Abstract
Free full text

Evidence for N-Glycan Shielding of Antigenic Sites during Evolution of Human Influenza A Virus Hemagglutinin
Abstract
After the emergence of influenza A viruses in the human population, the number of N-glycosylation sites (NGS) in the globular head region of hemagglutinin (HA) has increased continuously for several decades. It has been speculated that the addition of NGS to the globular head region of HA has conferred selective advantages to the virus by preventing the binding of antibodies (Ab) to antigenic sites (AS). Here, the effect of N-glycosylation on the binding of Ab to AS in human influenza A virus subtype H3N2 (A/H3N2) was examined by inferring natural selection at AS and other sites (NAS) that are located close to and distantly from the NGS in the three-dimensional structure of HA through a comparison of the rates of synonymous (dS) and nonsynonymous (dN) substitutions. When positions 63, 122, 126, 133, 144, and 246 in the globular head region of HA were non-NGS, the dN/dS was >1 and positive selection was detected at the AS located near these positions. However, the dN/dS value decreased and the evidence of positive selection disappeared when these positions became NGS. In contrast, dN/dS at the AS distantly located from the positions mentioned above and at the NAS of any location were generally <1 and did not decrease when these positions changed from non-NGS to NGS. These results suggest that the attachment of N-glycans to the NGS in the globular head region of HA prevented the binding of Ab to AS in the evolutionary history of human A/H3N2 virus.
INTRODUCTION
Influenza virus is a member of the family Orthomyxoviridae, which contains a segmented and negative-stranded RNA genome in an enveloped virion. This virus is classified into types A to C, and influenza A virus has caused four global pandemics in humans during the last hundred years (18, 22). Influenza A virus is further classified into subtypes based on the genetic and antigenic characteristics of hemagglutinin (HA) and neuraminidase (NA), which constitute envelope glycoproteins. To date, 16 HA (H1 to H16) and 9 NA (N1 to N9) subtypes have been identified, and influenza A viruses bearing H1N1 and H3N2 subtypes (A/H1N1 and A/H3N2 viruses) are currently cocirculating in humans.
Influenza A virus escapes from host immune responses by changing the antigenicity of HA and NA both gradually (antigenic drift) and abruptly (antigenic shift) (16). The HA consists of signal peptide, HA1 (amino acid positions 1 to 328 in the H3 numbering system), and HA2 (amino acid positions 330 to 550), and it forms trimers in virions (30). The ectodomain of an HA trimer can be divided into globular head and stem regions (30). The globular head region is composed of a part of HA1 (amino acid positions 58 to 272), while the stem region is composed of the other parts of HA1 and HA2. Antigenic sites (AS), which are targets of antibodies (Ab), constitute five epitopes (A to E) in A/H3N2 viruses. Epitopes A, B, D, and E reside in the globular head region, while epitope C is in the stem region of HA1 (24, 29).
Although the escape from host immune responses occurs through changes in amino acids at AS that are recognized by Ab (14, 18, 23), it has been proposed that the attachment of an oligosaccharide to the N-glycosylation site (NGS), which is the Asn residue of the sequon (Asn-Xaa-Ser/Thr, where Xaa is any amino acid except for Pro), in the globular head region also contributes to the escape (1, 21, 27, 28). This is based on observations in experimental studies that some N-glycans attached to the globular head region interfere with the binding of Ab to AS by masking the surface of HA (1, 21, 27, 28).
In the evolutionary studies of A/H1N1 and A/H3N2 viruses, it has been observed that the number of NGS in the globular head region has increased continuously for several decades after the emergence of these viruses in the human population (4, 6–8, 11, 32). It was speculated that the addition of NGS to the globular head region conferred selective advantages to the viruses by preventing the binding of Ab to AS. However, this hypothesis has been questioned, because the gain of NGS in the globular head region of HA did not appear to influence the amino acid variation at AS, and no correlation was observed between the transitions of antigenic clusters and gains or losses of NGS during the evolution of human A/H3N2 virus (4, 8).
If N-glycans attached to the globular head region contribute to preventing the binding of Ab to AS, it is expected that natural selection for amino acid substitutions at AS is weakened (strengthened) when AS that are not covered (covered) by N-glycans become covered (not covered) through gains (losses) of NGS. Natural selection for amino acid substitutions may be identified from a comparison of the rate of nonsynonymous substitution (dN) to that of synonymous substitution (dS). Positive, negative, and no selection are inferred when the dN/dS value is >1, <1, and 1, respectively. It has been observed that dN/dS at AS is often >1, whereas that at other sites (NAS) is usually <1 in HA of human A/H1N1 and A/H3N2 viruses (20, 23, 24, 31).
The purpose of the present study was to examine the effect of gains and losses of NGS on dN/dS at AS and NAS that are covered and not covered by N-glycans during the evolution of human A/H3N2 virus.
MATERIALS AND METHODS
Sequences.
All (2,158) complete HA-coding nucleotide sequences of A/H3N2 viruses isolated from humans in the period between 1968 and 2010 were retrieved from the Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) on 15 June 2010. After removing the sequences containing undetermined nucleotides and minor gaps, 1,903 HA sequences were used for the following analysis.
Phylogenetic tree.
A multiple alignment of HA sequences was made using the computer program MAFFT (12). The optimum model of nucleotide substitution for these sequences was selected using MODELTEST (17) with PAUP (version 4.0). The general time-reversible model with a gamma distribution for the rate of heterogeneity among sites (G) and invariable sites (I) (GTR+G+I) (gamma shape parameter = 1.3549, proportion of invariable sites = 0.2696, and transition/transversion rate ratio [κ] = 10) and the transversional model with G and I (TVM+G+I) (gamma shape parameter = 1.3543, proportion of invariable sites = 0.2695, and κ = 9) were selected as the most appropriate models by the hierarchical likelihood ratio test (hLRT) and Akaike information criterion (AIC), respectively.
Since TVM is a special case of GTR, a phylogenetic tree of human A/H3N2 virus was constructed using 1,903 complete HA-coding nucleotide sequences by the neighbor-joining (NJ) method with the evolutionary distances measured assuming GTR+G+I using PAUP. The HA sequence of A/duck/Hong Kong/7/1975 (accession no. CY006026) was used as the outgroup to root the phylogenetic tree. After constructing the phylogenetic tree, the branch lengths were reestimated by the maximum likelihood (ML) method with GTR+G+I.
Reconstruction of ancestral HA nucleotide sequences.
For each interior node of the phylogenetic tree, the ancestral HA nucleotide sequence was reconstructed by the maximum parsimony method using PAML (9, 10). In the HA amino acid sequences obtained by translating the HA nucleotide sequences at the interior and exterior nodes, the Asn residue of the sequon was considered an NGS. For each of the amino acid positions where an NGS was observed at some nodes of the phylogenetic tree, the state of N-glycosylation (whether the amino acid position was an NGS or a non-NGS) was inferred for each branch by referring to the state of the nodes located at the ends of the branch. Each node of the branch was identified as ancestral or descendant because the phylogenetic tree was rooted. For each branch, the amino acid position was inferred to be an NGS (non-NGS) when the position was an NGS (non-NGS) at both nodes. A gain (loss) of an NGS was inferred to have occurred at the branch when the position was a non-NGS (NGS) at the ancestral node and an NGS (non-NGS) at the descendant node.
Prediction of 3D structures for N-glycans.
The three-dimensional (3D) structures for the variants of N-glycans were constructed using SWEET (5). The structures of sugar chains were inferred using formulae for N-glycans that were reported to be attached to the HA of A/H1N1 viruses (15) (see Table S1 in the supplemental material). The distance between the NGS and each amino acid position in the 3D structure of HA was computed using the structure of trimeric HA for A/Memphis/102/72 (Protein Data Bank [PDB] no. 3HMG). Amino acid positions located within 10 or 15 Å from the NGS were assumed to be covered by N-glycans, whereas those located farther than 10 or 15 Å away were assumed not to be covered by N-glycans (see Results). The AS (NAS) of HA1 located within 10 and 15 Å from NGS are denoted AS10c (NAS10c) and AS15c (NAS15c), respectively, and the AS (NAS) of HA1 located farther than 10 and 15 Å away are denoted AS10uc (NAS10uc) and AS15uc (NAS15uc), respectively. When an amino acid site was located within 10 or 15 Å of more than one NGS, that site was categorized as AS10c, AS15c, NAS10c, or NAS15c of an NGS that was generated first during evolution. The amino acid sites that were possibly covered by any NGS during evolution were eliminated from AS10uc, AS15uc, NAS10uc, and NAS15uc of all NGS, resulting in the same set of amino acid sites included in AS10uc, AS15uc, NAS10uc, and NAS15uc for all NGS. The amino acid positions constituting sequons during any period of evolution were eliminated from all categories.
Estimation of dN/dS at AS and NAS.
For each amino acid position that was found to be an NGS during some period in the evolutionary history of human A/H3N2 virus, dS and dN at AS10c, AS15c, AS10uc, AS15uc, NAS10c, NAS15c, NAS10uc, and NAS15uc were estimated for the branches where the position was an NGS and a non-NGS separately. When an amino acid site in AS10c, AS15c, NAS10c, and NAS15c of a particular position was considered to possibly be covered by an N-glycan attached to another position on some branches where the former position was a non-NGS, these branches were eliminated from the computation of dN and dS for the non-NGS branches of the former position. In addition, three branches connecting HA sequences derived from human infections of triple-reassortant A/H3N2 viruses were excluded from all computations of dS and dN (Fig. 1) (2).
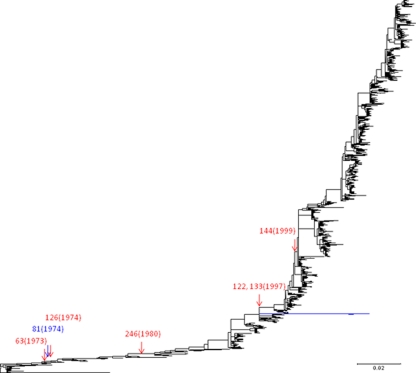
Gains and losses of NGS in the globular head region of HA in the evolutionary history of human A/H3N2 virus. The phylogenetic tree was constructed for 1,904 HA-coding nucleotide sequences by the NJ method with GTR+G+I. The red and blue arrows indicate the trunk branches where gains and losses of NGS in the globular head region occurred, respectively, with the amino acid positions of NGS indicated. The numbers in parentheses indicate the earliest (for gains) or most recent (for losses) year when the virus with the NGS was isolated. The long branches colored in blue were excluded from the computation of dS and dN, because the HA sequences at the ends were derived from human infections of triple-reassortant A/H3N2 viruses.
The dS and dN values were computed as Sd/S and Nd/N, respectively, where Sd and Nd denote the numbers of synonymous and nonsynonymous differences summed over the branches of sites included in each category, and S and N denote the numbers of synonymous and nonsynonymous sites averaged over the branches of sites included in each category with the weight proportional to the branch length. The numbers of synonymous and nonsynonymous differences as well as the numbers of synonymous and nonsynonymous sites for each branch of individual codon sites were obtained by comparing the sequences at the ancestral and descendant nodes using the modified Nei-Gojobori method with κ = 10. Since positions 165 and 285 appeared to be NGS throughout the evolutionary history of human A/H3N2 virus, the analysis of dS and dN was not conducted with these positions.
Statistical test.
The effect of gains and losses of NGS on dN/dS was examined at AS and NAS that can be covered and not covered by N-glycans during evolution. At AS10c, AS15c, AS10uc, AS15uc, NAS10c, NAS15c, NAS10uc, and NAS15uc of amino acid positions that were found to be NGS during some evolutionary period, the average dN/dS value during the evolutionary history of A/H3N2 virus was estimated using all branches of the phylogenetic tree irrespective of whether the position under consideration was an NGS or a non-NGS. The average dN/dS value was used to obtain the expected values of Sd and Nd for the branches where the position was an NGS and a non-NGS separately. The null hypothesis of equal dN/dS for NGS and non-NGS branches was tested by the chi-square test with one degree of freedom using the expected and observed values of Sd and Nd for both branches.
Natural selection operating at AS10c, AS15c, AS10uc, AS15uc, NAS10c, NAS15c, NAS10uc, and NAS15uc was examined for the branches where the position under consideration was an NGS and a non-NGS separately. The proportions of S and N were used to obtain the expected values of Sd and Nd (25), and the null hypothesis of selective neutrality was tested by the chi-square test with one degree of freedom. Positive and negative selection were inferred when the observed values of Nd and Sd were significantly greater than the expected values of Nd and Sd, respectively.
RESULTS
Size of N-glycans attached to HA.
Two types of N-glycans, high-mannose and complex oligosaccharides, are known to be attached, in various forms, to HA (13, 15). To infer the amino acid positions possibly covered by N-glycans in the 3D structure of HA, the maximum distance between sugar molecules within N-glycans was measured using formulae for N-glycans that were reported to be attached to HA (15). The maximum distance obtained was 16.78 to 34.63 Å (average, 25.4 Å) (see Table S1 in the supplemental material). If we assume that the shape of N-glycans is approximately spherical, the positions located within 8 to 17 Å of an NGS are considered to be covered by N-glycans. Therefore, the amino acid positions located within 10 or 15 Å of an NGS were assumed to be covered by N-glycans, whereas those located farther than 10 and 15 Å away were assumed to not be covered in the present study. The AS (NAS) located within 10 and 15 Å of an NGS are denoted AS10c (NAS10c) and AS15c (NAS15c), respectively, and the AS (NAS) located farther than 10 or 15 Å away are denoted AS10uc (NAS10uc) and AS15uc (NAS15uc), respectively (see Materials and Methods for details).
Gains and losses of NGS in HA of human A/H3N2 virus.
A phylogenetic tree was constructed for 1,903 complete HA-coding nucleotide sequences of A/H3N2 viruses isolated from humans in the period between 1968 and 2010 (Fig. 1), and the ancestral nucleotide sequence was inferred at each interior node. When the nucleotide sequences at the interior and exterior nodes were translated into amino acid sequences, some of them contained NGS at amino acid positions 63, 81, 122, 126, 133, 144, 165, and 246 in the globular head region and at positions 6, 7, 8, 22, 38, 45, 276, 285, 483, and 498 in the stem region (Fig. 2). N-glycans appeared to cover AS when they were attached to positions 45, 63, 81, 122, 126, 133, 144, 165, 246, 276, and 285 (Table 1).
Table 1
Amino acid sites possibly covered by N-glycan in the 3D structure of trimeric HA
| Antigenic sitea and NGSb | Amino acid site (epitope: amino acid position[s]) possibly covered by N-glycanc |
|---|---|
| AS10c | |
    45 45 | C: 44, 45, 46, 47, 297, 312 |
    63 63 | D: 96; E: 62, 63, 75, 78, 91, 92, 94 |
    81 81 | A: 150; D: 117, 121; E: 78, 80, 81, 82, 83 |
    122 122 | A: 122, 124, 150; D: 121 |
    126 126 | A: 124, 126, 130, 132, 168; B: 128, 129, 164, 165; D: 167 |
    133 133 | A: 131, 132, 133, 135, 146, 152; B: 155 |
    144 144 | A: 135, 137, 138, 140, 142, 143, 144, 145, 146 |
    246 246 | B: 163, 164, 165, 186; D: 201, 203, 212, 216, 217, 218, 219, 244, 246, 247, 248 |
    276 276 | C: 50, 51, 53, 54, 275, 276, 278 |
| AS15c | |
    45 45 | C: 44, 45, 46, 47, 48, 294, 297, 299, 300, 305, 307, 308, 309, 310, 311, 312 |
    63 63 | D: 96; E: 59, 62, 63, 67, 75, 78, 80, 82, 87, 88, 91, 92, 94 |
    81 81 | A: 122, 150, D: 117, 121, 177, 179; E: 57, 59, 67, 78, 80, 81, 82, 83, 87, 260, 261 |
    122 122 | A: 122, 124, 126, 132, 133, 150, 152, 168; D: 121, 167, 170, 172, 174, 176, 177, 179; E: 80, 81 |
    126 126 | A: 122, 124, 126, 130, 131, 132, 133, 152, 168; B: 128, 129, 155, 157, 163, 164, 165; D: 167, 244, 246, 247 |
    133 133 | A: 122, 124, 126, 130, 131, 132, 133, 135, 137, 140, 142, 144, 145, 146, 150, 152; B: 128, 129, 155, 156, 157, 158, 194 |
    144 144 | A: 133, 135, 137, 138, 140, 142, 143, 144, 145, 146; D: 96, 226; E: 75 |
    246 246 | A: 126, 130, 168; B: 128, 129, 163, 164, 165, 186, 187, 188, 190, 197; D: 167, 182, 201, 203, 209, 212, 213, 214, 215, 216, 217, 218, 219, 227, 228, 229, 242, 244, 246, 247, 248 |
    276 276 | C: 48, 50, 51, 53, 54, 273, 275, 276, 278, 279, 280; E: 57, 59 |
The occurrences of gains and losses of NGS that may cover AS in the evolutionary history of human A/H3N2 virus was inferred from the analysis of nucleotide sequences at the interior and exterior nodes of the phylogenetic tree. When A/H3N2 virus emerged in the human population in 1968, only positions 81 and 165 were NGS in the globular head region of HA (19). The NGS at position 165 has been maintained throughout the evolutionary history of the virus. However, position 81 became a non-NGS in 1974 (Fig. 1). The NGS generated at positions 63, 122, 126, 133, 144, and 246 have been retained during evolution. In the stem region of HA, position 285 has been maintained as an NGS throughout the evolutionary history of the virus. The NGS at position 276 reverted to a non-NGS 3 years after its generation in 1993. At position 45, the gains and losses of NGS occurred sporadically since 1997.
Effect of N-glycosylation on natural selection at AS and NAS.
N-glycans appeared to cover AS when they were attached to positions 45, 63, 81, 122, 126, 133, 144, 165, 246, 276, and 285, as indicated above. For each of these amino acid positions, the effect of N-glycosylation on natural selection at AS was examined by computing dN/dS at AS10c, AS15c, AS10uc, and AS15uc for the branches where the amino acid position under consideration was an NGS and a non-NGS separately. When these positions were non-NGS, dN/dS values at AS10c and AS15c were estimated to be 0.42 to 2.25 and 0.36 to 2.20, respectively (Table 2). (Note that dN/dS was incalculable for some cases because dS = 0.) Positive selection was detected at AS10c of position 144 (P = 0.03 by χ2 test). When the positions mentioned above became NGS, dN/dS at AS10c and AS15c generally decreased to 0.11 to 1.54 and 0.23 to 1.47, respectively, and no positive selection was detected. A reduction of dN/dS was observed at AS10c of 3 out of 5 positions and at AS15c of 6 out of all 6 positions where the comparison of dN/dS between the NGS and non-NGS branches could be made, and the latter observation was statistically significant (P = 0.03 by binomial test). Furthermore, the reduction of dN/dS at AS15c of position 144 was statistically significant (P < 0.05 by χ2 test).
Table 2
dN/dS values estimated at antigenic sites possibly covered by N-glycan
| Antigenic site,a NGS class,b and NGSc | dN/dSd | |
|---|---|---|
| Non-NGS | NGS | |
| AS10c | ||
    Maintained Maintained | ||
        63 63 | NA | 1.26 |
        122 122 | NA | NA |
        126 126 | 0.73 | 0.11 |
        133 133 | 0.43 | 0.44 |
        144 144 | 2.25 | 1.54 |
        246 246 | 1.01 | 0.82 |
        Avg Avg | 1.30 | 0.82 |
    Lost Lost | ||
        45 45 | 0.95 | NA |
        81 81 | 0.42 | 0.50 |
        276 276 | 1.52 | NA |
        Avg Avg | 1.07 | 0.94 |
| AS15c | ||
    Maintained Maintained | ||
        63 63 | NA | 1.47 |
        122 122 | 0.40 | 0.39 |
        126 126 | 0.99 | 0.66 |
        133 133 | 2.20 | 1.21 |
        144* 144* | 1.93 | 0.74 |
        246 246 | 0.97 | 0.33 |
        Avg** Avg** | 1.42 | 0.73 |
    Lost Lost | ||
        45 45 | 0.36 | NA |
        81 81 | 0.54 | 0.23 |
        276 276 | 1.75 | NA |
        Avg Avg | 0.70 | 0.45 |
The NGS analyzed above may be classified into two groups according to whether they were maintained (positions 63, 122, 126, 133, 144, and 246) or lost (positions 45, 81, and 276) during the evolution of human A/H3N2 virus. When the average dN/dS values at AS10c and AS15c for the positions included in each class was computed for the NGS and non-NGS branches separately, it was observed that the dN/dS value for the NGS branches generally was smaller than that for the non-NGS branches (Table 2). In particular, the difference was statistically significant at AS15c of the class of positions maintained as NGS during evolution (P < 0.01 by χ2 test).
For AS10uc and AS15uc, the dN/dS values were estimated to be 0.45 to 0.62 and 0.39 to 0.75, respectively, when the positions 45, 63, 81, 122, 126, 133, 144, 246, and 276 were non-NGS (Table 3). Negative selection was detected in most cases, and no positive selection was detected at all. When these positions became NGS, the dN/dS values at AS10uc and AS15uc did not change to any large extent (they were 0.32 to 0.73 and 0.47 to 0.76, respectively), except for AS15uc of position 276, where an increase in dN/dS (from 0.67 to 2.48) was observed. Negative selection again was detected in most cases.
Table 3
dN/dS values estimated at antigenic sites possibly not covered by N-glycan
| Antigenic site,a NGS class,b and NGSc | dN/dSd | |
|---|---|---|
| Non-NGS | NGS | |
| AS10uc | ||
    Maintained Maintained | ||
        63 63 | 0.45 | 0.56 |
        122 122 | 0.62 | 0.52 |
        126 126 | 0.52 | 0.56 |
        133 133 | 0.60 | 0.52 |
        144 144 | 0.60 | 0.51 |
        246 246 | 0.56 | 0.55 |
        Avg Avg | 0.59 | 0.54 |
    Lost Lost | ||
        45 45 | 0.56 | 0.32 |
        81 81 | 0.56 | 0.49 |
        276 276 | 0.54 | 0.73 |
        Avg Avg | 0.55 | 0.57 |
| AS15uc | ||
    Maintained Maintained | ||
        63 63 | 0.39 | 0.73 |
        81 81 | 0.73 | 0.47 |
        122 122 | 0.74 | 0.66 |
        126 126 | 0.72 | 0.70 |
        133 133 | 0.75 | 0.67 |
        144 144 | 0.65 | 0.76 |
        246 246 | 0.53 | 0.73 |
        Avg Avg | 0.67 | 0.71 |
    Lost Lost | ||
        45 45 | 0.69 | NA |
        81 81 | 0.73 | 0.47 |
        276* 276* | 0.67 | 2.48 |
        Avg Avg | 0.70 | 0.91 |
It should be noted that the dN/dS value at AS may be influenced by natural selection operating not only on the specific function of AS but also on the HA as a whole. To eliminate the possibility that the reduction of dN/dS at AS10c and AS15c observed above reflected a change in functional constraints on the entire region of HA, e.g., due to a change in the effective population size of human A/H3N2 virus, dN/dS was also computed for NAS10c, NAS15c, NAS10uc, and NAS15uc.The dN/dS values obtained at NAS generally were much smaller than those obtained at AS, and negative selection was detected in most cases whether the amino acid positions 45, 63, 81, 122, 126, 133, 144, 246, and 276 were non-NGS or NGS (see Tables S2 and S3 in the supplemental material). Although a significant difference in dN/dS was observed between NGS and non-NGS branches in some cases, dN/dS was usually greater in the former branches than in the latter, which was the opposite of the pattern observed at AS10c and AS15c.
DISCUSSION
After the emergence of A/H3N2 virus in the human population in 1968, NGS were generated at amino acid positions 63, 122, 126, 133, 144, and 246 in the globular head region of HA. Since the glycosidic linkage of oligosaccharides may be flexible, N-glycans may have multiple conformations in solution (26). In addition, the structure of N-glycans may vary among NGS in HA (13, 15). For these reasons, two different distances from NGS, 10 and 15 Å, were used as the possible ranges of amino acid positions covered by N-glycans in the present study. When positions 63, 122, 126, 133, 144, and 246 were non-NGS, the dN/dS value was >1 on average for both AS10c and AS15c and positive selection was detected at AS10c of position 144, suggesting that AS10c and AS15c of these positions contained target sites of Ab. However, the dN/dS values at AS10c and AS15c decreased, and the evidence of positive selection disappeared when the positions mentioned above became NGS. In contrast, the dN/dS values at AS10uc and AS15uc generally were <1 and did not change to any large extent whether the positions mentioned above were NGS or non-NGS. In addition, the dN/dS values at NAS10c, NAS15c, NAS10uc, and NAS15uc often were greater when these positions were NGS than when they were non-NGS, which was the opposite of the pattern observed at AS10c and AS15c. These results indicate that natural selection for amino acid substitutions at AS10c and AS15c was weakened due to the gain of NGS in the globular head region of HA, supporting the hypothesis that the N-glycans attached to the NGS in the globular head region of HA prevented the binding of Ab to AS in the evolutionary history of human A/H3N2 virus.
The results obtained in the present study may contradict those obtained in a previous study, where a reduction of amino acid variability was not observed after gains of NGS in the globular head region at ±5 amino acid sites from the NGS in the linear sequence of HA for human A/H3N2 virus (8). There are several differences between the present and previous studies that may have caused conflicting results. First, in the present study, amino acid sites possibly covered by N-glycans were identified using the distance from NGS in the 3D structure of trimeric HA, taking into account the size of N-glycans, whereas in the previous study, the ±5 amino acid sites from NGS were assumed to be covered by N-glycans. In addition, the AS and NAS were analyzed separately in the present study, whereas they were mixed in the previous study. It is therefore likely that the effect of N-glycans on natural selection for amino acid substitutions at AS could be detected more efficiently and specifically in the present study than in the previous study. Second, in the present study, the change in natural selection for amino acid substitutions was measured using dN/dS, whereas the number of different amino acids was used in the previous study. Since the amino acid variability is more strongly influenced by the mutation rate than the dN/dS value, it is likely that the change in natural selection was more reliably identified in the present study than in the previous study.
The N-glycans attached to NGS at positions 45 and 276 in the stem region of HA appeared to have covered AS in the evolutionary history of human A/H3N2 virus. However, the NGS at these positions have been generated only sporadically and have not been maintained for a long evolutionary period. It is known that natural selection for amino acid substitutions operates strongly around the receptor binding pocket (RBP), which is located in the globular head region of HA, because Ab may interfere with viral entry into the host cell more efficiently when they bind to the amino acid positions closely located to the RBP (24, 29). Indeed, positive selection was detected at AS10c of amino acid position 144, which was located close to the RBP, and the NGS generated in the globular head region appeared to be maintained longer than those generated in the stem region during evolution. It is therefore conceivable that N-glycans attached to the globular head region of HA were advantageous to the virus by preventing the neutralization by Ab, whereas those attached to the stem region were less advantageous.
Furthermore, the NGS at amino acid position 81, which was located in the globular head region, also was lost during evolution. This event apparently occurred after the generation of NGS at position 63, which was located adjacent to position 81 in the 3D structure of HA. The dN/dS values at AS10c and AS15c of position 63 were incalculable when this position was a non-NGS. However, experimental studies have shown that N-glycosylations at positions 63 and 81 both block the binding of Ab to AS (1, 21). Since some of the AS that were likely to be covered by N-glycans attached to positions 63 and 81 overlapped (29), and since the NGS at position 63 has been maintained after its generation, it is speculated that the generation of NGS at position 63 compensated for the advantageous effect of N-glycosylation at position 81 by shielding the overlapping amino acid sites from the access of Ab, which may have allowed for the loss of the NGS at position 81 after the generation of the NGS at position 63.
ACKNOWLEDGMENTS
We thank two anonymous reviewers for valuable comments.
Y.K. was supported by a JSPS Research Fellowship for Young Scientists (21-4991).
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.06147-11
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/86/7/3446.full.pdf
Citations & impact
Impact metrics
Article citations
Exploiting the Affimer platform against influenza A virus.
mBio, 15(8):e0180424, 22 Jul 2024
Cited by: 0 articles | PMID: 39037231 | PMCID: PMC11323568
Potential pandemic risk of circulating swine H1N2 influenza viruses.
Nat Commun, 15(1):5025, 13 Jun 2024
Cited by: 3 articles | PMID: 38871701
Cancer cells and viruses share common glycoepitopes: exciting opportunities toward combined treatments.
Front Immunol, 15:1292588, 01 Mar 2024
Cited by: 0 articles | PMID: 38495885 | PMCID: PMC10940920
Review Free full text in Europe PMC
Combinatorial immune refocusing within the influenza hemagglutinin RBD improves cross-neutralizing antibody responses.
Cell Rep, 42(12):113553, 13 Dec 2023
Cited by: 2 articles | PMID: 38096052 | PMCID: PMC10801708
Coronaviruses spike glycoprotein endodomains: The sequence and structure-based comprehensive study.
Protein Sci, 32(11):e4804, 01 Nov 2023
Cited by: 1 article | PMID: 37833239 | PMCID: PMC10599102
Go to all (56) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - CY006026
Protein structures in PDBe
-
(3 citations)
PDBe - 3HMGView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A broadly neutralizing human monoclonal antibody directed against a novel conserved epitope on the influenza virus H3 hemagglutinin globular head.
J Virol, 88(12):6743-6750, 02 Apr 2014
Cited by: 35 articles | PMID: 24696468 | PMCID: PMC4054378
N-linked glycans on influenza A H3N2 hemagglutinin constrain binding of host antibodies, but shielding is limited.
Glycobiology, 25(1):124-132, 16 Sep 2014
Cited by: 10 articles | PMID: 25227423
Isolation of an Egg-Adapted Influenza A(H3N2) Virus without Amino Acid Substitutions at the Antigenic Sites of Its Hemagglutinin.
Jpn J Infect Dis, 71(3):234-238, 27 Apr 2018
Cited by: 1 article | PMID: 29709975
H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation.
Hum Vaccin Immunother, 14(8):1840-1847, 14 May 2018
Cited by: 82 articles | PMID: 29641358 | PMCID: PMC6149781
Review Free full text in Europe PMC