Abstract
Free full text

The extended PP1 toolkit: designed to create specificity
Associated Data
Abstract
Protein Ser/Thr phosphatase-1 (PP1) catalyzes the majority of eukaryotic protein dephosphorylation reactions in a highly regulated and selective manner. Recent studies have identified an unusually diversified PP1 interactome with the properties of a regulatory toolkit. PP1-interacting proteins (PIPs) function as targeting subunits, substrates and/or inhibitors. As targeting subunits, PIPs contribute to substrate selection by bringing PP1 into the vicinity of specific substrates and by modulating substrate specificity via additional substrate docking sites or blocking substrate-binding channels. Many of the nearly 200 established mammalian PIPs are predicted to be intrinsically disordered, a property that facilitates their binding to a large surface area of PP1 via multiple docking motifs. These novel insights offer perspectives for the therapeutic targeting of PP1 by interfering with the binding of PIPs or substrates.
PP1 and the challenge of specificity
Protein phosphorylation represents one of the most common post-translational modifications in eukaryotes. It affects 30–70% of all cellular proteins and some cellular processes, such as the entry into mitosis, are associated with thousands of phosphorylation events [1,2]. In mammals, phosphorylation reactions are catalyzed by ~500 protein kinases [3]. The majority of these phosphorylation events are highly dynamic owing to their ability to be rapidly reversed by protein phosphatases. Whereas the numbers of protein tyrosine kinases and phosphatases are well balanced (~100 each), intriguingly, the mammalian genome encodes only ~40 protein Ser/Thr phosphatases to offset ~400 Ser/Thr kinases [4]. This discrepancy raises the key question of how do so few protein Ser/Thr phosphatases reverse the actions of this large number of protein kinases in a specific and regulated manner? The emerging consensus is that the diversity of protein Ser/Thr phosphatases is, in decisive contrast to Ser/Thr kinases, not achieved primarily by gene duplication, but rather by their unparalleled ability to form stable protein–protein complexes. This property results in the accumulation of an abundant number of phosphatase holoenzymes, each with its own substrate and mode of regulation. This concept has been well illustrated for protein phosphatases-1 (PP1) and -2A (PP2A), which belong to the phosphoprotein phosphatase (PPP) superfamily of protein Ser/Thr phosphatases, and together account for more than 90% of the protein phosphatase activity in eukaryotes [4,5]. Recent data suggest that mammals contain as many as 650 distinct PP1 complexes and approximately 70 PP2A holoenzymes [6], indicating that PP1 catalyzes the majority of protein dephosphorylation events in eukaryotic cells.
In this review, we discuss recently acquired insights that help to explain how PP1 functions in a specific and regulated manner. First, we address the broad substrate specificity of the free catalytic subunit and discuss how its action is controlled by a substrate-targeting and inhibitory toolkit. Next, we discuss how these PP1-interacting proteins (PIPs) form stable complexes with PP1 via degenerate docking motifs, often in the context of a structurally disordered interaction domain. Finally, we highlight the molecular mechanisms of substrate selection and holoenzyme regulation, and explore how structural insights can be used to develop PP1 as a therapeutic target.
The substrate specificity of the catalytic subunit
All members of the PPP superfamily (PP1, PP2A (PP2), PP2B (PP3) and PP4-7) have catalytic cores that share the same structural fold and catalytic mechanism [7]. Differences between these enzymes reside mainly in the solvent-exposed loops that determine the shape and charge of the surface, and hence the affinity for ligands. For example, the catalytic site of PP1 is conspicuously surrounded by acidic residues [8,9]. This feature likely explains why PP1 dephosphorylates the β-subunit of phosphorylase kinase much faster than the more acidic α-subunit, a property that has been widely used to biochemically differentiate PP1 from other protein Ser/Thr phosphatases [10]. Another unique feature of PP1 is that it poorly dephosphorylates short peptides modeled after its physiological substrates, demonstrating that, unlike many protein kinases, PP1 does not recognize a consensus sequence surrounding the phosphorylated residue. Instead, efficient substrate binding depends on docking motifs for PP1 surface grooves that are remote from the active site. Under controlled buffer conditions, the free PP1 catalytic subunit has an exceptionally broad substrate specificity. Bacterially expressed mammalian PP1 even acts as a protein tyrosine phosphatase and can dephosphorylate small molecules such as p-nitrophenylphosphate [8]. However, the catalytic subunit that is purified from mammalian tissues does not act on these atypical substrates. This finding suggests that the native enzyme is more selective, possibly because the metals that are incorporated in the active site (Fe2+ and Zn2+) differ from those of the bacterially expressed enzyme (Mn2+) (Figure 1). Mammalian genomes contain three PP1-encoding genes that together encode four distinct catalytic subunits: PP1α, PP1β/δ and the splice variants PP1γ1 and PP1γ2 [4,11,12], which differ mainly in their extremities. However, the free PP1 isoforms exhibit a similarly broad substrate specificity.

Surface representation of the structure of PP1α (PDB ID 1FJM). (a) The active site of PP1 (green) contains two metal ions (pink spheres) and lies at the Y-shaped intersection of three substrate-binding grooves; the hydrophobic (blue), the acidic (orange), and the C-terminal (red). (b) A 130° rotation of a, showing binding sites for the PP1-docking motifs RVxF (purple), SILK (cyan) and MyPhoNE (wheat).
Physiological substrates of PP1 can be classified into two groups on the basis of their affinity for the catalytic subunit. Some substrates (e.g. the tumor suppressor BRCA1) have high-affinity docking sites for PP1 and form stable heterodimeric complexes with the phosphatase even when they are dephosphorylated (see Table S1 in the supplementary material online). By contrast, other substrates (e.g. glycogen phosphorylase) establish only weak interactions with the catalytic subunit, as suggested by the nearly complete inhibition of their dephosphorylation by physiological salt concentrations and their inability to form stable complexes with PP1 [13]. The efficient in vivo dephosphorylation of the latter substrates requires PIPs that provide additional substrate docking sites or increase the local substrate concentration by tethering the phosphatase to substrate-containing compartments. Thus, substrate selection by PP1 clearly depends on phosphatase docking motifs and subcellular targeting subunits which, together, constitute the substrate-targeting and -specifying toolkit of PP1.
The PP1 protein interactome
Most proteins interact with a limited number of ligands, but a small proportion of proteins, termed hubs, have many partners [14]. Party hubs interact with many of their ligands simultaneously, whereas date hubs bind their distinct partners at different times or locations. PP1 isozymes can be classified as date hubs because they form stable complexes with numerous proteins but only a few proteins can interact simultaneously (see Table S1 in the supplementary material online). PP1-interacting proteins were originally identified using classical biochemical approaches as well as yeast two-hybrid screens. More recently, in silico screenings based on stringent definitions of the five-residue RVxF-type PP1-docking motif, combined with a biochemical validation procedure, have led to a near doubling of the PP1 interactome [15,16]. Novel PP1 complexes also have been identified by affinity chromatography with covalently bound microcystin-LR, a potent small-molecule inhibitor of PPP phosphatases, in combination with the selective elution of PP1-bound proteins by competition with a synthetic RVxF-type docking peptide [17]. Yet another set of PP1 complexes has been identified using antibody arrays [18]. However, these latter two approaches do not differentiate between direct and indirect interaction partners. At present, approximately 180 mammalian genes are known to encode direct PP1 interactors (see Table S1 in the supplementary material online), but the real number is almost certainly much higher. Indeed, a bioinformatics-assisted screen recovered only about one-third of the previously known mammalian PIPs with an RVxF motif [15], indicating that about 450 genes, instead of the currently validated 150, are likely to encode this type of PIP. In addition, unbiased screens suggest that ~30% of PIPs do not have a functional RVxF motif. Collectively, these data suggest that PP1 forms stable complexes with as many as 650 mammalian proteins.
Mutual control of PP1 and PIPs
Although only a minority of PP1 complexes have been functionally analyzed, some recurring themes have emerged. Some PIPs are PP1 substrates, and their controlled dephosphorylation serves a regulatory function. Conversely, many PIPs control PP1 by acting as substrate-targeting subunits or inhibitors. Interestingly, a subset of PIPs are both substrates and regulators for associated PP1.
Substrates
More than a dozen vertebrate PIPs have been identified as PP1 substrates (see Table S1 in the supplementary material online). Some of these substrate PIPs are selectively dephosphorylated on a single site, whereas others are dephosphorylated rather indiscriminately at multiple positions [19–21]. Nearly half of the known substrate PIPs are enzymes. They are often activated by dephosphorylation, as is the case for BRCA1, an E3 ubiquitin ligase, focal adhesion kinase (FAK), the protein phosphatase CDC25C and caspase 2 [20–23]. By contrast, PP1 maintains the associated protein kinases NEK2 and Aurora-A in an inactive state [24]. Dephosphorylation by PP1 stabilizes the transcription factor Ikaros [25] and regulates the binding of ligands to various PIPs [26–29].
Some substrate-PIPs also regulate PP1 function. Inhibitor-2 and the protein kinase-C potentiated inhibitor (CPI-17) are both substrates and potent inhibitors of PP1 [19,30], and protein kinase NEK2 as well as the membrane-targeting protein TIMAP are dephosphorylated by PP1 but also target other proteins in the complex for PP1-mediated dephosphorylation [31,32]. Thus, PP1 has diverse effects on substrate PIPs, with some of these substrates functioning as PP1 regulators. One can envisage that the stable association between PP1 and a subset of its substrates has facilitated the evolution of such a reciprocal relationship, although it cannot be excluded that some PIPs originated as PP1 regulators and became substrates only later in evolution.
Substrate-targeting proteins
Many PIPs contain specific domains that mediate the binding of PP1 to specific cellular compartments or macromolecular complexes (see Table S1 in the supplementary material online). Indeed, PIPs can target PP1 to such diverse structures as the plasma membrane (e.g. integrin αIIB), mitochondria (e.g. URI), endoplasmic reticulum (e.g. the stress-induced protein GADD34), glycogen particles (e.g. G-subunits), the actin cytoskeleton (e.g. spinophilin, also called neurabin-2), chromatin (e.g. Repo-man) and nucleoli (e.g. NOM1). The targeting by PIPs brings PP1 into close proximity to specific subsets of substrates; the associated increased local substrate concentration is sufficient to increase the dephosphorylation rate by up to several orders of magnitude [33].
The concept of multi-targeting emerges from the ability of some PIPs to target PP1 to various signaling complexes, and hence to function as signal integrators. Multi-targeting can be explained by the presence of multiple targeting domains or by competition between substrates for a single, multifunctional targeting domain. Among the best studied PIPs that harbor multiple targeting domains are GADD34, the myosin phosphatase targeting subunit MYPT1, PNUTS, and spinophilin (Figure 2). GADD34 promotes the PP1-mediated dephosphorylation of the translation regulator eIF2α, the GTP-regulatory proteins TSC1 and TSC2, TGFβ receptor 1 as well as I-κB kinase (IKK) [34–37]. The respective targeting to the latter two substrates is mediated by the GADD34-binding adaptor proteins SMAD7 and CUEDC2. MYPT1 contains binding sites for the retinoblastoma protein, polo-like kinase-1, and a number of actin-binding proteins, including myosin, merlin and moesin, which are all PP1 substrates [38–40]. PNUTS targets PP1 to the γ-aminobutyric acid receptor, transcription factor LCP1 and telomeric protein TRF2 [41–43]. In addition to its actin-binding domain, spinophilin contains three other substrate-targeting domains: an interaction domain for G protein-coupled receptors, a PDZ domain that recruits soluble (e.g. the ribosomal protein kinase p70S6K) and integral membrane proteins (e.g. glutamate and ryanodine receptors) and a C-terminal coiled-coil domain that mediates the binding to doublecortin [44–47]. NIPP1 is a targeting PIP with a single, multifunctional substrate-targeting domain [48,49]. It possesses a phosphothreonine-binding forkhead-associated domain that binds various putative PP1 substrates involved in transcription, pre-mRNA splicing and cell-cycle regulation.
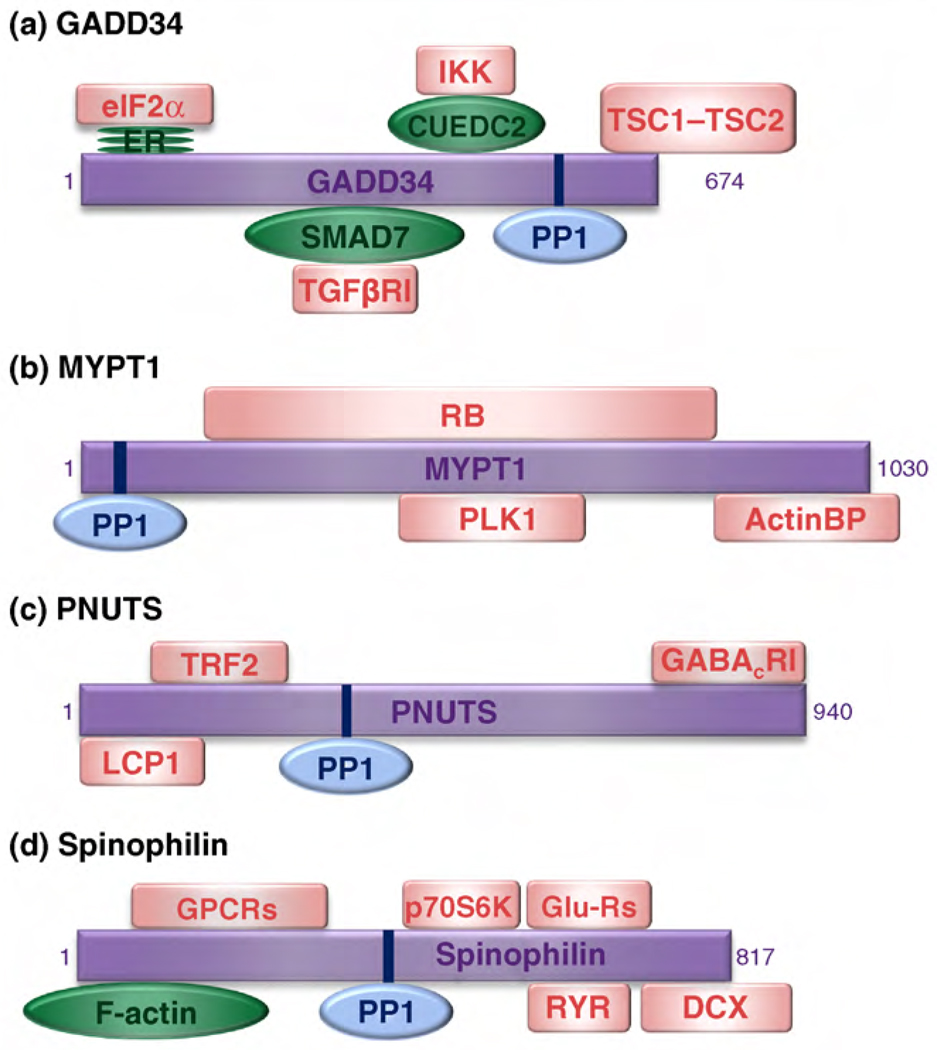
PP1-interacting proteins with multiple substrate-targeting domains. The figure shows selected PIPs that can act as signal integrators because they have multiple substrate-targeting domains. (a) GADD34, (b) MYPT1, (c) PNUTS and (d) spinophilin target PP1 (blue) either directly to its substrates (pink squares) or via adaptor proteins (green ovals). Some of the substrates (LCP1, TRF2, GABACR1 and GPCRs) are still hypothetical. Abbreviations: ER, endoplasmic reticulum; RB, retinoblastoma protein; ActinBP, actin-binding proteins (including myosin, merlin and moesin); GPCRs, G protein-coupled receptors; Glu-Rs, glutamate receptors; RYR, ryanodine receptors.
Substrate-specifiers and inhibitors
More than half of all PIPs inhibit PP1 when glycogen phosphorylase is used as a substrate [15]. Most of these PIPs are poor inhibitors, but some substrate and targeting PIPs, including GADD34 [50], the neurabins [51], PNUTS [52] and NIPP1 [53], are inhibitory in the low nanomolar range. Nevertheless, these proteins are better defined as substrate specifiers than as inhibitors, because they selectively inhibit the dephosphorylation of only a subset of substrates, including glycogen phosphorylase. In addition, at least some of these PIPs have an opposite effect, enhancing specific activity towards the PP1 physiological substrates, as has been best illustrated for the glycogen targeting G-subunits and MYPT1 [12,54].
Nonetheless, some PIPs are true PP1 inhibitors because they block access to the active site and inhibit the dephosphorylation of all substrates. These PIPs constitute the PP1 inhibitory toolkit. Some PIPs, including Inhibitor-1, CPI-17 and their paralogues are inhibitory only when phosphorylated, functioning as pseudosubstrates [11,19]. Similarly, a phosphorylated domain of the targeting PIP MYPT1 acts as a pseudosubstrate inhibitor [55]. The inhibitory activity of other PIPs, including Inhibitor-2 and Inhibitor-3, does not require prior phosphorylation. These inhibitors often function as a second or third noncatalytic subunit of PP1 complexes. However, there is some variation in the architecture of these holoenzymes. In vivo, Inhibitor-3 and CPI-17 paralogues appear to be holoenzyme-specific, as they are present in PP1 complexes containing SDS22 and MYPT1, respectively [19,56]. In these complexes, the targeting and inhibitory subunits have non-overlapping PP1-docking sites. By contrast, Inhibitor-2 functions as an inhibitory subunit of various PP1 holoenzymes, including complexes containing NEK2, spinophilin and Aurora-A [57,58]. Because each of these targeting PIPs, as well as Inhibitor-2, has a functional RVxF motif as one of several PP1-docking sites, they must compete for the RVxF-binding groove within the complex. Although a similar reasoning applies to the complex of PP1 containing GADD34 and Inhibitor-1, this complex is stabilized additionally by interactions between GADD34 and Inhibitor-1 [50].
PP1-docking motifs
Most PIPs contain four to eight residue docking sequences that combine to create a large interaction surface for PP1. On average, a docking motif occupies ~425Å2 of the total PP1 surface, creating an ~850Å2 interaction surface [30,59–61]. Assuming that the entire surface is involved in the binding of PIPs, PP1 has up to 30 non-overlapping PIP-binding sites, much fewer than the number of distinct PIPs (see Table S1 in the supplementary material online). Therefore, PIPs must share PP1-docking motifs, a conclusion that is supported by substantial experimental evidence. However, PIPs differ in the number and the type of their PP1-docking sites, thus enabling a combinatorial control of PP1 [54]. In addition, PP1-docking motifs are degenerate and sequence variants show considerable differences in their affinity for PP1. These structural insights suggest that small-molecule compounds that compete with specific (combinations of) docking motifs for binding to PP1 can be used to functionally disrupt subsets of PP1 holoenzymes and may have a therapeutic potential (Box 1).
The primary PP1-docking motif, commonly referred to as the RVxF motif, is present in about 70% of all PIPs. It generally conforms with the consensus sequence K/R K/R V/I x F/W, where x is any residue other than Phe, Ile, Met, Tyr, Asp, or Pro (see Table S1 in the supplementary material online) [15]. Often, this motif is flanked N-terminally by basic residues and C-terminally by acidic residues. The RVxF sequence binds in an extended conformation to a PP1 hydrophobic groove that is 20Å away from the active site (Figure 1). Binding of this motif does not change the PP1 conformation and functions only to anchor the PIPs to PP1 [30,59–62]. However, this binding event is essential, as it brings PP1 into close proximity with its PIPs and promotes secondary interactions that contribute to PP1 isoform selection and determines the activity and substrate specificity of the holoenzyme [30,51,54,60]. The contribution of the RVxF motif to the binding of PP1 is interactor-dependent; although it is essential for the binding of many PIPs, some PIPs still interact strongly with PP1 in the presence of an excess of a synthetic RVxF peptide or despite alterations in their RVxF motif [54]. Surprisingly, some established RVxF variants, such as the KSQKW sequence of Inhibitor-2, deviate considerably from the consensus RVxF sequence [30]. Thus, the true diversity of RVxF motifs remains elusive, suggesting that the PP1 interactome could be even larger than currently estimated.
A PP1-docking sequence that is present in seven of the known vertebrate PP1 interactors is the so-called SILK-motif (see Table S1 in the supplementary material online), which contains the consensus sequence G/S I L R/K [15]. Always positioned N-terminal to the RVxF sequence, it binds in a hydrophobic groove on the opposite face of the PP1 active site (Figure 1). Identical with the RVxF motif, it does not change the conformation of PP1, but instead fulfills an anchoring function [15,30,62]. MYPT1 contains an N-terminal PP1 interaction motif that adheres to the consensus sequence R x x Q V/I/L K/R x Y/W, where x can be any residue [15,61]. This motif, referred to as the myosin phosphatase N-terminal element or MyPhoNE, is also present in six other PIPs, again always N-terminal to the RVxF sequence (see Table S1 in the supplementary material online). The MyPhoNE motif of MYPT1 lies within a five-turn α-helix, which faces hydrophobic residues in a shallow hydrophobic cleft on PP1 (Figure 1), and contributes to substrate selection.
Some PIPs, including MYPT1 [61] and the neurabins [51], interact with PP1 in an isoform-dependent manner, suggesting that they possess isoform-specific docking sites. Because the PP1 isoforms differ mainly at the N- and C-termini, these represent obvious binding places for specific docking sequences. This is certainly the case for MYPT1, which contains eight tandem ankyrin repeats that interact with the PP1β/δ C-terminus. The interaction centers around two tyrosine residues that are exclusive to the PP1β/δ isoform [61]. However, recent mutagenesis studies suggest that the N-terminal MyPhoNE motif might also contribute to isoform selection [63]. Surprisingly, neurabins do not interact with PP1 N- or C-termini; thus, the basis of their isoform selectivity is unclear [60]. Although an RVxF flanking sequence in spinophilin has been identified as an auxiliary PP1γ1-selectivity determinant [51,64], the available structural data have not disclosed an underlying mechanism [60].
Structural insights into PP1–PIP interactions
In their unbound form, full-length PIPs or their PP1-interacting domains often do not fold into a typical three-dimensional protein structure, hampering their structural characterization. Nonetheless, the few available structures have yielded crucial insights into substrate selection and inhibition.
PIPs as intrinsically disordered proteins
Recent structural studies identified a subset of PIPs, including DARPP-32, Inhibitor-2 and spinophilin, that are highly disordered in their unbound state and do not assume a well defined three-dimensional structure [60,65]. These findings place them in the group of intrinsically unstructured (IUPs) or disordered (IDPs) proteins (Box 2). Their intrinsic flexibility enables them to form extensive and unique interactions with PP1 through several docking motifs, as illustrated by crystal structures of PP1 complexes ([30,60,61], Figure 3). However, there are distinct differences between the unbound forms of these intrinsically disordered PIPs. Detailed analysis has shown that DARPP-32, Inhibitor-2 and spinophilin display different degrees of unstructured character in solution. Whereas the spinophilin PP1-interacting domain lacks any preferred secondary or tertiary structure, distinct preferred conformations can be identified in DARPP-32 and Inhibitor-2 [60,65]. The structural preferences of PIPs in their unbound form could help to drive the formation of complexes withPP1 through conformational selection. The flexibility of these proteins might play a larger role than simply enabling extensive interactions surfaces with PP1, as~70%of Inhibitor-2 remains flexible in the PP1-bound state. In this manner, the residual flexible region might form additional or new binding sites for other proteins.
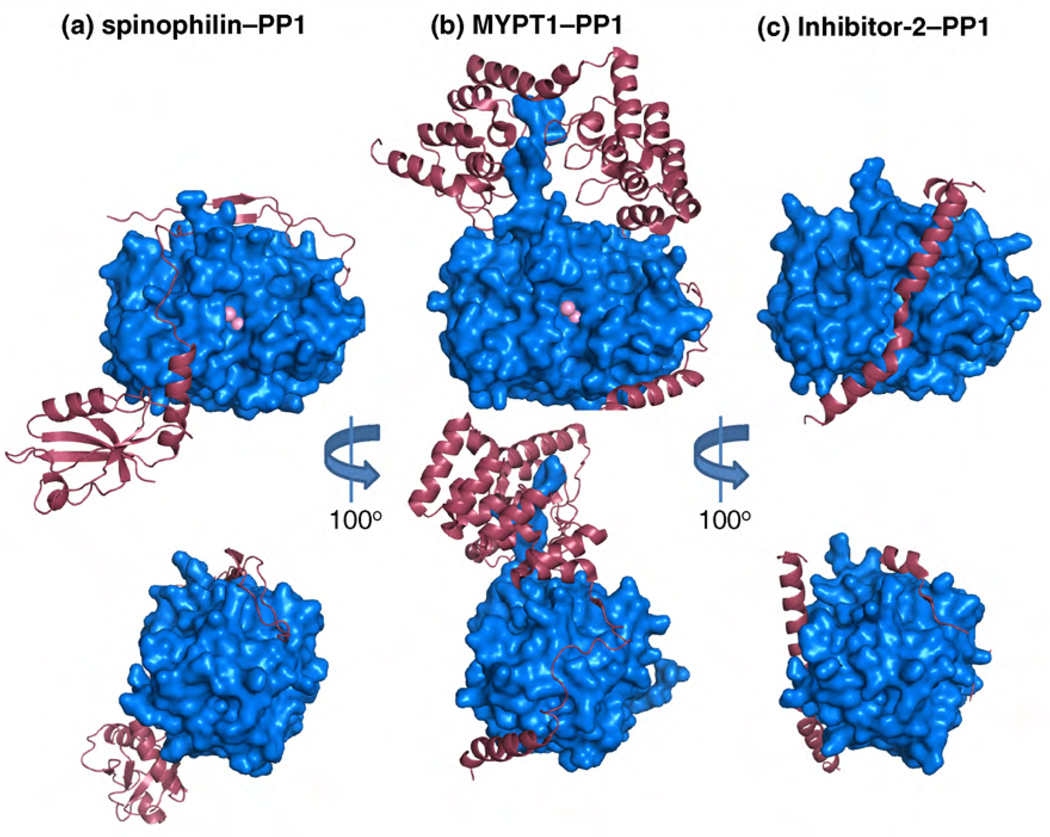
Crystal structures of protein–PP1 complexes. The regulatory proteins are shown as pink ribbons and PP1 as a blue surface representation. Top images are centered around the active site of PP1 whereas the bottom images have been rotated 100° to highlight the interaction within the RVxF docking motif. (a) The crystal structure of spinophilin417–583–PP1α7–330 (PDB ID 3EGG). (b) The crystal structure of MYPT11–299–PP1δ (PDB ID 1S70). (c) The crystal structure of Inhibitor-2–PP1γ (PDB ID 2O8A). These PP1 interacting proteins share only the common RVxF motif interaction. All other interactions are unique for each holoenzyme.
So, how many PIPs have an intrinsically disordered PP1-interaction domain? A bioinformatics analysis using the IUPRED program [66], which scores for the occurrence of disorder-inducing amino acids that are typically enriched in IDPs (Box 2), predicts that the PP1 interaction domain of about two-thirds of all RVxF-type PIPs is disordered in a region of at least 100 residues (see Table S1 in the supplementary material online). In nearly half of these IDPs, the unstructured region flanks both sides of the RVxF sequence. It seems likely that the high percentage of intrinsically disordered PIPs is crucial for the creation of extensive interaction areas upon PP1 binding and thus for the formation of unique PP1 holoenzymes.
Structural basis of substrate selection
PP1 contains three potential substrate-binding grooves: the hydrophobic, the acidic and the C-terminal groove. These grooves form a Y-shape surface that intersects at the PP1 active site (Figure 1). The recently described structure of the spinophilin–PP1 complex revealed that spinophilin blocks the C-terminal substrate-binding groove ([60], Figure 3a). Follow-up mutational and biochemical analysis showed that this interaction plays a crucial role in substrate selection by the spinophilin–PP1 complex. Owing to the obstruction of the C-terminal substrate groove, the activity of the spinophilin–PP1 complex is restricted to specific substrates. Thus, this mode of regulation allows PP1 to dephosphorylate substrates that exclusively bind the acidic and the hydrophobic grooves, while blocking the dephosphorylation of those that require interaction with residues in the C-terminal groove.
At first glance, a similar substrate-binding groove modification cannot be identified in the MYPT1–PP1 structure (Figure 3b). However, C-terminal to the RVxF interaction site, MYPT1 has eight tandem ankyrin repeats that interact with the PP1 C-terminus, extending the PP1 acidic substrate-binding groove, which might play a crucial role in the recruitment of MYPT1–PP1 holoenzyme substrates. However, a biochemical analysis suggests that the MYPT1 N-terminal domain, comprising the MyPhoNE motif, participates in the positive selection of substrates [61].
Structural basis of PP1 inhibition
PP1 is potently inhibited by some small-molecule toxins, including microcystin-LR, nodularin-R and tautomycin, which all block its active site [9,67]. CPI-17, an inhibitor of the MYPT1–PP1 complex, is a structured protein and must be phosphorylated to become a potent inhibitor [19]. Recent findings show that CPI-17 undergoes a significant structural modification upon phosphorylation, which allows the phosphorylated residue to become exposed to the surface and primed for inhibition of the holoenzyme [68]. Inhibitor-1 and its paralogues are also phosphorylation-dependent inhibitors and, although structural information is lacking, it is apparent that its inhibitory activity requires the phosphorylated residue to be pushed into the active site of PP1. By contrast, Inhibitor-2 does not require phosphorylation to inhibit PP1. The structure of the PP1–Inhibitor-2 complex revealed three crucial interaction sites [30], involving an RVxF motif, a SILK-sequence and a long, kinked α-helix of Inhibitor-2 (Figure 3c). This α-helix binds the acidic and hydrophobic substrate-binding grooves and, in doing so, covers the active site, preventing all PP1 activity. In addition, although Inhibitor-2 binding does not induce a conformational change of PP1, it does trigger the release of one of two metals that are essential for catalysis.
Enhanced selectivity through regulation of PIPs
The exquisite specificity of PP1 in vivo is explained by the structural design and diversity of its toolkit, and by various regulatory mechanisms that impinge on PIPs (Figure 4). Some PIPs are expressed in a cell type-dependent manner, accounting for cell type-specific PP1 activity [4,5,11]. Recent data show that the concentration of several PIPs is controlled by regulated proteolysis [69–72]. Moreover, many signaling pathways interfere with the affinity of specific PIPs for PP1. For example, phosphorylation of Ser/Thr residues in or near RVxF-type docking sequences is often associated with a reduced binding affinity for the RVxF-binding channel [54]. Signaling can also result in the recruitment or release of inhibitory PIPs [11,19,73]. Another PIP control mechanism involves positive or negative allosteric regulation by metabolites or other proteins [74,75]. The PP1-mediated dephosphorylation of some substrate- PIPs is restrained through regulated masking of the phosphorylated residues by 14-3-3 proteins [20,23,76–78]. Finally, PP1–PIP complexes are highly dynamic and different PIPs compete for the same PP1 binding sites [77,78]. Ultimately, the concentration and PP1-binding affinities of PIPs determines which PP1 holoenzymes are formed.
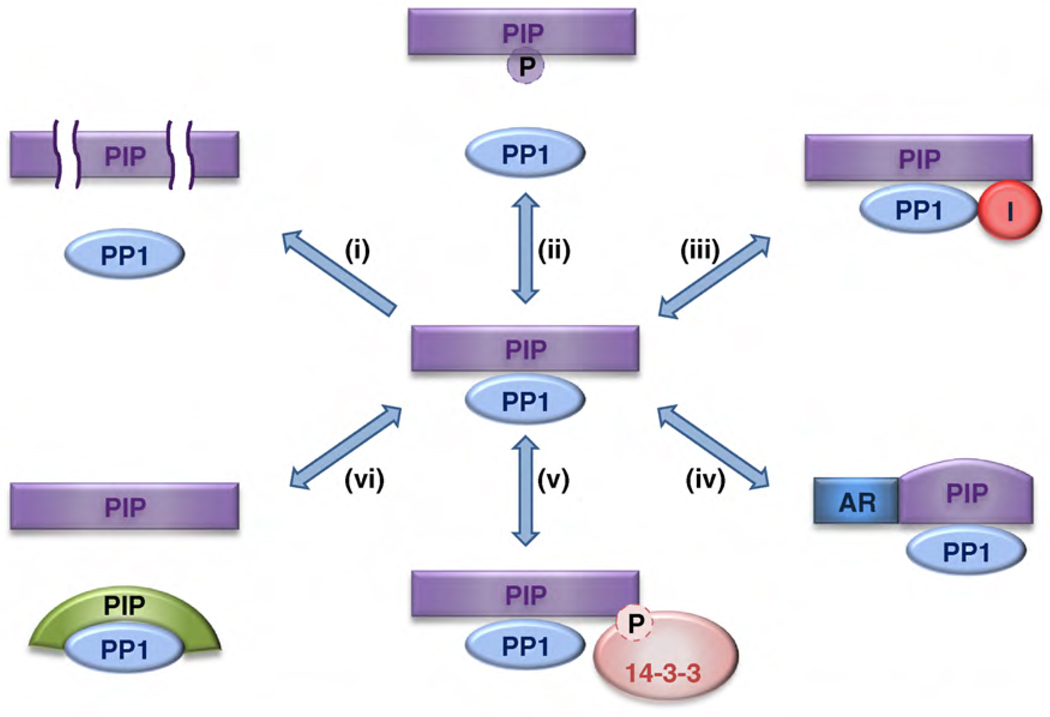
PIP-mediated regulation of PP1 holoenzymes. The figure gives an overview of regulatory mechanisms that affect PIPs, and hence PP1 function. (i) Controlled proteolysis of PIPs. (ii) Dissociation of holoenzymes by the phosphorylation of PIPs. (iii) Recruitment or dissociation of inhibitory PIPs. (iv) Allosteric regulation by the binding of metabolites or proteins to PIPs. (v) Substrate masking by 14-3-3 protein binding. (vi) Competition between PIPs for the same binding sites on PP1. Abbreviations: AR, allosteric regulator; I, inhibitor; P, phosphate.
Concluding remarks and future perspectives
The view of PP1 as an imprecise housekeeping enzyme, which was based largely on assays with the purified catalytic subunit, is no longer tenable. Indeed, PP1 is a highly specific and regulated phosphatase, owing to the unusual diversity and structural design of its regulatory toolkit. There could be as many distinct PP1 complexes as there are protein Ser/Thr kinases, suggesting that both types of enzymes have a similarly restricted substrate specificity at the holoenzyme level. Clearly, much more work is required to uncover the true diversity of the PP1 interactome. In the coming years, the determination of additional atomic resolution three-dimensional structures of PP1 complexes should yield much needed novel insight into the PIP–PP1 interaction and substrate selection mechanisms. This will be a daunting task because many PIPs are intrinsically disordered, rendering crystallization of these holoenzymes extremely difficult. However, as only structural data will reveal how PP1 can be developed as a therapeutic target by interfering with its binding to PIPs and substrates, it is a goal worth pursuing.
Acknowledgements
M.B. is supported by a Concerted Research Action (GOA/10/016) and by the National Science Foundation - Flanders (grant G.0487.08). W.P. is the Manning Assistant Professor for Medical Science at Brown University and is supported by NIH grant R01NS056128. The authors thank Dr R. Page and Dr E. Van Ael for critical comments on this manuscript.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.tibs.2010.03.002.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.tibs.2010.03.002
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3131691?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.tibs.2010.03.002
Article citations
Protein phosphatase-1 regulates the binding of filamin C to FILIP1 in cultured skeletal muscle cells under mechanical stress.
Sci Rep, 14(1):27348, 09 Nov 2024
Cited by: 0 articles | PMID: 39521905 | PMCID: PMC11550807
CAVPENET Peptide Inhibits Prostate Cancer Cells Proliferation and Migration through PP1γ-Dependent Inhibition of AKT Signaling.
Pharmaceutics, 16(9):1199, 12 Sep 2024
Cited by: 0 articles | PMID: 39339236 | PMCID: PMC11434739
SZ-685C inhibits the growth of non-functioning pituitary adenoma by down-regulating miR-340-3p and inducing autophagy.
Heliyon, 10(17):e37230, 30 Aug 2024
Cited by: 0 articles | PMID: 39286117 | PMCID: PMC11402753
Molecular mechanism of IKK catalytic dimer docking to NF-κB substrates.
Nat Commun, 15(1):7692, 03 Sep 2024
Cited by: 0 articles | PMID: 39227404 | PMCID: PMC11371828
gp78-regulated KAP1 phosphorylation induces radioresistance in breast cancer by facilitating PPP1CC/PPP2CA ubiquitination.
iScience, 27(9):110847, 30 Aug 2024
Cited by: 0 articles | PMID: 39297166 | PMCID: PMC11409047
Go to all (324) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (4)
-
(1 citation)
PDBe - 2O8AView structure
-
(1 citation)
PDBe - 3EGGView structure
-
(1 citation)
PDBe - 1S70View structure
-
(1 citation)
PDBe - 1FJMView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The PP1 binding code: a molecular-lego strategy that governs specificity.
FEBS J, 280(2):584-595, 21 Mar 2012
Cited by: 185 articles | PMID: 22360570
Review
Biogenesis and activity regulation of protein phosphatase 1.
Biochem Soc Trans, 45(1):89-99, 01 Feb 2017
Cited by: 53 articles | PMID: 28202662
Review
Interactor-guided dephosphorylation by protein phosphatase-1.
Methods Mol Biol, 1053:271-281, 01 Jan 2013
Cited by: 9 articles | PMID: 23860659
Review
Dissecting the sequence determinants for dephosphorylation by the catalytic subunits of phosphatases PP1 and PP2A.
Nat Commun, 11(1):3583, 17 Jul 2020
Cited by: 22 articles | PMID: 32681005 | PMCID: PMC7367873
Funding
Funders who supported this work.
NIGMS NIH HHS (1)
Grant ID: T32 GM007601
NINDS NIH HHS (3)
Grant ID: R01 NS056128
Grant ID: R01NS056128
Grant ID: R01 NS056128-04





