Abstract
Free full text

Ebola virus entry requires the cholesterol transporter Niemann-Pick C1
Infections by the Ebola (EboV) and Marburg (MarV) filoviruses cause a rapidly fatal hemorrhagic fever in humans for which no approved antivirals are available1. Filovirus entry is mediated by the viral spike glycoprotein (GP), which attaches viral particles to the cell surface, delivers them to endosomes, and catalyzes fusion between viral and endosomal membranes2. Additional host factors in the endosomal compartment are likely required for viral membrane fusion. However, despite considerable efforts, these critical host factors have defied molecular identification3,4,5. Here we describe a genome-wide haploid genetic screen in human cells to identify host factors required for EboV entry. Our screen uncovered 67 mutations disrupting all six members of the HOPS multisubunit tethering complex, which is involved in fusion of endosomes to lysosomes6, and 39 independent mutations that disrupt the endo/lysosomal cholesterol transporter protein Niemann-Pick C1 (NPC1)7. Cells defective for the HOPS complex or NPC1 function, including primary fibroblasts derived from human Niemann-Pick type C1 disease patients, are resistant to infection by EboV and MarV, but remain fully susceptible to a suite of unrelated viruses. We show that membrane fusion mediated by filovirus glycoproteins and viral escape from the vesicular compartment requires the NPC1 protein, independent of its known function in cholesterol transport. Our findings uncover unique features of the entry pathway used by filoviruses and suggest potential antiviral strategies to combat these deadly agents.
We have developed haploid genetic screens to gain insight into biological processes relevant to human disease8,9. Here we use this approach to explore the filovirus entry pathway at unprecedented level of detail. To interrogate millions of gene disruption events for defects in EboV entry, we used a replication-competent vesicular stomatitis virus bearing the EboV glycoprotein (rVSV-GP-EboV)10. Although this virus replicates in most cell lines, it inefficiently killed near-haploid KBM7 cells (Figure S1C). In an unsuccessful attempt to induce pluripotency in KBM7 cells by expression of OCT4, SOX-2, c-MYC and KLF411, we obtained HAP1 cells (Figure S1A). HAP1 cells grew adherently and no longer expressed hematopoietic markers (Figure S1B). The majority of these cells in early passage cultures were haploid for all chromosomes, including chromosome 8 (which is diploid in KBM7 cells). Unlike KBM7 cells, HAP1 cells were susceptible to rVSV-GP-EboV (Figure S1C) allowing screens for filovirus host factors.
We used a retroviral gene-trap vector9 to mutagenize early-passage HAP1 cells. To generate a control dataset, we mapped ~800,000 insertions using deep sequencing (Table S1). Next, we selected rVSV-GP-EboV-resistant cells, expanded them as a pool, and mapped insertion sites. Enrichment for mutations in genes was calculated by comparing a gene’s mutation frequency in resistant cells to that in the control dataset (Figure S2). We identified a set of genes enriched for mutations in the rVSV-GP-EboV-resistant cell population (Figure 1A, S3 and Table S2). Nearly all of these candidate host factors are involved in the architecture and trafficking of endo/lysosomal compartments. Gratifyingly, our screen identified cathepsin B (CatB), the only known host factor whose deletion inhibits EboV entry5. Further inspection showed that mutations were highly enriched in all 6 subunits of the homotypic fusion and vacuole protein-sorting (HOPS) complex (VPS11, VPS16, VPS18, VPS33A, VPS39 and VPS41), for which we identified 67 independent mutations. The HOPS complex mediates fusion of endosomes and lysosomes6 and affects endosome maturation12,13. The identification of all members of the HOPS complex demonstrates high, and possibly saturating, coverage of our screen. We also identified factors involved in biogenesis of endosomes (PIKFYVE, FIG4)14, lysosomes (BLOC1S1, BLOC1S2)15, and in targeting of luminal cargo to the endocytic pathway (GNPTAB)16. The strongest hit was the Niemann-Pick disease locus NPC1, encoding an endo/lysosomal cholesterol transporter7. NPC1 also affects endosome/lysosome fusion and fission17, calcium homeostasis18 and HIV-1 release19.
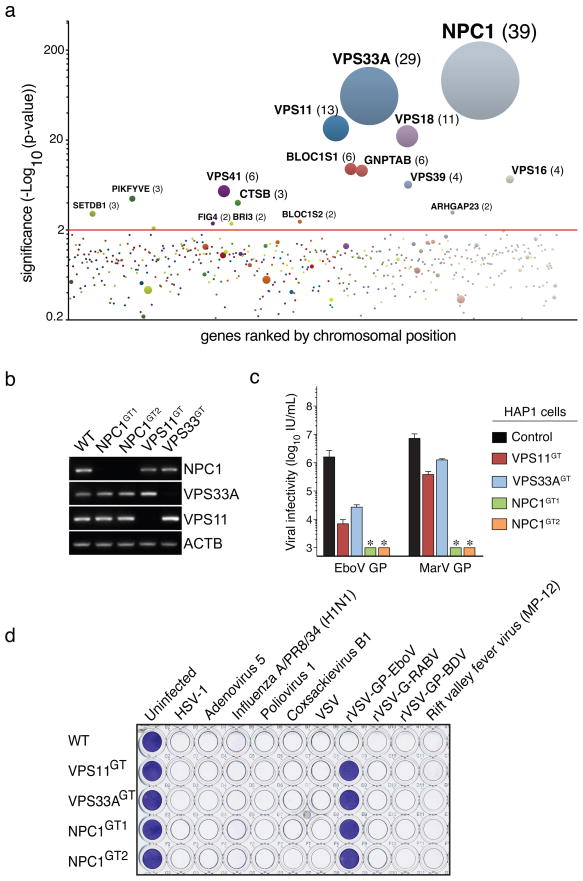
a, Genes enriched for gene-trap insertions in the rVSV-GP-EboV-selected cell population compared to unselected control cells. Circles represent genes and their size corresponds to the number of independent insertions identified in the rVSV-GP-EboV selected population. Genes are ranked on the X-axis based on chromosomal position. b, RT-PCR analysis of the expression levels of NPC1, VPS33A and VPS11 in mutant clones. c, Infectivity of VSV pseudotyped with the indicated filovirus glycoproteins. Means ± standard deviation (SD) (n=3) are shown. EboV, Ebola virus (Zaire), MarV, Marburg virus. *below detection limit. d, HAP1 clones were infected with viruses including recombinant VSV viruses carrying rabies or Borna disease virus glycoproteins (rVSV-G-RABV and rVSV-GP-BDV) and stained with crystal violet.
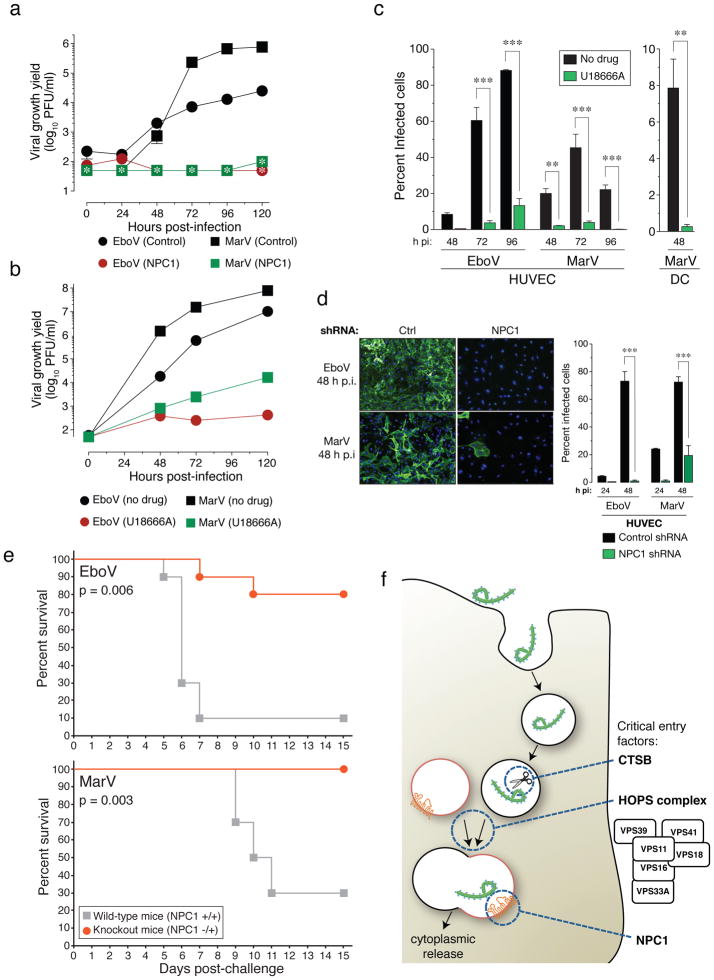
a, NPC1 patient fibroblasts were exposed to EboV or MarV at a multiplicity of infection (MOI) of 0.1. Supernatants were harvested and yields of infectious virus were measured. *below detection limit. b, Vero cells treated with DMSO or U18666A (20 μM) were infected with EboV or MarV at an MOI of 0.1 and yields of infectious virus were measured. c, Human peripheral blood monocyte-derived dendritic cells (DC) and umbilical-vein endothelial cells (HUVEC) were infected in the presence or absence of U18666A at an MOI of 3 and the percentage of infected cells was determined by immunostaining. d, HUVEC were transduced with lentiviral vectors expressing a non-targeting shRNA (Ctrl) or an shRNA targeting NPC1, infected with EboV or MarV at an MOI of 3 and the percentage of infected cells was determined. Representative images of cells are also shown: green, viral antigen; blue, nuclear counterstain. For panels a–d, Means ±SD are shown (n=2 to 3). In panels a–b, error bars are not visible because they are within the symbols. For panels c–d, ** p-value < 0.01, *** p-value < 0.001. e, Survival of NPC1+/+ and NPC1−/+ mice (n=10 for each group) inoculated i.p. with ~1000 pfu of mouse-adapted EboV or MarV. f, A proposed hypothetical model for the roles of CatB, the HOPS complex, and NPC1 in Ebola virus entry.
We subcloned the resistant cell population to obtain clones deficient for VPS11 and VPS33A, and NPC1 (Figure S4A, B and Figure 1B). These mutants displayed marked resistance to infection by rVSV-GP-EboV and VSV pseudotyped with EboV or MarV GP (Figure 1C and Figure S4C). Cells lacking a functional HOPS complex or NPC1 were nonetheless fully susceptible to infection by a large panel of other enveloped and nonenveloped viruses, including VSV and recombinant VSV bearing different viral glycoproteins (Figure 1D and S5). The susceptibility of HAP1 clones to rVSV-GP-EboV infection was restored by expression of the corresponding cDNAs (Figure S6A, B, C).
Loss of NPC1 causes Niemann-Pick disease, a neurovisceral disorder characterized by cholesterol and sphingolipid accumulation in lysosomes7. We tested susceptibility of patient primary fibroblasts to filovirus GP-dependent infection. NPC1-mutant cells were infected poorly or not at all by rVSV-GP-EboV and VSV pseudotyped with filovirus GP proteins (Figure 2A, B), and infection was restored by expression of wild type NPC1 (Figure 2C).
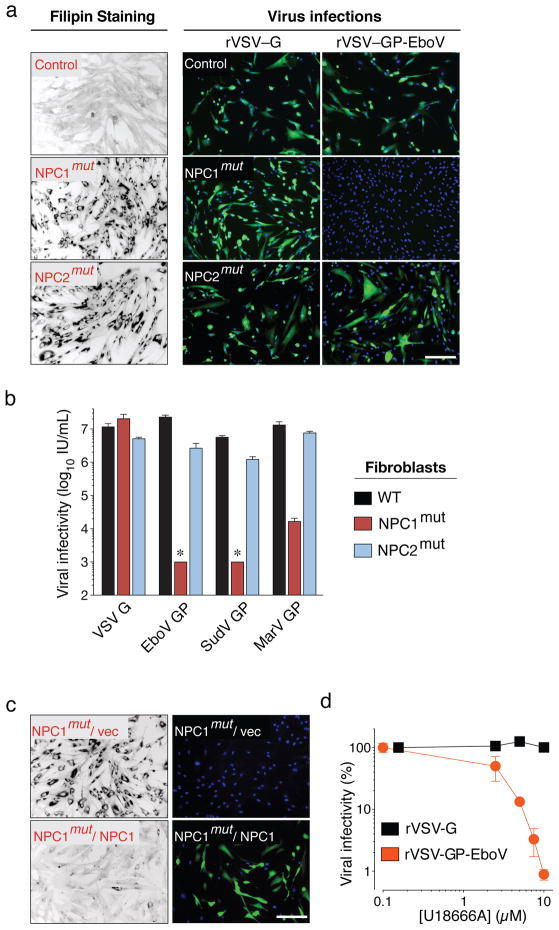
a, Primary skin fibroblasts from a healthy individual and patients carrying homozygous mutations in NPC1 or NPC2 were stained with filipin, or challenged with rVSV-G or rVSV-GP-EboV. Filipin-stained (black) and infected cells (green) were visualized by fluorescence microscopy. Filipin-stained images were inverted for clarity. Blue indicates Hoechst nuclear counterstain. b, Infectivity of VSV pseudotyped with the indicated viral glycoproteins in control and Niemann-Pick fibroblasts. *below detection limit. c, NPC1 patient fibroblasts expressing empty vector or human NPC1 were stained with filipin or challenged with rVSV-GP-EboV. d, Infectivity of rVSV-G and rVSV-GP-EboV in Vero cells preincubated for 30 minutes with the indicated concentrations of U18666A. Scale bars, 200 μm (a, c). Means ±SD (n=3 to 6) are shown (b, d).
Mutations in NPC2 cause identical clinical symptoms and phenocopy defects in lipid transport20. Surprisingly, NPC2-mutant fibroblasts derived from different patients were susceptible to filovirus GP-dependent infection (Figure 2A and Figure S7), despite a similar accumulation of cholesterol in NPC2- and NPC1-mutant cells (Figure 2B). Moreover, cholesterol clearance from NPC1-null cells by cultivation in lipoprotein-depleted growth medium did not confer susceptibility (Figure S8). Therefore, resistance of NPC1-deficient cells to rVSV-GP-EboV is not caused by defects in cholesterol transport per se.
Filoviruses display broad mammalian host and tissue tropism21,22. To determine if NPC1 is generally required for filovirus GP-mediated infection, we used NPC1-null Chinese hamster ovary (CHO) cells. Loss of NPC1 conferred complete resistance to viral infection (Figure S6D) that was reversed by expression of human NPC1 (Figure S6E). Certain small molecules such as U18666A23 and the antidepressant imipramine24 cause a cellular phenotype similar to NPC1 deficiency possibly by targeting NPC123. Prolonged U18666A treatment was reported to modestly inhibit VSV25. However, we found that brief exposure of Vero cells and HAP1 cells to U18666A or imipramine potently inhibited viral infection mediated by EboV GP but not VSV or rabies virus G (Figure 2D, S9, and S10). Because U18666A inhibits rVSV-GP-EboV infection only when added at early time points, it likely affects entry rather than replication (Figure S10). Thus, NPC1 has a critical role in infection mediated by filovirus glycoproteins that is conserved in mammals and likely independent of NPC1’s role in cholesterol transport.
Filoviruses bind to one or more cell-surface molecules2,26,27 and are internalized by macropinocytosis28,29. In VPS33A- and NPC1-mutant cells, we observed no significant differences in binding or internalization of Alexa 647-labeled rVSV-GP-EboV (Figure 3A, Figure S11 and Figure S12A). Similar results were obtained by flow cytometry using fluorescent EboV virus-like particles (Figure S12B). Moreover, bullet-shaped VSV particles were readily observed by electron microscopy at the cell periphery and within plasma membrane invaginations resembling nascent macropinosomes (Figure 3B). Finally, VPS33A- and NPC1-null cells were fully susceptible to vaccinia virus entry by macropinocytosis (Figure S13). Thus, GP-mediated entry is not inhibited at viral attachment or early internalization steps in NPC1- or HOPS-defective cells, suggesting a downstream defect.
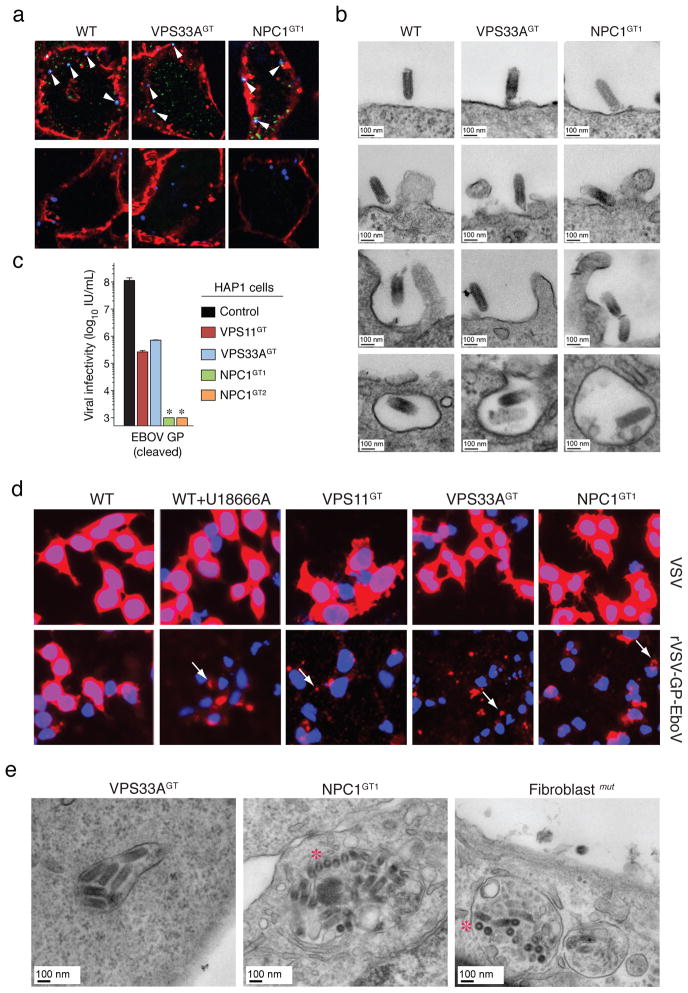
a, Viral particles attach and internalize into HOPS-and NPC1-deficient cells. Indicated HAP1 clones were infected with Alexa 647-labeled rVSV-GP-EboV (blue) at 4°C. Non-internalized, bound viral particles (blue) were also stained with a GP-specific antibody (green) and the plasma membrane with Alexa 594-wheat germ agglutinin (red) (top panels). To assess viral internalization, cells were warmed to 37°C (bottom panels). Internalized viral particles (blue punctae) are resistant to acid-stripping and inaccessible to a GP antibody. b, Cells were inoculated with rVSV-GP-EboV and examined by transmission electron microscopy. Representative images of early entry steps are shown. c, In vitro-cleaved rVSV-GP-EboV cannot bypass the infection block observed in VPS11GT, VPS33AGT and NPC1GT cells. Infectivity of thermolysin-cleaved rVSV-GP-EboV in the indicated HAP1 clones. *below the limit of detection. d, Viral escape into the cytoplasm is blocked in HOPS complex- and NPC1-deficient cells. Wild type HAP1 cells treated with U18666A (10 μg/ml), and the indicated mutant clones were infected with VSV or rVSV-GP-EboV virus for 3h and processed for VSV M staining (red). Punctuate staining is indicated by arrows. e, Electron micrographs of rVSV-GP-EboV-infected VPS33A- and NPC1-deficient HAP1 cells and NPC1-deficient fibroblasts showing agglomerations of bullet-shaped VSV particles in vesicular compartments. All images were taken at 3 h post-inoculation. Asterisks highlight rVSV-GP-EboV particles in cross-section.
Cathepsin L (CatL)-assisted cleavage of EboV GP by CatB is required for viral membrane fusion3,5. Mutant HAP1 cells possess normal CatB/CatL activity (Figure S14B, C) and were fully susceptible to mammalian reoviruses, which utilize CatB or CatL for entry (Fig. S14D). Moreover, these cells remained refractory to in vitro-cleaved rVSV-GP-EboV particles (Figure 3C) that no longer required CatB/CatL activity within Vero cells (Figure S14A). Therefore the HOPS complex and NPC1 are likely required downstream of the initial GP proteolytic processing steps that generate a primed entry intermediate.
Finally, we used the intracellular distribution of the internal VSV M (matrix) protein as a marker for membrane fusion (Figure 3D). Cells were infected with native VSV or rVSV-GP-EboV and immunostained to visualize the incoming M protein. Endosomal acid pH-dependent entry of either virus into wild type HAP1 cells caused redistribution of the incoming viral M throughout the cytoplasm (Figure 3D) (Figure S15A). By contrast, only punctate, perinuclear M staining was obtained in drug-treated and mutant cells infected with rVSV-GP-EboV or rVSV-GP-MarV (Figure 3D and Figure S15B). Electron micrographs of mutant cells infected with rVSV-GP-EboV revealed agglomerations of viral particles within vesicular compartments (Figure 3E and S16A) containing LAMP-1 (Figure S16B), suggesting that fusion and uncoating of incoming virus is arrested. Similarly, U18666A treatment increased the number of viral particles in NPC1-and LAMP1-positive endosomes (Figure S17). Therefore, NPC1 and the HOPS complex are required for late step(s) in filovirus entry leading to viral membrane fusion and escape from the lysosomal compartment.
We next tested if infection by authentic EboV and MarV is affected in NPC1-mutant primary patient fibroblasts. Yields of viral progeny were profoundly reduced for both viruses in mutant cells (Figure 4A). Stark reductions in viral yield were also obtained in Vero cells treated with U18666A (Figure 4B). Moreover U18666A greatly reduced infection of human peripheral blood monocyte-derived dendritic cells and umbilical-vein endothelial cells (HUVEC) (Figure 4C), without affecting cell number or morphology (Figure S19). Finally, knockdown of NPC1 in HUVEC diminished infection by filoviruses (Figure 4D and S18). These findings indicate that NPC1 is critical for authentic filovirus infection.
We assessed the effect of NPC1 mutation in lethal mouse models of EboV and MarV infection. Heterozygous NPC1 (NPC1−/+) knockout mice and their wild type littermates were challenged with mouse-adapted EboV or MarV and monitored for 28 days. Whereas NPC1+/+ mice rapidly succumbed to infection with either filovirus, NPC1−/+ mice were largely protected (Figure 4E).
We have used global gene disruption in human cells to discover components of the unusual entry pathway used by filoviruses. Most of the identified genes affect aspects of lysosome function, suggesting that filoviruses exploit this organelle differently from all other viruses that we have tested (Figure 4F). The unanticipated role for the hereditary disease gene NPC1 in viral entry, infection, and pathogenesis may facilitate the development of anti-filovirus therapeutics.
Methods Summary
Adherent HAP1 cells were generated by the introduction of OCT4/SOX2/c-Myc and KLF4 transcription factors. 100 million cells were mutagenized using a retroviral gene-trap vector. Insertion sites were mapped for approximately 1% of the unselected population using parallel sequencing. Cells were infected with rVSV-GP-EboV and the resistant cell population was expanded. Genes that were statistically enriched for mutation events in the selected population were identified, and the roles of selected genes in filovirus entry were characterized.
Methods online
Cells
KBM7 cells and derivatives were maintained in IMDM supplemented with 10% FCS, L-glutamine, and penicillin streptomycin. Vero cells and primary human dermal fibroblasts (Coriell Institute for Medical Research) were maintained in DMEM supplemented with 10% FCS, L-glutamine, and penicillin streptomycin. Wild type and NPC1-null (CT43) Chinese hamster ovary (CHO) fibroblasts were maintained in DMEM-Ham’s F-12 medium (50–50 mix) supplemented with 10% FCS, L-glutamine, and penicillin streptomycin30.
To generate dendritic cells (DC), primary human monocytes were cultured at 37°C, 5% CO2, and 80% humidity in RPMI supplemented with 10% human serum, L-glutamine, sodium pyruvate, HEPES, penicillin-streptomycin, recombinant human granulocyte monocyte-colony stimulating factor (50 ng/ml) and recombinant human interleukin-4 (50 ng/ml)) for 6 days. Cytokines were added every two days by replacing half of the culture volume with fresh culture media. DC were collected on day 6, characterized by flow cytometry (see below) and utilized immediately. Human umbilical vein endothelial cells (HUVEC) were obtained from Lonza (Walkersville, MD) and maintained in endothelial grown medium (EGM; Lonza).
HAP1 cells were used for the haploid screen and fibroblasts or CHO cells were used for hit validation and functional studies. Vero cells are commonly used in studies of filovirus replication, because they are highly susceptible to infection. DC and HUVEC resemble cell types that are early and late targets of filovirus infection in vivo, respectively31,32.
Flow cytometry of DC
Human DC were treated with Fc-block (BD Pharmingen) prior to incubation with mouse anti-human CD11c-APC (BioLegend) and mouse anti-human CD209-PE or isotype controls. DC were washed and resuspended in PBS for flow cytometric analysis using a BD FACSCanto II flow cytometer (BD Biosciences). Data analysis was completed using FlowJo software. >95% of cells were routinely observed to be CD11c+, DC-SIGN+.
Viruses
Recombinant VSV expressing eGFP and EboV GP (rVSV-GP-EboV) was recovered and amplified as described10. Recombinant rVSV-GP-BDV was generously provided by Juan Carlos de la Torre. rVSV-G-RABV was generated by replacement of the VSV G ORF in VSV-eGFP33 with that of the SAD-B19 strain of rabies virus, and recombinant virus was recovered and amplified34. VSV pseudotypes bearing glycoproteins derived from EboV, Sudan virus, and MarV were generated as described35.
The following non-recombinant viruses were used: Adenovirus type 5 (ATCC), Coxsackievirus B1 (ATCC), Poliovirus 1 Mahoney (generously provided by Christian Schlieker), HSV-1 KOS (generously provided by Hidde Ploegh), Influenza A/PR8/34 (H1N1) (Charles Rivers), Rift valley fever virus MP-12 (generously provided by Jason Wojcechowskyj), and mammalian reovirus serotype 1, (generously provided by Max Nibert).
Generation of HAP1 cells
Retroviruses encoding SOX2, C-MYC, OCT4 and KLF4 were produced36. Concentrated virus was used to infect near haploid KBM7 cells in three consecutive rounds of spin-infection with an interval of 12 hours. Colonies were picked and tested for ploidy. One clonally derived cell line (referred to as HAP1) was further grown and characterized. Karyotyping analysis demonstrated that the majority of the cells (27/39) were fully haploid, a smaller population (9/39) was haploid for all chromosomes except chromosome 8, like the parental KBM7 cells. Less than 10% (3/39) was diploid for all chromosomes except for chromosome 8 that was tetraploid.
Haploid genetic screen
Gene trap virus was produced in 293T cells by transfection of pGT-GFP, pGT-GFP+1 and pGT-GFP+2 combined with pAdvantage, CMV-VSVG and Gag-pol. The virus was concentrated using ultracentrifugation for 1.5 h at 25,000 r.p.m. in a Beckman SW28 rotor. 100 million HAP1 cells were infected. A proportion of the cells was harvested for genomic DNA isolation to create a control dataset. For the screen, 100 million mutagenized cells were exposed to rVSV-GP-EboV at an MOI ~100. The resistant colonies were expanded and ~30 million cells were used for genomic DNA isolation.
Sequence analysis of gene trap insertion sites
Insertion sites were identified by sequencing the genomic DNA flanking gene trap proviral DNA as described before8. In short, a control dataset was generated containing insertion sites in mutagenized HAP1 cells before selection with rVSV-GP-EboV. Genomic DNA was isolated from ~40 million cells and subjected to a linear PCR followed by linker ligation, PCR and sequencing using the Genome Analyzer platform (Illumina). Insertions sites were mapped to the human genome and insertion sites were identified that were located in Refseq genes. Insertions in this control dataset comprise of ~400,000 independent insertions that meet this criteria (Table S1). To generate the experimental dataset, insertions in the mutagenized HAP1 cells after selection with rVSV-GP-EboV were identified using an inverse PCR protocol followed by sequencing using the Genome Analyzer. The number of inactivating mutations (i.e. sense orientation or present in exon) per individual gene was counted as well as the total number of inactivating insertions for all genes. Enrichment of a gene in the screen was calculated by comparing how often that gene was mutated in the screen compared to how often the genes carries an insertion in the control dataset. For each gene a p-value (corrected for false discovery rate) was calculated using the one-sided Fisher exact test (Table S2).
Characterization of the HAP1 mutant lines
Genomic DNA was isolated using Qiamp DNA mini kit (Qiagen). To confirm that the cells were truly clonal and to confirm the absence of the wild type DNA locus, a PCR was performed with primers flanking the insertion site using the following primers: (NPC-F1, 5′-GAAGTTGGTCTGGCGATGGAG-3′; NPC1-R2, 5′-AAGGTCCTGATCTAAAACTCTAG-3′; VPS 33 A–F 1, 5′-TGTCCTACGGCCGAGTGAACC-3′; VPS 33 A–R 1, 5′-CTGTACACTTTGCTCAGTTTCC-3′; VPS 11-F 1, 5′-GAAGGAGCCGCTGAGCAATGATG-3′; VPS 11-R 1, 5′-GGCCAGAATTTAGTAGCAGCAAC-3′. To confirm the correct insertion of the gene trap at the different loci a PCR was performed using the reverse (R1) primers of NPC1, VPS11 and VPS33A combined with a primer specific for the gene trap vector: PGT-F1; 5′-TCTCCAAATCTCGGTGGAAC-3′. To determine RNA expression levels of NPC1, VPS11 and VPS33A, total RNA was reverse transcribed using Superscript III (Invitrogen) and amplified using gene specific primers: (VPS 11: 5′-CTGCTTCCAAGTTCCTTTGC-3′ a n d 5′-AAGATTCGAGTGCAGAGTGG-3′; NPC1: 5′-CCACAGCATGACCGCTC-3′ and 5′-CAGCTCACAAAACAGGTTCAG-3′; VPS 33 A: 5′-TTAACACCTCTTGCCACTCAG-3′ and 5′-TGTGTCTTTCCTCGAATGCTG-3′.
Generation of stable cell populations expressing an NPC1-FLAG fusion protein
A human cDNA endoding NPC1 (Origene) was ligated in-frame to a triple FLAG sequence and the resulting gene encoding a C-terminally FLAG-tagged NPC1 protein was subcloned into the pBABE-puro retroviral vector37. Retroviral particles packaging the NPC1-FLAG gene or no insert were generated by triple transfection in 293T cells, and used to infect control and NPC1-deficient human fibroblasts and CHO lines. Puromycin-resistant stable cell populations were generated.
Cell viability assays for virus treatments
KBM7 and HAP1 cells were seeded at 10,000 cells per well in 96-well tissue culture plates and treated with the indicated concentrations of rVSV-GP-EboV. After three days cell viability was measured using an XTT colorimetric assay (Roche). Viability is plotted as percentage viability compared to untreated control. To compare susceptibility of the HAP1 mutants to different viruses, they were seeded at 10,000 cells per well and treated with different cytolytic viruses at a concentration that in pilot experiments was the lowest concentration to produce extensive cytopathic effects. Three days after treatment, viable, adherent cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) and stained with crystal violet.
VSV infectivity measurements
Infectivities of VSV pseudotypes were measured by manual counting of eGFP-positive cells using fluorescence microscopy at 16–26 h post-infection, as described previously5. rVSV-GP-EboV infectivity was measured by fluorescent-focus assay (FFA), as described previously10.
Filipin staining
Filipin staining to visualize intracellular cholesterol was done as described38. Cells were fixed with paraformaldehyde (3%) for 15 min at room temperature (RT). After three PBS washes, cells were incubated with filipin complex from Streptomyces filipinensis (Sigma-Aldrich) (50 μg/mL) in the dark for 1 h at RT. After three PBS washes, cells were visualized by fluorescence microscopy in the DAPI channel.
Measurements of cysteine cathepsin activity
Enzymatic activities of CatB and CatL in acidified postnuclear extracts of Vero cells, human fibroblasts, and CHO lines were assayed with fluorogenic peptide substrates Z-Arg-Arg-AMC (Bachem Inc., Torrance, CA) and (Z-Phe-Arg)2-R110 (Invitrogen) as described39. As a control for assay specificity, enzyme activities were also assessed in extracts pretreated with E-64 (10 μM), a broad-spectrum cysteine protease inhibitor, as previously described10. Active CatB and CatL within intact cells were labeled with the fluorescently-labeled activity-based probe GB111 (1 μM) and visualized by gel electrophoresis and fluorimaging, as described previously40.
Purification and dye conjugation of rVSV-GP-EboV
rVSV-GP-EboV was purified and labeled with Alexa Fluor 647 (Molecular Probes, Invitrogen Corporation) as described41 with minor modifications. Briefly, Alexa Fluor 647 (Molecular Probes, Invitrogen Corporation) was solubilized in DMSO at 10 mg/mL and incubated at a concentration of 31.25 μg/ml with purified rVSV-GP-EboV (0.5 mg/ml) in 0.1 M NaHCO3 (pH 8.3) for 90 min at RT. Virus was separated from free dye by ultracentrifugation. Labeled viruses were resuspended in NTE (10 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA) and stored at −80°C.
Virus binding/internalization assay
Cells were inoculated with an MOI of 200–500 of Alexa 647-labeled rVSV-GP-EboV at 4°C for 30 min to allow binding of virus to the cell surface. Cells were subsequent fixed in 2% paraformaldehyde (to examine virus binding) or following a 2 h incubation at 37°C and an acid wash to remove surface-bound virus. The cellular plasma membrane was labeled by incubation of cells with 1 μg/mL Alexa Fluor 594 wheat germ agglutinin (Molecular Probes, Invitrogen) in PBS for 15 min at RT. External virus particles were detected using a 1:2000 dilution of antibody 265.1, a mouse monoclonal specific for Ebola GP. The GP antibodies were detected by Alexa 488-conjugated goat anti-mouse secondary antibody (Molecular Probes, Invitrogen). After washing with PBS, cells were mounted onto glass slides using Prolong Antifade Reagent (Invitrogen, Molecular Probes). Fluorescence was monitored with a epifluorescence microscope (Axiovert 200M; Carl Zeiss, Inc.; Thornwood, NY) and representative images were acquired using Slidebook 4.2 software (Intelligent Imaging Innovations; Denver, CO)41,42.
VSV M protein-release assay
Cells grown on 12 mm coverslips coated with poly-D-lysine (Sigma-Aldrich) were pre-treated with 5 μg/ml puromycin for 30 min and inoculated with rVSV at an MOI of 200–500 in the presence of puromycin. After 3 h, cells were washed once with PBS and fixed with 2% paraformaldehyde in PBS for 15 min at RT. To detect VSV M protein, fixed cells were incubated with a 1:7500 dilution of monoclonal antibody 23H12 (kind gift of Doug Lyles43), in PBS containing 1% BSA and 0.1 % Triton X-100 for 30 min at RT. Cells were washed three times with PBS, and the anti-M antibodies were detected using a 1:750 dilution of Alexa 594-conjugated goat anti-mouse secondary antibodies. Cells were counter-stained with DAPI to visualize nuclei. Cells were washed three times and mounted onto glass slides after which M localization images were acquired using a Nikon TE2000-U inverted epifluorescence microscope (Nikon Instruments, Inc.; Melville, NY). Representative images were acquired with Metamorph software (Molecular Devices).
Electron microscopy
Confluent cell monolayers in 6-well plates were inoculated with rVSV-GP-EboV at a MOI of 200–500 for 3 h. Cells were fixed for at least 1 h at RT in a mixture of 2.5% glutaraldehyde, 1.25% paraformaldehyde and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4). Samples were washed extensively in 0.1 M sodium cacodylate buffer (pH 7.4) and treated with 1% osmiumtetroxide and 1.5% potassiumferrocyanide in water for 30 min at RT. Treated samples were washed in water, stained in 1% aqueous uranyl acetate for 30 min, and dehydrated in grades of alcohol (70%, 90%, 2×100%) for 5 min each. Cells were removed from the dish with propyleneoxide and pelleted at 3,000 rpm for 3 min. Samples were infiltrated with Epon mixed with propyleneoxide (1:1) for 2 h at RT. Samples were embedded in fresh Epon and left to polymerize for 24–48 h at 65°C. Ultrathin sections (about 60–80 nm) were cut on a Reichert Ultracut-S microtome and placed onto copper grids. For preparation of cryosections the virus-inoculated cells were rinsed once with PBS and removed from the dish with 0.5 mM EDTA in PBS. The cell suspension was layered on top of an 8% paraformaldehyde cushion in an eppendorf tube and pelleted for 3 min at 3.000 rpm. The supernatant was removed and fresh 4% paraformaldehyde was added. After 2 h incubation, the fixative was replaced with PBS. Prior to freezing in liquid nitrogen the cell pellets were infiltrated with 2.3 M sucrose in PBS for 15 min. Frozen samples were sectioned at −120°C and transferred to formvar-carbon coated copper grids. Grids were stained for lysosomes with a mouse monoclonal antibody raised against LAMP1 (H4A3; Santa Cruz Biotechnology, Inc.). The LAMP1 antibodies were visualized with Protein-A gold secondary antibodies. Contrasting/embedding of the labeled grids was carried out on ice in 0.3% uranyl acetete in 2% methyl cellulose. All grids were examined in a TecnaiG2 Spirit BioTWIN mission electron microscope and images were recorded with an AMT 2k CCD camera.
Authentic filoviruses and infections
Vero cells were pretreated with culture medium lacking or containing U18666A (20 μM) for 1 h at 37°C. VERO cells and primary human dermal fibroblasts were exposed to EboV-Zaire 1995 or MarV-Ci67 at an MOI of 0.1 for 1 h. Viral inoculum was removed and fresh culture media with or without drug was added. Samples of culture supernatants were collected and stored at −80°C until plaque assays were completed.
DC were collected and seeded in 96-well poly-d-lysine coated black plates (Greiner Bio-One) at 5×104 cells per well or in 6 well plates at 106 cells per well in culture media and incubated overnight at 37°C. They were pretreated with medium lacking or containing U18666A as described above. DC were exposed to EboV-Zaire 1995 or MarV-Ci67 at an MOI of 3 for 1 h. Virus inoculum was removed and fresh culture media with or without drug was added. Uninfected cells with or without drug served as negative controls. Cells were incubated at 37°C and fixed with 10% formalin at designated times. HUVEC were seeded in 96-well poly-d-lysine coated black plates at 5 × 104 cells per well in culture media, treated with U18666A, infected, and processed as described above for DC.
Cytotoxicity analysis
DC and HUVEC were seeded in 96-well plates. Following overnight incubation at 37°C, U18666A was added at the same concentrations used for the viral infection studies. Cells in culture media without drug served as the untreated control. At indicated times post treatment, an equal volume of Cell Titer-Glo Reagent (Promega) was added to wells containing cells in culture media. Luminescence was measured using a plate reader.
Plaque assays for titration of filoviruses
Tenfold serial dilutions of culture supernatants or serum were prepared in modified Eagle medium with Earle’s balanced salts and nonessential amino acids (EMEM/NEAA) plus 5% heat -inactivated fetal bovine serum. Each dilution was inoculated into a well of a 6-well plate containing confluent monolayers of Vero 76 cells. After adsorption for 1 hour at 37°C monolayers were overlaid with a mixture of 1 part of 1% agarose (Seakem) and 1 part of 2X Eagle basal medium (EBME), 30mM Hepes buffer and 5% heat- inactivated fetal bovine serum. Following incubation at 37°C, 5% CO2, 80% humidity for 6 days, a second overlay with 5% Neutral Red was added. Plaques were counted the following day, and titers were expressed as PFU/ml.
Analysis of filovirus-infected cultures by immunofluorescence
Formalin-fixed cells were blocked with 1% bovine serum albumin solution prior to incubation with primary antibodies. EboV-infected cells and uninfected controls were incubated with EboV GP-specific monoclonal antibodies 13F644 or KZ5245. MarV-infected cells and uninfected controls were incubated with MarV GP-specific monoclonal antibody 9G4. Cells were washed with PBS prior to incubation with either goat anti-mouse IgG or goat anti-human IgG conjugated to Alexa 488. Cells were counterstained with Hoechst stain (Molecular Probes®), washed with PBS and stored at 4°C.
Image analysis
Images were acquired at 9 fields/well with a 10× objective lens on a Discovery-1 high content imager (Molecular Devices) or at 6 fields/well with a 20× objective lens on an Operetta (Perkin Elmer) high content device. Discovery-1 images were analyzed with the “live/dead” module in MetaXpress software. Operetta images were analyzed with a customized scheme built from image analysis functions present in Harmony software.
Animals and filovirus challenge experiments
Mouse-adapted EboV has been described46. Mouse-adapted MarV Ci67 was provided by Sina Bavari47. Female and male BALB/c NPC1+/− mice and BALB/c NPC1 +/+ mice (5 to 8 week old) were obtained from Jackson Laboratory (Bar Harbor, ME). Mice were housed under specific-pathogen-free conditions. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adhered to principles stated in the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996). The facility where this research was conducted is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. For infection, mice were inoculated i.p. with a target dose of 1000 pfu (30,000 X the 50% lethal dose) of mouse-adapted EboV or mouse-adapted MarV Ci67 virus in a biosafety level 4 laboratory. Mice were observed for 28 days after challenge by study personnel and by an impartial third party. Daily observations included evaluation of mice for clinical symptoms such as reduced grooming, ruffled fur, hunched posture, subdued response to stimulation, nasal discharge, and bleeding. Serum was collected from surviving mice to confirm virus clearance. Back titration of the challenge dose by plaque assay determined that EboV-infected mice received 900 pfu/mouse and MarV-infected mice received 700 pfu/mouse.
RNA interference
Lentiviral vectors expressing an shRNA specific for NPC1 (Sigma-Aldrich; clone# TRCN0000005428; sequence CCACAAGTTCTATACCATATT) or a nontargeting control shRNA (Sigma-Aldrich; SHC002; sequence CAACAAGATGAAGAGCACCAA) were packaged into HIV-1 pseudotype virus by transfection in HEK-293T cells and lentivirus-containing supernatants were harvested at 36h and 48 h post-transfection and centrifuged onto HUVEC in 12-well plates in the presence of 6 μg/mL polybrene at 2500 rpm, 25°C for 90 min. HepG2 cells were transduced as above but without the centrifugation step. Cells were subjected to puromycin selection 24 h after the last lentiviral transduction (HepG2, 1 μg/mL; HUVEC, 1.5 μg/mL) for 48–72 h prior to harvest for experiments. The level of NPC1 knockdown was assessed by SDS-polyacrylamide gel electrophoresis of cell extracts and immunoblotting with an α-NPC1 polyclonal antibody (Abcam).
EboV Replicon Assay
EboV support plasmids were created by cloning the NP, VP35, VP30 and L genes from cDNA (generously provided by Elke Mühlberger48) into pGEM3 (Promega) and the mutant pL-D742A plasmid was generated by Quik-Change site-directed mutagenesis (Stratagene). Truncated versions of the EboV non-coding sequence were generated by overlap-extension PCR and appended to the eGFP ORF. The replicon pZEm was prepared as described previously49. The replicon RNA sequence is flanked on the 5′ end by a truncated T7 promoter with a single guanosine nucleotide and on the 3′ end by the HDV ribozyme sequence and T7 terminator. The transcribed replicon RNA consists of the following EboV Zaire sequences (Genbank accession AF086833): [5′] – single guanosine nt −176 nt genomic 5′ terminus – 55 nt L mRNA 3′ UTR – eGFP ORF (antisense orientation) – 100 nt NP mRNA 5′ UTR – 155 nt genomic 3′ terminus [3′]. The viral replicon assay was performed as described previously49 except that U18666A (20 μg/mL) was included in the supplemented DMEM where indicated. Images were collected directly from 6-cm dishes with a Zeiss Axioplan inverted fluorescent microscope.
Supplementary Material
1
2
3
4
5
6
Acknowledgments
We would like to thank M. Kielian, H. Ploegh, V. Prasad, and D. Sabatini for critical reading of the manuscript and valuable advice, C. Guimaraes, V. Blomen and T. Peterson for helpful suggestions, M. Bogyo for providing the CatB/CatL activity probe (GB111), T.-Y. Chang for his gift of NPC1-null CHO cells, D. Lyles for the antibody to VSV M, M. Nibert for providing reovirus, J. de la Torre for providing rVSV-GP-BDV, J. Wojcechowskyj for providing RVF, E. Mühlberger for providing Ebola cDNA and M. Ericsson for support with electron microscopy. This research was supported by NIH grants R01 AI088027 (K.C.), AI081842 and U54 AI057159 (NERCE-BEID) (S.P.W.), and R21 HG004938 (T.R.B.), and by the U.S. Army Project, #CBM.VAXPLAT.05.10.RD.005 (J.M.D.). T.R.B. was additionally supported by the Whitehead Fellows Program. S.P.W. is a recipient of a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award. A.W. was additionally supported by NIH-funded training programs T32 GM007288 and T32 AI070117 at the Albert Einstein College of Medicine. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Footnotes
Author contributions
K.C., S.P.W., T.R.B., and J.M.D. contributed equally to this study. The study was conceived by K.C., S.P.W. and T.R.B. J.E.C. and T.R.B. devised and implemented the haploid genetic screen, generated the HAP1 cells and identified hits by deep sequencing and cell cloning. P.D.C. carried out karyotype analysis on the HAP1 line. K.C. created and characterized the rVSV-GP-EboV virus used in the screen. A.M.G. created the rVSV-G-RABV. J.E.C., G.O., and K.C. performed entry and infection experiments with the HAP1 cells. A.C.W. and K.C. carried out entry and infection experiments with rVSVs in human fibroblasts, CHO and Vero cells. N. M. and K.C. carried out RNAi experiments with primary cells. M.R. was involved in experimental strategy and design and performed entry and infection experiments by high-resolution fluorescence and electron microscopy. N.M. carried out VLP entry experiments and P.J.K the replicon assay. A.C.W. performed the cysteine cathepsin enzyme assays. A.S.H., A.I.K and J.M.D. performed the infection and animal challenge experiments with the authentic viral agents. G.R. performed fluorescence microscopy and image analysis with filovirus-infected cell cultures. J.E.C., K.C., S.P.W., and T.R.B. wrote the paper.
Competing financial interests
J.E.C., M.R., S.P.W., K.C. and T.R.B. have filed a patent on filovirus host factors identified in this study and T.R.B. is a co-founder of Haplogen, an early-stage company involved in haploid genetic approaches.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature10348
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3175325?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Characterizing Marine Medaka (Oryzias melastigma) Haploid Embryonic Stem Cells: A Valuable Tool for Marine Fish Genetic Research.
Animals (Basel), 14(18):2739, 21 Sep 2024
Cited by: 0 articles | PMID: 39335329 | PMCID: PMC11428384
Cholesterol and Cholesterol-Lowering Medications in COVID-19-An Unresolved Matter.
Int J Mol Sci, 25(19):10489, 29 Sep 2024
Cited by: 0 articles | PMID: 39408818 | PMCID: PMC11477656
Review Free full text in Europe PMC
Emerging and reemerging infectious diseases: global trends and new strategies for their prevention and control.
Signal Transduct Target Ther, 9(1):223, 11 Sep 2024
Cited by: 0 articles | PMID: 39256346 | PMCID: PMC11412324
Review Free full text in Europe PMC
Discovery of Thiophene Derivatives as Potent, Orally Bioavailable, and Blood-Brain Barrier-Permeable Ebola Virus Entry Inhibitors.
J Med Chem, 67(18):16381-16402, 09 Sep 2024
Cited by: 0 articles | PMID: 39248591 | PMCID: PMC11440591
Lipid Selectivity of Membrane Action of the Fragments of Fusion Peptides of Marburg and Ebola Viruses.
Int J Mol Sci, 25(18):9901, 13 Sep 2024
Cited by: 0 articles | PMID: 39337389 | PMCID: PMC11432738
Go to all (825) article citations
Other citations
Wikipedia (Showing 7 of 7)
- https://en.wikipedia.org/wiki/Marburg_virus
- https://en.wikipedia.org/wiki/MAb114
- https://en.wikipedia.org/wiki/Niemann–Pick_disease
- https://en.wikipedia.org/wiki/Sean_Whelan_(scientist)
- https://en.wikipedia.org/wiki/NPC1
- https://en.wikipedia.org/wiki/KBM-7_cells
- https://en.wikipedia.org/wiki/Zaire_ebolavirus
Show less
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AF086833
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Filoviruses Use the HOPS Complex and UVRAG To Traffic to Niemann-Pick C1 Compartments during Viral Entry.
J Virol, 94(16):e01002-20, 30 Jul 2020
Cited by: 6 articles | PMID: 32493822 | PMCID: PMC7394885
Direct Intracellular Visualization of Ebola Virus-Receptor Interaction by In Situ Proximity Ligation.
mBio, 12(1):e03100-20, 12 Jan 2021
Cited by: 4 articles | PMID: 33436438 | PMCID: PMC7844541
Ebola virus entry requires the host-programmed recognition of an intracellular receptor.
EMBO J, 31(8):1947-1960, 06 Mar 2012
Cited by: 224 articles | PMID: 22395071 | PMCID: PMC3343336
Filovirus entry: a novelty in the viral fusion world.
Viruses, 4(2):258-275, 07 Feb 2012
Cited by: 61 articles | PMID: 22470835 | PMCID: PMC3315215
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHGRI NIH HHS (2)
Grant ID: R21 HG004938-01
Grant ID: R21 HG004938
NIAID NIH HHS (9)
Grant ID: AI081842
Grant ID: R01 AI081842
Grant ID: R56 AI081842
Grant ID: T32 AI070117
Grant ID: R01 AI081842-03
Grant ID: R01 AI088027-03
Grant ID: U54 AI057159
Grant ID: R01 AI088027
Grant ID: U54 AI057159-09
NIGMS NIH HHS (1)
Grant ID: T32 GM007288





