Abstract
Free full text

Evolution and Disorder
Abstract
The evolution of disordered proteins or regions of proteins differs from ordered proteins because of the differences in their sequence composition, intramolecular contacts and function. Recent assessments of disordered protein evolution at the sequence, structural and functional levels support this hypothesis. Disordered proteins have a different pattern of accepted point mutations, exhibit higher rates of insertions and deletions, and generally, but not always, evolve more rapidly than ordered proteins. Even with these high rates of sequence evolution, a few examples have shown that disordered proteins maintain their flexibility under physiological conditions, and it is hypothesized that they maintain specific structural ensembles.
Introduction
Intrinsically disordered proteins (IDPs) perform essential functions in organisms from viruses to vertebrates, and their improper functioning is responsible for numerous disease states in humans, including some cancers and neurodegenerative diseases [1-6]. These proteins or regions of proteins do not form a compact globular structure and populate broad conformational ensembles that are characterized by molecular motions on multiple timescales [7]. IDPs have a different amino acid composition than ordered proteins, and they often are composed of low complexity sequences [8,9]. The various functions of IDPs range from flexible linkers, whose sole functional constraint appears to be the maintenance of flexibility, to displaying protein modification sites, in which the sites are functionally important, to molecular recognition, in which large regions of the IDP interacts with other proteins [2,4,5].
The sequences of IDPs are expected to be less constrained because they do not form intramolecular, long-range contacts [10]. However, the sequence evolution of some regions within IDPs may be under selection for specific functions. For instance, protein modification sites may be constrained because changing these sites will have a deleterious effect on signaling events [11]. In addition, regions involved in protein-protein interactions are possibly constrained by intermolecular contacts [12]. Additionally, recent evolutionary studies suggest that function is not the sole factor constraining protein evolution [13]. Therefore, looking at the evolution of IDPs not only provides information about their evolution, but about genome evolution in general.
Accepted point mutations differ between disordered and ordered proteins
One way to describe how proteins evolve over time is to compare the sequences of homologous proteins, where homology depends upon the sequences having shared a common ancestor. From these comparisons, models of protein evolution can be inferred based upon the frequency with which different amino acids occur at the same position among the homologues. These models reflect the point mutations that are accepted by evolution because they have a neutral or positive effect upon protein function [14]. Evolutionary models are commonly used to identify or align homologous proteins, however, they also provide information about how proteins evolve.
Two recent studies show that there are differences in the accepted point mutations between disordered and ordered proteins. One study compared evolutionary models inferred from disordered and ordered protein sequences [15]. From these models, it was shown that disordered sequences have a greater likelihood of changing and that the changes are non-conservative compared with ordered sequences. The second study compared the conservation of secondary structure and intrinsic disorder when sequences were evolved under three evolutionary models [16]. This study showed that when disordered protein is evolved by order-biased evolutionary models, the disordered regions are not conserved. Two of the models used in this study were well-known substitutions matrices, PAM120 and BLOSUM62, the third model chose substitutions based on the frequency of the amino acids within the dataset being evolved. Each of these models reflect the evolution of ordered proteins. PAM120 was explicitly based upon protein sequences for which structures had been determined [14]. BLOSUM62 is implicitly based upon structured protein due to the removal of ambiguous alignments that contained gaps [17], when insertions and deletions occur more often in IDPs (Figure 1). In the third model based on amino acid frequencies, the frequencies were taken from the entire protein, not just the disordered region of the protein, which again biases the model away from disordered sequence [18]. Given that these models are biased toward evolutionary changes found in ordered protein, it is not surprising that disordered regions were lost. Additionally, studies of conserved disordered domains indicate that disordered regions, if not their sequences, are maintained over various evolutionary time scales [12,19-24]. Hence, disordered proteins evolve differently from ordered proteins to maintain their disordered structure.
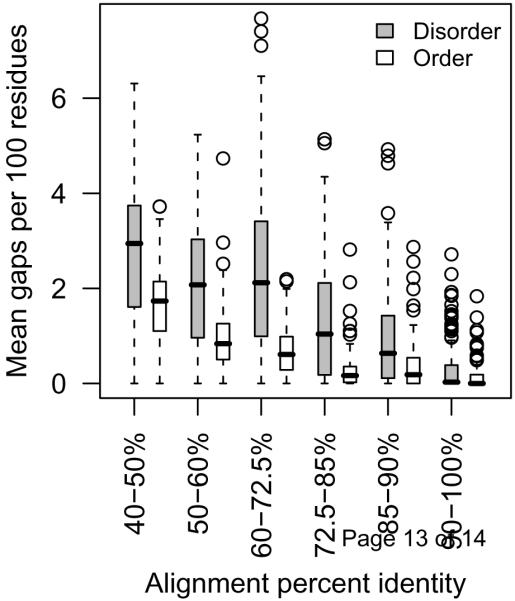
Disordered proteins have more insertions and/or deletions than ordered proteins. Homologues within families of structurally-characterized disordered regions or completely ordered proteins were aligned [15], and the alignments were compared at various levels of percent identity for the number of gaps of any length within the alignment. The box plots show median (dark bar), quartiles ( grey boxes), 1.5x the interquartile range (dashed lines) and means that were outside of this range (circles). There are significantly more gaps within disordered regions of proteins than within the ordered proteins at all levels of percent identity (Wilcoxon rank sum tests, p<0.01). (A. K. Johnson, MS Thesis: Comparing models of evolution between ordered and disordered proteins, University of Idaho, 2008.)
Rates of evolution are generally but not always high in disordered proteins
Another way to describe how proteins evolve over time is to determine the rate at which their amino acid sequences change. An early study of a limited number of proteins containing both ordered and disordered regions showed that the disordered regions of most of these proteins evolve more rapidly than their ordered regions [25]. Because this study compared evolutionary rates within a protein, differences due to transcription and translation were held constant. Other early studies indicated that disordered proteins evolve rapidly by repeat expansions, especially in single amino acid repeats [26,27].
These early studies are supported by recent comparisons among the genomes of several organisms. Genomic studies necessarily rely upon prediction of disorder and order, since relatively few disordered proteins have been characterized structurally. These genomic studies indicate that disordered proteins, in general, evolve rapidly when compared with ordered proteins, although specific instances of slowly-evolving disordered proteins are found [24,28-30]. Indeed, a significant fraction of Pfam domains are predicted to be disordered, and the sequence conservation of these domains is similar to that of Pfam domains predicted to be ordered [21,31]. Identifying structural or biological factors to explain why various disordered regions exhibit high or low evolutionary rates remains a challenging, unsolved problem.
Several recent genomic studies describing biophysical factors that correlate with rates of protein evolution may help in solving this challenging problem. One factor that is found consistently in these studies is the generally slow rate of evolution for genes that are highly expressed, and this slow rate is at both the DNA and protein sequence levels [32]. Recent studies have shown that disordered proteins are very tightly regulated and their levels of gene expression are quite low, which correlates well with high rates of evolution for genes with low expression levels [33,34]. One question that can be asked is what the causative factors are in these correlations. In the case of genes with high expression levels, the slow rates of evolution are hypothesized to be due to the high cost of overcoming protein misfolding [13]. Since disordered proteins do not fold, misfolding is not a pressure that would act on these proteins no matter what their level of expression. Additionally, there appears to be strong selection to reduce the negative consequences of large concentrations of disordered protein by tightly controlling their expression at all levels of transcription, translation and protein degradation [33]. These results suggest that the correlation between low gene expression levels and high rates of evolution may be due to weakly-expressed, rapidly-evolving disordered proteins. Conversely, those disordered proteins that do not evolve rapidly may be less common, highly-expressed proteins.
Evolution of dynamic behavior
Do the large differences in the sequences of homologous IDPs lead to differences in dynamic behavior? To date only two studies have experimentally characterized the dynamic behavior of an IDP family [19,23]. In a study from our groups, NMR relaxation experiments were used to investigate the dynamic behavior of a conserved linker domain from the 70 kDa subunit of replication protein A (RPA70). We showed that dynamic behavior is conserved in the face of negligible sequence conservation. We also showed that dynamic behavior is under selection, even for very flexible IDPs that do not appear to interact with other proteins. In the study by Gage and co-workers, mesophilic and thermophilic variants of the anti-sigma factor, FlgM, were analyzed using circular dichroism. In this study, the thermophilic variant exhibited significant structure (resembling a molten globule) at mesophilic temperatures, and this structure was successively eliminated as temperature was increased. By contrast the mesophilic variant shows no evidence for structure at mesophilic temperatures. It is unclear what molecular features are responsible for this behavior.
The dynamic behavior of IDPs is a poorly understood molecular property that appears to be necessary for function and is probably under multiple levels of evolutionary selection. To understand the evolution of dynamic behavior it is necessary to determine realistic structural ensembles of IDPs from the same protein families. Intrinsically disordered proteins form ensembles of structures that can have a broad range of compactness, secondary structure content, and conformational dynamics. A general model based on local interactions has been proposed to describe the structural ensembles of intrinsically disordered proteins [35-37]. In this model, ensembles are generated using a database of dihedral angles that are observed in the loop regions of folded proteins. Ensembles generated using this method can predict residual dipolar couplings and small-angle X-ray scattering data for intrinsically disordered proteins with high accuracy. The results from both of these studies suggest that any conformational biasing observed in the unfolded state is defined locally. Interestingly, one of the same groups had to modify the method described above to accurately model intrinsically-disordered β-synuclein [38]. The approach was modified to account for the presence of long-range structure by selecting models that satisfied long-range distance restraints. Transient long-range structure was also detected for the intrinsically disordered transactivation domain of the tumor-suppressor protein, p53 and the microtubule-associated protein, tau [39-41]. The functional consequences of the transient long-range structure observed for some intrinsically disordered proteins have not been determined. However, the presence of transient long-range structure is consistent with the notion that intrinsically disordered proteins have been selected to perform functions that require them to maintain a specific structural ensemble. It would be very surprising if all of the different selective pressures acting on intrinsically disordered proteins resulted in a single category of structural ensemble.
Evolution of functional sites within IDPs
One important function of IDPs is displaying sites for protein modification, for example, protein phosphorylation sites [5,42]. The evolutionary characteristics of IDPs, especially their rapid rate of evolution and their propensity for insertions and deletions (Figure 1), may result in these modification sites evolving differently over time than if they were in ordered regions. On the most recent time scale measured by single nucleotide polymorphisms within a species, phosphorylated serines and threonines seem to evolve at the same rate as their non-phosphorylated counterparts [43]. In comparisons of more distantly-related species, such as among mammals, phosphorylated sites are more conserved than non-phosphorylated sites, but phosphorylated sites within IDPs evolve more rapidly than sites in ordered regions [11,43]. In the most distant comparisons, such as between fungi and flies, the positions of phosphorylated sites within homologous IDPs shift, conserving a functional site somewhere within the IDP [44,45]. Thus the often rapid rate of evolution and the greater proportion of insertions and deletions in IDPs influence the perceived rate of evolution of phosphorylation sites at greater divergence times.
Evolution of coupled folding and binding
The most commonly observed function of IDPs is the folding of disordered segments when they bind to other proteins and/or DNA. This coupling of folding and binding is thought to balance the specificity and affinity of protein-protein and protein-DNA interactions, which is essential for the fidelity of many biological processes. If binding is not specific, then the wrong interactions will occur but specific binding requires an extensive molecular interface, which results in a high affinity. If the affinity is too high, then the interaction will last too long and processing will be disrupted. One solution to this problem of balancing specificity and affinity was the evolution of coupled folding and binding [46-48]. When folding and binding are coupled, one (or both) of the interacting partners maintains a state of high conformational entropy when not bound. Some of this conformational entropy is lost during binding because a more ordered structure is formed. This entropy loss reduces the overall affinity of the interaction and this affinity loss can be offset by additional compensating interactions in the bound state, leading to an increase in specificity. Highly specific interactions with modest affinities can be accommodated with this mechanism. Recent theoretical studies lend support to this hypothesis but a rigorous experimental test has not been performed [49].
The term “coupled folding and binding” suggests that both folding and binding occur simultaneously. This is probably not the case and there are at least two step-wise mechanisms, referred to as conformational selection (CS) and induced folding (IF), used to describe the process [48,50]. Both mechanisms assume that an equilibrium distribution of structures exist and some of these structures resemble the bound state and some of these structures do not resemble the bound state (Figure 2). An IDP is said to bind by CS if the structures that resemble the bound state occur at a high enough probability to selectively interact with the binding partner. By contrast, the IDP will bind by IF if there is a very low population of structures that resemble the bound state and all structures are competent for binding in a low affinity state. In this case, just about any structure in the ensemble can bind, and this binding is followed by a conformational rearrangement to form the specific contacts necessary for the final bound state. These two mechanisms are not mutually exclusive and represent two extreme cases of what is possible. In reality most IDP mechanisms probably share some features of both pathways. We propose that the conformational flexibility of the free IDP will determine the probability for a particular pathway. If the flexibility is high, then IF is more probable. If the flexibility is low, then CS is more probable.
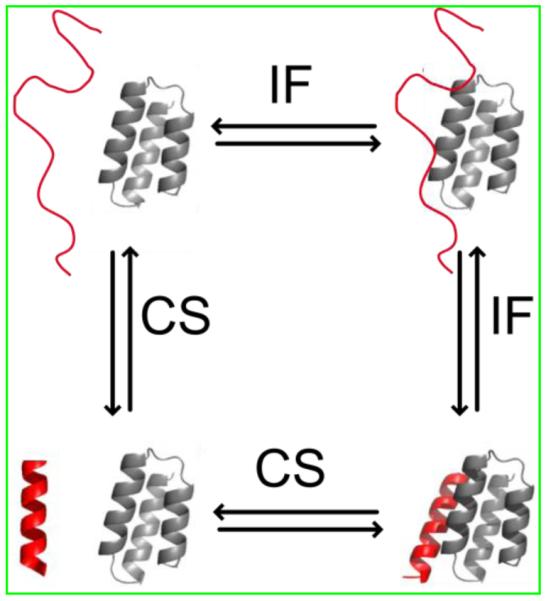
Thermodynamic cycle showing the relationship between induced folding (IF) and conformational selection (CS) binding mechanisms. A representative IDP structure is shown in red. This structure can either bind with low affinity and then fold into the native structure (IF) or can fold into the native structure and then bind (CS). Figure adapted from [54].
Only a few groups have rigorously investigated the mechanism for coupled folding and binding reactions and these studies support the hypothesis that the relative flexibility of the free state of an IDP will determine whether the binding mechanism is induced fit or conformational selection. In a study by Sugase and Wright, the interaction between the phosphorylated kinase inducible activation domain (pKID) of the transcription factor CREB and the KIX domain of the CREB binding protein was investigated using relaxation dispersion [51]. Relaxation dispersion is a type of NMR experiment that allows one to define the on and off rates for the binding of individual amino acids. This information can be combined with knowledge of the conformational flexibility in the free state to distinguish between binding mechanisms that are predominantly induced fit or conformational selection. Interestingly, the results of this study showed evidence for both binding mechanisms in the two different alpha helices that form when pKID binds to KIX. In another study by Levy and Woylnes, a computational approach was used to simulate transition state ensembles for protein-protein interactions where folding and binding are coupled and where folding precedes binding [52]. While this study did not strictly distinguish between conformational selection and induced fit binding mechanisms, it did show a strong correlation between the flexibility of the free state and the kinetics of binding. Binding occurs more rapidly when the flexibility of the free state is high, which is consistent with the induced fit binding mechanism, and more slowly when the flexibility of the free state is low, which is consistent with the conformational selection binding mechanism. A relationship between the flexibility of the free state and binding mechanism was also observed for different mutants of staphylococcal nuclease [53]. In this study, a single point mutant was used to switch the binding mechanism from conformational selection to induced fit. More work is required to determine whether there is a connection between IDP flexibility and their binding mechanisms.
Conclusions
Although disordered proteins often evolve at a fast rate and have different accepted point mutations, the conservation of their flexibility and function indicate that these proteins have important physiological roles in all organisms from viruses to vertebrates. As more comparisons are made among homologous disordered regions and proteins, the evolutionary constraints on flexibility and disordered protein functions, such as coupled folding and binding, will be elucidated. Aside from understanding the evolution of disordered proteins, per se, it is important to study the role of disordered proteins in the context of evolutionary theories that were developed to explain the evolution of ordered proteins.
Acknowledgements
CJB is supported by the National Institutes of Health (P20RR16448). GWD is supported by the National Science Foundation (Award # 0939014) and the American Cancer Society (RSG-07-289-01-GMC). AKD has been supported by the National Institutes of Health (R01LM007688 and R02GM071714) and more recently by the National Science Foundation (MCB-0849803).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.sbi.2011.02.005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3112239?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.sbi.2011.02.005
Article citations
Positive Selection Drives the Evolution of the Structural Maintenance of Chromosomes (SMC) Complexes.
Genes (Basel), 15(9):1159, 03 Sep 2024
Cited by: 0 articles | PMID: 39336750 | PMCID: PMC11431564
Intrinsically disordered sequences can tune fungal growth and the cell cycle for specific temperatures.
Curr Biol, 34(16):3722-3734.e7, 31 Jul 2024
Cited by: 0 articles | PMID: 39089255
Evolutionary analysis of ZAP and its cofactors identifies intrinsically disordered regions as central elements in host-pathogen interactions.
Comput Struct Biotechnol J, 23:3143-3154, 02 Aug 2024
Cited by: 0 articles | PMID: 39234301 | PMCID: PMC11372611
Evolution of Virus-like Features and Intrinsically Disordered Regions in Retrotransposon-derived Mammalian Genes.
Mol Biol Evol, 41(8):msae154, 01 Aug 2024
Cited by: 1 article | PMID: 39101471 | PMCID: PMC11299033
Modulation of biophysical properties of nucleocapsid protein in the mutant spectrum of SARS-CoV-2.
Elife, 13:RP94836, 28 Jun 2024
Cited by: 6 articles | PMID: 38941236
Go to all (177) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Deciphering the cause of evolutionary variance within intrinsically disordered regions in human proteins.
J Biomol Struct Dyn, 35(2):233-249, 04 May 2016
Cited by: 4 articles | PMID: 26790343
Ensembles from Ordered and Disordered Proteins Reveal Similar Structural Constraints during Evolution.
J Mol Biol, 431(6):1298-1307, 05 Feb 2019
Cited by: 4 articles | PMID: 30731089
Protein flexibility and intrinsic disorder.
Protein Sci, 13(1):71-80, 01 Jan 2004
Cited by: 190 articles | PMID: 14691223 | PMCID: PMC2286519
Protein dynamism and evolvability.
Science, 324(5924):203-207, 01 Apr 2009
Cited by: 488 articles | PMID: 19359577
Review
Funding
Funders who supported this work.
American Cancer Society (4)
Grant ID: R01LM007688
Grant ID: R02GM071714
Grant ID: RSG-07-289-01-GMC
Grant ID: MCB-0849803
NCRR NIH HHS (3)
Grant ID: P20 RR016448
Grant ID: P20 RR016448-08
Grant ID: P20RR16448
NIGMS NIH HHS (2)
Grant ID: R01 GM071714-04
Grant ID: R01 GM071714
NLM NIH HHS (3)
Grant ID: R01 LM007688
Grant ID: R01 LM007688-04
Grant ID: R01LM007688
National Institutes of Health (1)
Grant ID: P20RR16448
National Science Foundation (1)
Grant ID: 0939014




