ) has led to important insight to the mechanism of MPYS function. Previous studies suggest that MPYS function as an adaptor protein that recruits IRF3 upon viral infection8. However, we found that the loss-of-function HAQ mutant has similar IRF3 association as WT-MPYS (Supplementary Fig. 1a). Furthermore, though the R293Q SNP is adjacent to a MPYS Traf2 binding site, it also has similar Traf2 binding as WT-MPYS (Supplementary 1b). Thus the IFNβ defect in those SNPs is not easily explained by the adaptor model of MPYS. Instead, we found that SNP-adjacent cysteines, Cys-292 and C88xxC91, are critical for IFNβ stimulation. We speculate that these SNPs may influence the function of adjacent critical cysteines, thus affect IFNβ stimulation.
In our cohorts, the HAQ/HAQ homozygosity accounts for ~3% of the total population (33 out of 1074). Interestingly, according to the frequency data in the dbSNPs, the H71-Q293 homozygosity frequency is almost 9 times higher in Asian (Han Chinese and Japanese) (~15%) than in European (~1.7%). Meanwhile, the H232/H232 homozygosity in Asian is about 1~2%, which is similar to what we found in American in this study (Table 1). This suggests that the HAQ haplotype, but not the H232, may be under natural selection.
MPYS is important for host defense against RNA and DNA viruses in mice 6, 7, thus the HAQ homozygous individuals may be susceptible to viral infection. Conversely, MPYS is also required for Listeria monocytogenes-induced IFNβ response in fibroblasts and macrophages6. Since Type I IFN mediates the susceptibility to Listeria monocytogenes infection11, these HAQ individuals may be resistant to Listeria monocytogenes infection. Furthermore, since MPYS mediates the adjuvant activity of DNA vaccines7, these HAQ individuals may be poor recipients for DNA vaccinations.
In conclusion, we have identified a novel MPYS SNP haplotype (HAQ) that is defective in transmission of signals leading to IFNβ production. Future studies should be directed at understanding the role of HAQ in various human infectious diseases.
Materials and Methods
MPYS SNP Genotyping
Human DNA samples from cohort I were collected at Duke University Medical Center (IRB protocol 1048) and used at National Jewish Health (IRB protocol HS2255). Human DNA samples from cohort II were collected and used at University of Washington, Division of Pulmonary and Critical Care Medicine (IRB protocol 03-8666). The genotyping was performed using custom Taqman probes and 2xTaqman genotyping mix on the Applied Biosystems 7900 Sequence Detection System following standard manufacturer’s protocols. Each 15-μl reaction contained 15ng genomic DNA. Genotypes were determined using the SDS 2.3 software (Applied Biosystems). All human studies were performed in compliance with the US Department of Health and Human Services Guide and approved by related Institutional Review Boards.
MPYS exon sequencing
Primers were designed to amplify exons (1–2), exons (3-4-5), exon 6, exon7a and exon (7b-3′UTR) (Supplementary Table 2). PCR were performed with PrimeSTAR DNA polymerase (TaKaRa, R010A) under the following conditions: 98°C 1 min, 32 cycles of 98°C 10 sec, 60°C 5 sec and 72°C 1 min, then 72°C for 5 min. The PCR products were purified using the QIAquick PCR Purification kit (28106) and directly sequenced using Agencourt Genomic Service.
Cell Culture
The 293MT cells were sub-cultured from 293GT12 cells in DMEM (GIBCO, cat: 11965), 5% FBS (Biosource, 200p-500HI), sodium pyruvate (GIBCO 11360, 1mM), HEPES buffer (GIBCO 15630-080, 10mM) and 2-ME (50μM) (Life Technologies, Gaithersburg MD). Sub-cultured 293GT cells that stimulate IFNβ production following MPYS overexpression were selected as 293MT cells.
Reporter Gene Assay
Cells were seeded in 24-well dishes (~1 × 105/ml) and transfected the following day using Effectene transfection reagent kit (Qiagen, 301427). Reporter assays were performed as previous described 13. All experiments were repeated at least three times and results are representative.
Measuring IFNβ production
Human IFNβ production was measured using the Verikine™ Human IFN-β ELISA kit (PBL interferon source, 41410). Briefly, 293MT cells were cultured with 500μl DMEM medium in 24 wells plate as before. MPYS DNA was transfected into the cells as above. After 24 hrs, the medium was collected. The ELISA was performed using pre-coated plate following manufacturer’s protocols. The IFNβ detection limit is 50pg/ml to 4000pg/ml.
Immunoprecipitation
Cells were lysed in 0.33% CHAPS buffer (150 mM NaCl, 10 mM Tris pH 7.5, 10 mM sodium pyrophosphate, 2 mM Na3VO4, 10 mM NaF, 0.4 mM EDTA, 1 mM PMSF, and 1 μg/ml each of aprotinin, α1-antitrypsin, and leupeptin) at 4°C for 1h. Cell lysates were centrifuged at 12,000g at 4°C for 10 min. Immunoprecipitation was done in the lysates with indicated Ab-conjugated Sepharose beads.
Listeria monocytogenes infection
Cells were infected with Listeria monocytogenes strain 10403S at multiplicity of five bacteria/cell. Cells were washed at 1 h after infection. Live bacteria were killed by addition of gentamicin. Supernatant is collected after 18h after infection.
Intracellular staining
Cells were harvested and washed in FACS buffer (PBS with 2% FCS, 0.05% sodium azide and 0.2μg/ml 2.4g2 Fc-receptor blocking Ab). Cells were then re-suspended in BD Cytofix/Cytoperm™ buffer (BD Bioscience, 554722) for 20 min at RT. BD Perm/Wash buffer™ (BD Bioscience, 554723) was added into the cell suspension. Cells were collected and washed with BD Perm/wash buffer™ again. Cells were then suspended in BD Perm/wash buffer™ containing staining Ab for 15 min at RT. Cells were collected and washed with BD Perm/wash buffer™ twice, then analyzed using a FACSCalibur. Data were analyzed by Flow-Jo software (Tree Star, Inc., San Carlos, CA).
Bone-marrow derived macrophages reconstitution
BMM from STING−/− mice were reconstituted with MSCV-GFP construct expressing MPYS, HAQ or vec. Briefly, MSCV-MPYS-GFP, MSCV-HAQ-GFP or MSCV-Vec-GFP construct was transfected into Amphotropic phoenix cells to make retrovirus. After 48 hrs, supernatant were collected and used to spin-infect the bone-marrow cells as before14. The bone-marrow cells were then cultured in DMEM medium containing 20% FCS,10% L-CSF, 1mM sodium pyruvate, 2mM L-glutamine, 100 units/ml penicillin, 100μg/ml streptomycin, and 50μM 2-ME. After 7 days, cells were examined for F4/80 expression. GFP positive cells were then sorted and used for experiments.
Acknowledgments
This work was supported by NIH 3R01AI062739-05S209 (J.C.C), 5R01AI062739-05 (J.C.C), HL089807-01 (M.M.W) and NIH U19 ES11375 (D.A.S. and I.V.Y.) J.C.C is Ida and Cecil Green Professor of Immunology. We thank Dr. Barber GN for providing STING−/− Femurs. For the human studies, we gratefully acknowledge all participants.
Footnotes
Supplementary information is available at Genes and Immunity website.
Conflict of Interests
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/gene.2010.75
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/gene201075.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The common Sting1 HAQ, AQ alleles rescue CD4 T cellpenia, restore T-regs, and prevent SAVI (N153S) inflammatory disease in mice.
Elife, 13:RP96790, 18 Sep 2024
Cited by: 1 article | PMID: 39291958 | PMCID: PMC11410371
A DNA-Modularized STING Agonist with Macrophage-Selectivity and Programmability for Enhanced Anti-Tumor Immunotherapy.
Adv Sci (Weinh), 11(32):e2400149, 19 Jun 2024
Cited by: 1 article | PMID: 38898748 | PMCID: PMC11348061
Population-enriched innate immune variants may identify candidate gene targets at the intersection of cancer and cardio-metabolic disease.
Front Endocrinol (Lausanne), 14:1286979, 21 Mar 2024
Cited by: 0 articles | PMID: 38577257 | PMCID: PMC10991756
Review Free full text in Europe PMC
The germ theory revisited: A noncentric view on infection outcome.
Proc Natl Acad Sci U S A, 121(17):e2319605121, 05 Apr 2024
Cited by: 2 articles | PMID: 38578984 | PMCID: PMC11047106
At the Crossroads of the cGAS-cGAMP-STING Pathway and the DNA Damage Response: Implications for Cancer Progression and Treatment.
Pharmaceuticals (Basel), 16(12):1675, 01 Dec 2023
Cited by: 1 article | PMID: 38139802 | PMCID: PMC10747911
Review Free full text in Europe PMC
Go to all (72) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (3)
- (2 citations) dbSNP - rs7380824
- (2 citations) dbSNP - rs11554776
- (2 citations) dbSNP - rs1131769
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides.
PLoS One, 8(10):e77846, 21 Oct 2013
Cited by: 122 articles | PMID: 24204993 | PMCID: PMC3804601
The Common R71H-G230A-R293Q Human TMEM173 Is a Null Allele.
J Immunol, 198(2):776-787, 07 Dec 2016
Cited by: 40 articles | PMID: 27927967 | PMCID: PMC5225030
Multiple Homozygous Variants in the STING-Encoding TMEM173 Gene in HIV Long-Term Nonprogressors.
J Immunol, 200(10):3372-3382, 09 Apr 2018
Cited by: 11 articles | PMID: 29632140
TMEM173 variants and potential importance to human biology and disease.
Genes Immun, 20(1):82-89, 01 May 2018
Cited by: 60 articles | PMID: 29728611 | PMCID: PMC6212339
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHLBI NIH HHS (2)
Grant ID: R01 HL089807
Grant ID: HL089807-01
NIAID NIH HHS (5)
Grant ID: R01 AI062739-05
Grant ID: 3R01AI062739-05S209
Grant ID: R01 AI062739
Grant ID: 5R01AI062739-05
Grant ID: R01 AI062739-05S2
NIEHS NIH HHS (2)
Grant ID: U19 ES011375
Grant ID: U19 ES11375





