Abstract
Free full text

The Lin28/let-7 axis regulates glucose metabolism
SUMMARY
The let-7 tumor suppressor microRNAs are known for their regulation of oncogenes, while the RNA-binding proteins Lin28a/b promote malignancy by blocking let-7 biogenesis. In studies of the Lin28/let-7 pathway, we discovered unexpected roles in regulating metabolism. When overexpressed in mice, both Lin28a and LIN28B promoted an insulin-sensitized state that resisted high fat diet-induced diabetes, whereas muscle-specific loss of Lin28a and overexpression of let-7 resulted in insulin resistance and impaired glucose tolerance. These phenomena occurred in part through let-7-mediated repression of multiple components of the insulin-PI3K-mTOR pathway, including IGF1R, INSR, and IRS2. The mTOR inhibitor rapamycin abrogated the enhanced glucose uptake and insulin-sensitivity conferred by Lin28a in vitro and in vivo. In addition, we found that let-7 targets were enriched for genes that contain SNPs associated with type 2 diabetes and fasting glucose in human genome-wide association studies. These data establish the Lin28/let-7 pathway as a central regulator of mammalian glucose metabolism.
INTRODUCTION
There is an emerging consensus that metabolic disease and malignancy share common biological mechanisms. Reprogramming towards glycolytic metabolism can increase a cancer cell’s ability to generate biomass, a phenomenon termed the “Warburg Effect” (Denko, 2008; Engelman et al., 2006; Gao et al., 2009; Guertin and Sabatini, 2007; Laplante and Sabatini, 2009; Vander Heiden et al., 2009; Yun et al., 2009). Likewise, many genes identified in type 2 diabetes (T2D) genome wide association studies (GWAS) are proto-oncogenes or cell cycle regulators (Voight et al., 2010). MicroRNAs (miRNAs) are also emerging as agents of metabolic and malignant regulation in development and disease (Hyun et al., 2009; Peter, 2009). The let-7 miRNA family members act as tumor suppressors by negatively regulating the translation of oncogenes and cell cycle regulators (Johnson et al., 2005; Lee et al., 2007; Mayr et al., 2007; Kumar et al., 2008). Widespread expression and redundancy among the well-conserved let-7 miRNAs raise the question of how cancer and embryonic cells are able to suppress this miRNA family to accommodate rapid cell proliferation. In human cancers, loss of heterozygosity, DNA methylation, and transcriptional suppression have been documented as mechanisms to reduce let-7 (Johnson et al., 2005; Lu et al., 2007). Another mechanism for let-7 downregulation involves the RNA-binding proteins Lin28a and Lin28b (collectively referred to as Lin28a/b), which are highly expressed during normal embryogenesis and upregulated in some cancers to potently and selectively block the maturation of let-7 (Heo et al., 2008; Newman et al., 2008; Piskounova et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). By repressing the biogenesis of let-7 miRNAs and in some cases through direct mRNA binding and enhanced translation (Polesskaya et al., 2007; Xu and Huang, 2009; Xu et al., 2009; Peng et al., 2011), Lin28a/b regulate an array of targets involved in cell proliferation and differentiation in the context of embryonic cells, stem cells, and cancer.
Little is known about the in vivo function of the Lin28/let-7 axis. The pathway was first revealed in a screen for heterochronic mutants in C. elegans, where loss of lin-28 resulted in precocious vulval differentiation and premature developmental progression (Ambros and Horvitz, 1984; Moss et al., 1997; Nimmo and Slack, 2009), whereas loss of let-7 led to reiteration of larval stages and delayed differentiation (Abbott et al., 2005; Reinhart et al., 2000). We previously showed that Lin28a gain of function promotes mouse growth and delays sexual maturation, recapitulating the heterochronic effects of lin-28 and let-7 in C. elegans, as well as the height and puberty phenotypes linked to human genetic variation at the Lin28b locus identified in GWAS (Zhu et al., 2010). The conservation of Lin28 and let-7’s biochemical and physiological functions throughout evolution suggests an ancient mechanism for Lin28 and let-7’s effects on growth and developmental timing.
In this report we found that both Lin28a and LIN28B transgenic mice were resistant to obesity and exhibited enhanced glucose tolerance. In contrast, muscle-specific Lin28a knockout and inducible let-7 transgenic mice displayed glucose intolerance, suggesting that the Lin28/let-7 pathway plays a specific and tightly regulated role in modulating glucose metabolism in mammals. In vitro experiments revealed that Lin28a enhances glucose uptake via an increase in insulin-PI3K-mTOR signaling due in part to the derepression of multiple direct let-7 targets in the pathway, including IGF1R, INSR, IRS2, PIK3IP1, AKT2, TSC1 and RICTOR. Experiments with the mTOR-specific inhibitor rapamycin demonstrate that Lin28a regulates growth, glucose tolerance, and insulin sensitivity in an mTOR-dependent manner in vivo. In addition, analysis of T2D and fasting glucose whole genome associations suggests a genetic connection between multiple genes regulated by let-7 and glucose metabolism in humans. These metabolic functions for Lin28a/b and let-7 in vivo provide a mechanistic explanation for how this pathway might influence embryonic growth, metabolic disease and cancer.
RESULTS
Lin28a Tg mice are resistant to obesity and diabetes
We previously described a tetracycline-inducible Lin28a transgenic mouse that showed leaky constitutive Lin28a expression in the absence of induction (See Experimental Procedures, “Lin28a Tg” mice: Zhu et al., 2010). In that study, we showed that these mice cleared glucose more efficiently during glucose and insulin tolerance testing (GTT and ITT), classic metabolic tests used for the characterization of whole animal glucose handling. Given that young Lin28a Tg mice exhibited enhanced glucose metabolism, we tested if old Lin28a Tg mice were also resistant to age-induced obesity. Compared to Lin28a Tg mice, wild-type mice fed a normal diet gained significantly more fat mass with age (Figure 1A). Dual Energy X-ray Absorptiometry scans showed increased percentage lean mass and reduced percentage body fat in the Lin28a Tg mice (Figure 1B,C). To rule out behavioral alterations, we measured activity over three days in isolation cages and found no differences in horizontal activity, O2/CO2 exchange, and food/water intake between wild-type and Tg mice (Figure S1A,B). To determine if these mice were resistant to HFD-induced obesity, we fed mice a diet containing 45% kcals from fat, and observed resistance to obesity in the Lin28a Tg mice (Figure 1D). Lin28a Tg mice consumed as much high-fat food as their wild-type littermates, ruling out anorexia (data not shown). Furthermore, we inquired if Lin28a Tg mice were resistant to HFD-induced diabetes and found that they had markedly improved glucose tolerance and insulin sensitivity under HFD conditions (Figure 1E,F). Lin28a Tg mice also showed resistance to HFD-induced hepatosteatosis (Figure 1G). Taken together, leaky Lin28a expression in the muscle, skin and connective tissues (Zhu et al., 2010) protected against obesity and diabetes in the context of aging and HFD.
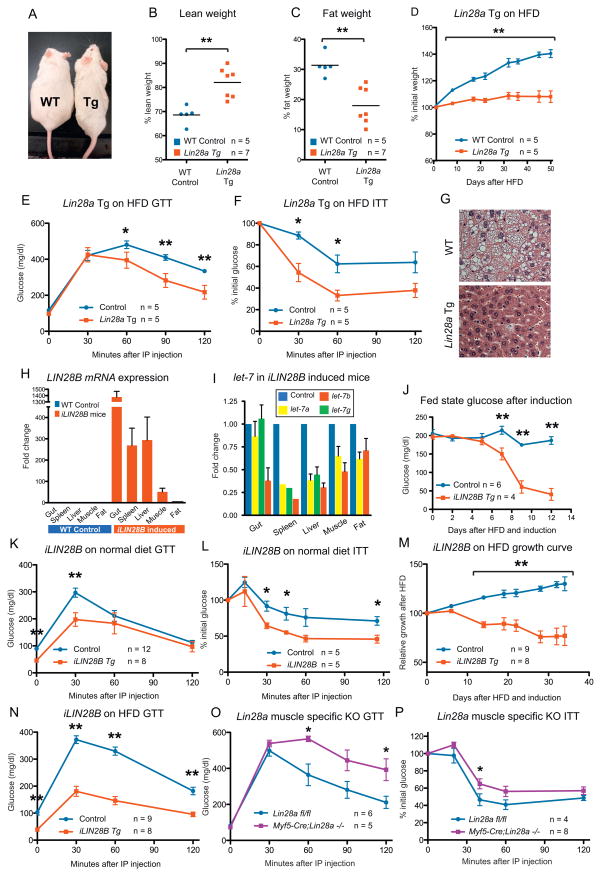
(A) Aged wild-type (left) and Lin28a Tg mice (right) fed a normal diet, at 20 weeks of age.
(B) Percentage body fat and (C) lean mass as measured by DEXA.
(D) Weight curve of mice fed a HFD containing 45% kcals from fat.
(E) Glucose tolerance test (GTT) and (F) Insulin tolerance test (ITT) of mice on HFD.
(G) Liver histology of mice fed HFD.
(H) Human LIN28B mRNA expression in a mouse strain with dox inducible transgene expression (named iLIN28B).
(I) Mature let-7 expression in gut, spleen, liver, muscle and fat.
(J) Kinetics of fed state glucose change after induction.
(K) GTT and (L) ITT under normal diets.
(M) iLIN28B growth curve under HFD.
(N) GTT after 14 days of HFD and induction.
(O) GTT and (P) ITT of Myf5-Cre; Lin28afl/fl mouse.
Controls for Lin28a Tg mice are WT. Controls for iLIN28B Tg mice carry only the LIN28B transgene. Controls for muscle knockout mice are Lin28afl/fl mice. The numbers of experimental animals are listed within the charts.
iLIN28B Tg mice are likewise resistant to diabetes
Although Lin28a and Lin28b both block let-7 miRNAs, they are differentially regulated, resulting in distinct expression patterns during normal development and malignant transformation (Guo et al., 2006; Viswanathan et al., 2009). Given that LIN28B is overexpressed more frequently than LIN28A in human cancer, we sought to determine if LIN28B exerts a similar effect on glucose metabolism. Thus, we generated a mouse strain with human LIN28B driven by a tetracycline transactivator rtTA placed under the control of the Rosa26 locus (iLIN28B mouse) (See Experimental Procedures). After 14 days of treatment with the tetracycline analogue doxycycline (dox), high levels of LIN28B were induced and mature let-7’s were repressed in metabolically important organs (Figure 1H,I), resulting in hypoglycemia with an average fasting glucose of <50 mg/dL in induced mice compared to >150 mg/dL in control mice (p < 0.01). To determine the kinetics of this effect, we measured fed state glucose daily and noted falling glucose levels after 5 days (Figure 1J). Glucose and insulin tolerance tests on dox-induced animals on normal diets showed considerable improvements in glucose tolerance and insulin sensitivity (Figure 1K,L). In checking for islet β cell hyperactivity, we found that iLIN28B mice produced no more insulin than control littermates during glucose challenge (data not shown). Under HFD, we found that induced iLIN28B mice were surprisingly resistant to weight gain (Figure 1M) despite a trend towards increased food intake (9.9 vs. 4.8 g/mouse/day; p = 0.075). These mice continued to exhibit superior glucose tolerance after 14 days of dox induction under HFD (Figure 1N), when average weights were 34.5 ± 1.05 grams for controls and 27.1 ± 0.99 grams for iLIN28B mice, demonstrating that HFD had a strong obesogenic and diabetogenic effect on control but not on LIN28B induced animals. Unlike the Lin28a Tg mice, expression was not leaky in the iLIN28B mice (Figure 1H and and3F)3F) and uninduced mice exhibited no growth or glucose phenotypes (Figure S1C,D), making this a better model for inducible Lin28 hyperactivation. These data show that both Lin28 homologues have similar effects on glucose metabolism and obesity, suggesting that these effects are mediated through common mRNA or miRNA targets of the Lin28 family.

(A) Western blot analysis of Lin28a protein expression in C2C12 myoblasts infected with control pBabe or Lin28a overexpression vector, and mouse ESCs, with tubulin as the loading control.
(B) Quantitative PCR for let-7 isoforms in C2C12 myoblasts, normalized to sno142, after Lin28a overexpression.
(C) 2-deoxy-D-[3H] glucose uptake assay on 3-day-differentiated C2C12 myotubes with and without Lin28a overexpression, treated with DMSO, the PI3K inhibitor LY294002, and the mTOR inhibitor rapamycin for 24 hrs.
(D) Western blot analysis of the effects of Lin28a overexpression on PI3K-mTOR signaling in C2C12 myoblasts, under serum-fed (fed), 18 hr serum starved (SS) or insulin-stimulated (Ins) conditions. Insulin stimulation was performed in serum-starved myoblasts with 10 μg/mL insulin for 5 min. Prior to insulin stimulation, serum-starved myoblasts were treated with either DMSO or 20 ng/mL rapamycin for 1 hr.
(E) Western blot analysis of the effects of let-7f or control miRNA on PI3K-mTOR signaling in C2C12 myoblasts under serum-fed (fed), 18 hr serum starved (SS) or insulin-stimulated (Ins) conditions.
(F) Western blot analysis of the effects of LIN28B induction by dox on PI3K-mTOR signaling in quadriceps muscles in vivo (n = 3 iLIN28B Tg mice and 3 LIN28B Tg only mice).
(G) Insr and p-4EBP1 protein levels in wild-type and Lin28a muscle-specific knockout adults.
Lin28a is physiologically required for normal glucose homeostasis
We then asked if Lin28a is physiologically required for normal glucose metabolism in one specific adult tissue compartment, skeletal muscle, since previous studies have found low but significant levels of Lin28a expression in the muscle tissues of mice (Yang and Moss, 2003; Zhu et al, 2010). We generated a skeletal muscle-specific knockout of Lin28a by crossing the mouse myogenic factor 5 Cre recombinase strain (Myf5-Cre) to the Lin28afl/fl conditional loss of function mouse (See Experimental Procedures). These muscle-specific knockout mice showed impaired glucose tolerance (Figure 1O) and insulin resistance (Figure 1P) relative to wild-type littermates, demonstrating that Lin28a activity in skeletal muscles is required for normal glucose homeostasis. We analyzed miRNA expression in muscle tissue by qRT-PCR and found no significant difference in let-7 levels during adult (data not shown) or embryonic stages (Figure S1E), suggesting that Lin28a loss of function affects glucose homeostasis either through let-7-independent mRNA binding or through changes in the spatiotemporal distribution of let-7 miRNA. Together, these data show that Lin28 isoforms are important and essential regulators of glucose homeostasis.
iLet-7 mice are glucose intolerant
In addition to their ability to suppress let-7 biogenesis, Lin28a and Lin28b also regulate mRNA targets such as Igf2, HMGA1, OCT4, histones and cyclins through non-let-7 dependent mechanisms of mRNA binding and enhanced translation (Polesskaya et al., 2007; Xu and Huang, 2009; Xu et al., 2009; Peng et al., 2011). To test if altered let-7 expression might produce the opposite phenotypes of Lin28a/b gain of function, we generated a mouse strain in which let-7g can be induced with dox under the control of the Rosa26 locus (iLet-7 mouse, See Experimental Procedures). To ensure that endogenous Lin28 would not block pri- or pre-let-7g biogenesis, we used a chimeric let-7g species called let-7S21L (let-7g Stem, mir-21 Loop), in which the loop region of the precursor miRNA derives from mir-21 and cannot be bound by Lin28, thus allowing for let-7 processing despite Lin28 expression (Piskounova et al., 2008). Global transgene induction from three weeks of age onwards increases mature let-7g levels in liver (>50-fold), skin (>20-fold), fat (~4-fold) and muscle (~4-fold) (Figure 2A). This level of let-7 overexpression led to reduced body size and growth rates in induced animals (Figure 2B,C). Growth retardation was proportional and not manifested as preferential size reduction in any particular organs (Figure S2A). Similar to the iLIN28B mice, leaky expression was not detected and uninduced male mice exhibited no growth or glucose phenotypes (Figure S2B–D).
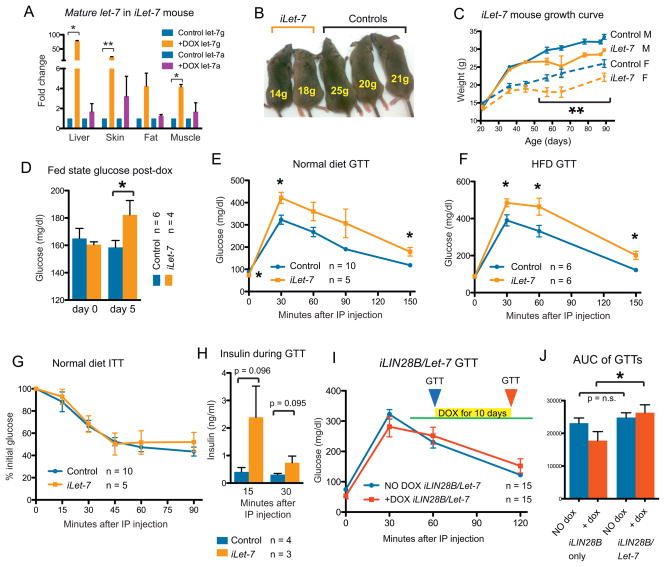
(A) let-7g and let-7a qRT-PCR in tissues of dox induced iLet-7 mice (n = 3) and controls (n = 3).
(B) Reduced size of induced animals.
(C) iLet-7 growth curve for males and females.
(D) Fed state glucose in iLet-7 mice induced for 5 days.
GTTs performed on mice fed with either (E) normal diet or (F) HFD.
(G) ITT on normal diet.
(H) Insulin production during a glucose challenge.
(I) GTT of LIN28B/Let-7 compound heterozygote mice before (blue) and after (red) induction with dox.
(J) Area under the curve (AUC) analysis for this GTT.
Controls for iLet-7 Tg mice carry either the Let-7 or Rosa26-M2rtTa transgene only. The numbers of experimental animals are listed within the charts.
After 5 days of let-7 induction, these iLet-7 mice produced an increase in fed state glucose (Figure 2D). GTT revealed glucose intolerance in mice fed normal (Figure 2E) or HFD (Figure 2F). Surprisingly, ITT failed to detect a difference in insulin sensitivity (Figure 2G). The decreased glucose tolerance in the setting of comparable insulin sensitivity suggested either decreased insulin production from islet β cells in response to glucose, or higher insulin secretion to compensate for peripheral insulin resistance. Thus, we measured insulin production following glucose challenge, and found that iLet-7 mice produced more insulin than controls (Figure 2H). These results demonstrated that broad overexpression of let-7 results in peripheral glucose intolerance and compensatory overproduction of insulin from islet β cells.
To test if let-7 induction could abrogate the glucose uptake phenotype of LIN28B overexpression, we crossed the iLIN28B to the iLet-7 inducible mice. After 10 days of induction, simultaneous induction of LIN28B and let-7g did not result in any differences in glucose tolerance (Figure 2I,J), in contrast to LIN28B or let-7g induction alone. Taken together, the opposing effects of Lin28 and let-7 expression on glucose regulation show that Lin28 overexpression influences metabolism in part by suppressing let-7, and that let-7 alone is sufficient to regulate glucose metabolism in vivo.
Insulin-PI3K-mTOR signaling is activated by Lin28a/b and suppressed by let-7
To dissect the molecular mechanism of the effects of Lin28 and let-7 on glucose regulation, we turned to the C2C12 cell culture system. Overexpression of Lin28a in C2C12 myoblasts resulted in protein levels of Lin28a similar to that observed in mouse embryonic stem cells (ESCs) (Figure 3A), and led to robust let-7 suppression (Figure 3B). In C2C12 myotubes differentiated for 3 days, Lin28a promoted Ser473 phosphorylation of Akt and Ser235/236 phosphorylation of S6 ribosomal protein, suggesting activation of the PI3K-mTOR pathway (Figure S3A). In this setting, Lin28a increased myotube glucose uptake by 50% (Figure 3C). Lin28a-dependent glucose uptake was abrogated by 24hr treatment with the PI3K/mTOR inhibitor LY294002 or the mTOR inhibitor rapamycin (Figure 3C), but not the MAPK/ERK inhibitor PD98059 (Figure S3B), demonstrating that Lin28a-dependent glucose uptake requires the PI3K-mTOR pathway.
To exclude myotube differentiation-dependent phenomena, we tested the effects of Lin28a on PI3K-mTOR signaling in undifferentiated myoblasts under serum-fed, serum-starved, and insulin-stimulated conditions (Figure 3D). In the serum-fed state, we found that Lin28a promoted the activation of PI3K/Akt signaling by increasing Akt phosphorylation at both Ser473 and Thr308, compared to the pBabe control. Furthermore, we found that Lin28a robustly increased the phosphorylation of mTORC1 signaling targets S6 and 4EBP1 in the serum-fed state. Serum-starvation for 18 hours abrogated the phosphorylation of Akt, S6 and 4EBP1, indicating that Lin28a-induction of PI3K-mTOR signaling requires exogenous growth factor stimulation. Upon insulin stimulation, Akt phosphorylation increased dramatically and, both phospho-S6 and phospho-4EBP1 levels were increased even further by Lin28a overexpression, suggesting that Lin28a increases the insulin-sensitivity of C2C12 myoblasts. Importantly, we found that rapamycin abrogated the Lin28a-induction of phospho-S6 and phospho-4EBP1 upon insulin stimulation, but did not affect let-7 levels (Figure S3C) or Lin28a itself (Figure 3D), indicating that the mTOR dependence is occurring downstream of Lin28a.
To test if the effects of Lin28a on insulin-PI3K-mTOR signaling are let-7-dependent, we transfected either mature let-7f duplex or a negative control miRNA into both Lin28a-overexpressing and pBabe control myoblasts (Figure S3D and Figure 3E). Because mature let-7 duplexes cannot be bound and inhibited by Lin28a protein, this experiment tests if PI3K-mTOR activation is occurring downstream of let-7. Transfection with control miRNA did not affect Lin28a-induction of the phosphorylation of Akt, S6, or 4EBP1 in serum-starved myoblasts upon insulin stimulation. Transfection with let-7f, however, attenuated the Lin28a-induction of phospho-Akt (Ser473), and abrogated the increase in S6 and 4EBP1 phosphorylation upon insulin stimulation in Lin28a-overexpressing myoblasts (Figure 3E). In pBabe control myoblasts, let-7 duplex still suppressed S6 and 4EBP1 phosphorylation in the serum-fed state, serum-starved, and insulin-stimulated conditions, relative to total S6 and 4EBP1 protein. The suppression of mTOR signaling by let-7 even in the absence of Lin28a implies that let-7 can act independently downstream of Lin28a. Together with data indicating that let-7 abrogates Lin28a-specific induction of p-Akt, p-S6 and p-4EBP1 upon insulin stimulation, this demonstrates that the effects of Lin28a on PI3K-mTOR signaling are at least in part due to let-7 and that Lin28 and let-7 exert opposing effects on PI3K-mTOR signaling.
To test if these effects of Lin28 on insulin-PI3K-mTOR signaling are also relevant in vivo, we examined the quadriceps muscles of iLIN28B mice and found that dox-induction led to increases in the phosphorylation of Akt (S473), S6 and 4EBP1, the targets of PI3K-mTOR signaling (Figure 3F). Furthermore, the Insulin-like growth factor 1 receptor (Igf1r) and the Insulin receptor (Insr) proteins were also upregulated in the muscles upon LIN28B induction, reinforcing the fact that Lin28a/b drives insulin-PI3K-mTOR signaling in C2C12 myoblasts and within mouse tissues. On the other hand, similar analysis of the Lin28a muscle-specific knockout mice revealed reduced Insr and p-4EBP1 expression (Figure 3G), demonstrating that Lin28a is both necessary and sufficient to influence glucose metabolism through the regulation of insulin-PI3K-mTOR signaling in vivo.
Lin28a/b and let-7 regulate genes in the insulin-PI3K-mTOR pathway
On the RNA level, Lin28a overexpression in C2C12 myoblasts leads to an increase in mRNA levels of multiple genes in the insulin-PI3K-mTOR signaling pathway (Figure S4A). Although both Lin28a suppression of let-7 and direct Lin28a binding to mRNAs could increase mRNA stability and thus increase mRNA levels, it is possible that these increases do not reflect direct interactions. To find direct targets, we performed a bioinformatic screen using the TargetScan 5.1 algorithm (Grimson et al., 2007), and found that 16 genes in the insulin-PI3K-mTOR pathway contained evolutionarily conserved let-7 binding sites in their respective 3′UTRs (Figure 4A,B). Next, we performed 3′ UTR luciferase reporter assays to determine if these genes were bona fide and direct targets of let-7. To do this, we generated luciferase reporters with twelve human 3′UTR fragments containing conserved let-7 sites. Luciferase reporter expression in human HEK293T cells after transfection of either mature let-7f duplex or a negative control miRNA demonstrated that the 3′ UTRs of INSR, IGF1R, IRS2, PIK3IP1, AKT2, TSC1 and RICTOR were targeted by let-7 for suppression (Figure 4C). Three-base mismatch mutations in the seed region of the let-7 binding sites abrogated let-7’s suppression of INSR, IGF1R and IRS2. To confirm that the luciferase reporters predicted actual changes in protein expression mediated by let-7, we assayed the endogenous expression of some of these proteins upon Lin28a/b overexpression. We found that an increase in Lin28a upregulated Irs2 (Figure 4D) in vitro, and that an increase in LIN28B upregulated Igf1r and Insr protein in skeletal muscles in vivo (Figure 3F). Conversely, INSR and IRS2 are reduced upon both let-7f transfection and LIN28B shRNA knockdown in HEK293T, demonstrating that these regulatory mechanisms hold in both mouse and human cells, and in the setting of both LIN28B gain and loss of function (Figure 4E). This establishes a direct mechanism for let-7’s repression and Lin28’s derepression of multiple components in the insulin-PI3K-mTOR signaling cascade.
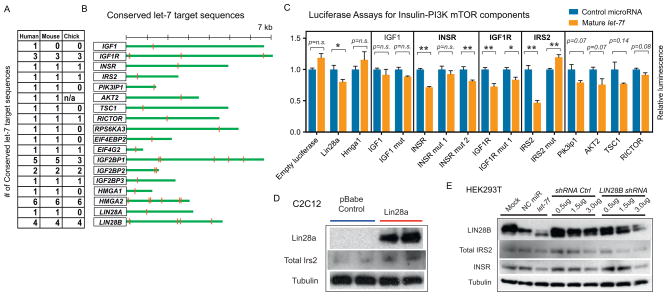
(A) Shown are the numbers of conserved let-7 binding sites within 3′UTRs found using the TargetScan algorithm.
(B) Putative let-7 binding sites in 16 genes of the insulin-PI3K-mTOR pathway and in Lin28a/b.
(C) 3′UTR luciferase reporter assays performed to determine functional let-7 binding sites. Bar graphs show relative luciferase reporter expression in human HEK293T cells after transfection of mature let-7f duplex normalized to negative control miRNA. Shown also are mutations in the seed sequence of the let-7 binding sites for INSR, IGF1R and IRS2.
(D) Western blot analysis of Lin28a, Irs2, and tubulin in C2C12 myoblasts with and without Lin28a overexpression.
(E) Western blot analysis of LIN28B, total IRS2, INSR and TUBULIN in HEK293T cells with either let-7f transfection or shRNA knockdown of LIN28B.
Previously, Lin28a has been shown to enhance Igf2 translation independently of let-7 (Polesskaya et al., 2007), offering an alternative mechanism by which Lin28a might activate the insulin-PI3K-mTOR pathway. To determine the relative contribution of this mechanism, we performed in vitro and in vivo loss of function experiments. Following siRNA knockdown of Igf2 in C2C12 (the efficacy of knockdown is shown in Figure S4B), we found only minimal changes in S6 and 4EBP1 phosphorylation (Figure S4C). In these C2C12 myotubes, glucose uptake was unaffected by Igf2 knockdown, but significantly decreased by let-7a (Figure S4D). In addition, we crossed the Lin28a Tg mice with Igf2 knockout mice and found that the absence of Igf2 did not abrogate enhanced glucose uptake, insulin sensitivity, or the anti-obesity effect mediated by Lin28a (Figure S4E–H). Taken together, these data indicate that the metabolic phenotypes we have observed are not solely due to the ability of Lin28a/b to promote translation of Igf02 mRNA, but do not rule out the possibility that Lin28a/b might modulate other mRNAs in the insulin-PI3K-mTOR signaling pathway.
mTOR mediates Lin28a’s enhancement of growth and glucose metabolism in vivo
Given that Lin28a activates the insulin-PI3K-mTOR pathway both in vitro and in vivo, we asked whether the metabolic effects of Lin28a in vivo could be abrogated by pharmacological inhibition of the mTOR pathway. To do this, we injected Lin28a Tg and wild-type littermates with rapamycin 3 times per week beginning when mice were 18 days old. Rapamycin abrogated the growth enhancement in Lin28a Tg mice at doses that had minimal growth suppressive effects on wild-type mice (Figure 5A,B), suggesting that Lin28a promotes growth in an mTOR-dependent manner. Selective suppression of Lin28a-driven growth was observed using several parameters: weight (Figure 5B,C), crown-rump length (Figure 5D), and tail width (Figure 5E). We also tested if the enhanced glucose uptake phenotype in vivo was likewise dependent on mTOR. Indeed, glucose tolerance testing showed that short-term rapamycin reversed the enhanced glucose uptake effect of Lin28a (Figure 5F,G) and reduced the insulin-sensitivity of Lin28a Tg mice to wild-type levels (Figure 5H,I). These data indicate that the glucose uptake, insulin sensitivity and animal growth phenotypes of Lin28a overexpression in vivo are dependent on mTOR signaling.
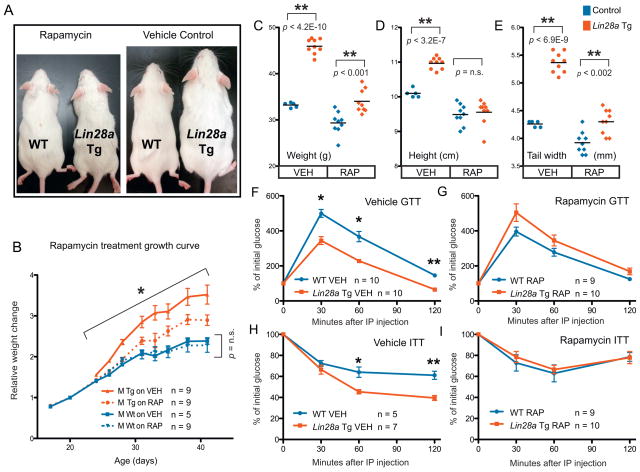
(A) Rapamycin (left 2 mice) and vehicle (right 2 mice) treated wild-type and Lin28a Tg mice shows relative size differences.
(B) Curves showing relative growth (normalized to weight on first day of treatment) for mice treated from 3 weeks to 6.5 weeks of age. Blue and red represent wild-type and Lin28a Tg mice, respectively. Solid and dotted lines represent vehicle and rapamycin treated mice, respectively. Growth was measured by several other parameters:
(C) weight, (D) crown-rump length or height, and (E) tail width.
(F) GTT performed after 2 doses of vehicle or (G) rapamycin.
(H) ITT performed after 1 dose of vehicle or (I) rapamycin.
Controls for Lin28a Tg mice are WT. The numbers of experimental animals are listed within the charts.
let-7 target genes are associated with type 2 diabetes in human GWAS
Finally, we sought to assess the relevance of the Lin28/let-7 pathway to human disease and metabolism, using human genetic studies of T2D and fasting glucose levels. Because the Lin28/let-7 pathway has not been previously implicated in T2D, we first asked whether any of the genes that lie in T2D association regions identified in T2D GWAS and meta-analyses (Voight et al., 2010) are known or predicted let-7 targets. We used TargetScan 5.1 to computationally predict let-7 targets (Grimson et al., 2007), and found that 14 predicted let-7 target genes lie in linkage disequilibrium to 39 validated common variant associations with T2D, including IGF2BP2, HMGA2, KCNJ11 and DUSP9 (strength of T2D association signals p < 4x10−9) (Table 1). Of the computationally predicted let-7 targets associated with T2D, IGF2BP1/2/3 and Hmga2 have been verified as let-7 targets in several studies (Boyerinas et al., 2008; Mayr et al., 2007). To validate the connection between Lin28 and GWAS candidate genes, we analyzed the expression of Igf2bp and Hmga family members in C2C12 cells with and without Lin28a overexpression, and observed increases in Igf2bp1, Igf2bp2, and Hmga2 mRNA following Lin28a overexpression (Figure 6A). To ensure that this was not a C2C12- or muscle-specific phenomenon, we confirmed the upregulation of these genes in 3T3 cells following human LIN28A or LIN28B overexpression on the mRNA (Figure 6B) and the protein level for the Igf2bp family (Figure 6C). We also observed increased expression of Igf2bp2 and Igf2bp3 (Figure 6D) in Lin28a Tg muscle, confirming this link in vivo.
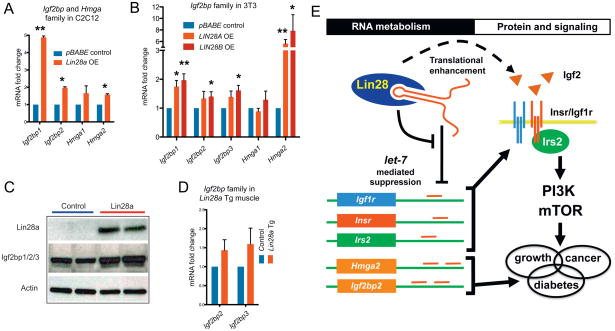
mRNA expression of Igf2bp and Hmga family members in (A) C2C12 with and without Lin28a overexpression and in (B) 3T3 cells with and without LIN28A or LIN28B overexpression.
(C) Western blot of NIH 3T3 cells with Lin28a overexpression showing Lin28a and Igf2bp1/2/3 protein levels (n = 3 biological replicates).
(D) Igf2bp2 and Igf2bp3 mRNA in Lin28a Tg muscle.
(E) Model of Lin28/let-7 pathway in glucose metabolism.
Table 1
The statistical enrichment for genes associated with T2D and fasting glucose among let-7 targets using the MAGENTA algorithm. The TargetScan algorithm was used to define the “All human let-7 targets” and the “Conserved let-7 targets” gene sets (http://www.targetscan.org/). mRNAs downregulation following let-7 overexpression (OE) was measured in primary human fibroblasts (Legesse-Miller et al., 2009), and protein downregulation following let-7 OE was measured in HeLa cells (Selbach et al., 2008). The enrichment cutoff used is the 75th percentile of all gene association scores in the genome. The enrichment fold is the ratio between the observed and expected number of genes above the enrichment cutoff. Genes linked to validated GWAS SNPs (39 SNPs for T2D and 14 SNPs for fasting glucose) were ordered according to the number of target gene sets they appear in and then alphabetically.
| let-7 target gene set | # genes analyzed† | Nominal enrichment p-value | Expected # genes above enrichment cutoff | Observed # genes above enrichment cutoff | Enrichment fold | # genes linked to validated GWAS SNPs | Genes linked to validated GWAS SNPs |
|---|---|---|---|---|---|---|---|
| Type 2 diabetes (DIAGRAM+ meta-analysis) | |||||||
| All targets predicted by TargetScan | 1763 | 0.036 | 441 | 462 | 1.05 | 14 | IGF2BP2, DUSP9, SLC5A6, TP53INP1, YKT6, ZNF512, HMGA2, KCNJ11, MAN2A2, MEST, NOTCH2, ZNF275, FAM72B, RCCD1 |
| Conserved targets predicted by TargetScan | 789 | 0.089 | 197 | 212 | 1.08 | 7 | IGF2BP2, DUSP9, SLC5A6, HMGA2, KCNJ11, MAN2A2, ZNF275 |
| Downregulated mRNAs following let-7 OE | 795 | 0.055 | 199 | 216 | 1.09 | 9 | IGF2BP2, DUSP9, SLC5A6, TP53INP1, YKT6, ZNF512, HHEX, IRS1, TLE4 |
| Downregulated mRNAs following let-7 OE + TargetScan | 502 | 0.061 | 126 | 140 | 1.11 | 6 | IGF2BP2, DUSP9, SLC5A6, TP53INP1, YKT6, ZNF512 |
| Downregulated proteins following let-7 OE | 97 | 1.0E-06* | 24 | 46 | 1.92 | 2 | IGF2BP2, CDKAL1 |
| Downregulated proteins following let-7 OE + TargetScan | 37 | 0.011 | 9 | 16 | 1.78 | 1 | IGF2BP2 |
| Fasting glucose (MAGIC meta-analysis) | |||||||
| All targets predicted by TargetScan | 1708 | 0.015 | 427 | 450 | 1.05 | 3 | CRY2, SLC2A2, GLIS3 |
| Conserved targets predicted by TargetScan | 759 | 0.042 | 190 | 207 | 1.09 | 1 | CRY2 |
| Downregulated mRNAs following let-7 OE | 750 | 1.0E-04* | 188 | 226 | 1.20 | 2 | CRY2, FADS1 |
| Downregulated mRNAs following let-7 OE + TargetScan | 484 | 0.013 | 121 | 141 | 1.17 | 1 | CRY2 |
| Downregulated proteins following let-7 OE | 96 | 0.632 | 24 | 23 | 0.96 | 0 | - |
| Downregulated proteins following let-7 OE +TargetScan | 35 | 0.245 | 9 | 11 | 1.22 | 0 | - |
We next asked whether there is a more widespread connection between T2D susceptibility and let-7 targets, in addition to the targets in validated T2D association regions (p < 5x10−8). To address this, we applied a computational method called MAGENTA (Meta-Analysis Gene-set Enrichment of variaNT Associations) (Segre et al., 2010) to GWAS meta-analyses of T2D and fasting glucose blood levels, and tested whether the distributions of disease or trait associations in predefined let-7 target gene sets are skewed towards highly ranked associations (including ones not yet reaching a level of genome-wide significance) compared to matched gene sets randomly sampled from the genome (Table 1). We tested three types of let-7 target definitions with increasing levels of target validation, from in silico predicted let-7 targets using TargetScan 5.1 (Grimson et al., 2007) to experimentally defined targets. For the latter, we used (i) a set of genes with at least one let-7 site in their 3′ UTR and whose mRNA was downregulated by let-7b overexpression in primary human fibroblasts (Legesse-Miller et al., 2009), and (ii) a set of genes whose protein levels were most strongly downregulated by let-7b overexpression in HeLa cells (Selbach et al., 2008). We first tested the let-7 target sets against the latest T2D meta-analysis of eight GWAS (called DIAGRAM+) (Voight et al., 2010), and found significant enrichment (Table 1). The enrichment rose from 1.05-fold for the broadest definition of let-7 targets predicted using TargetScan (~1800 genes; p = 0.036) to 1.92-fold for the experimentally validated target set based on protein level changes in response to let-7 overexpression (~100 genes; p = 1x10−6). In the latter case, an excess of about 20 genes regulated by let-7 at the protein level are predicted to contain novel SNP associations with T2D. Notably IGF2BP2, which is a canonical let-7 target that lies in a validated T2D association locus, was found in all types of let-7 target definitions in Table 1. Furthermore, the genes driving the T2D enrichment signals for the different let-7 target sets include both functionally redundant homologues of T2D-associated genes, such as IGF2BP1 (IGF2BP2), HMGA1 (HMGA2), DUSP12 and DUSP16 (DUSP9), and genes in the insulin-PI3K-mTOR pathway, including IRS2, INSR, AKT2 and TSC1 (best local SNP association p = 10−4 to 4*10−3).
We next tested for enrichment of let-7 target gene associations with fasting glucose levels, using data from the MAGIC (Meta-Analysis of Glucose and Insulin-related traits Consortium) study of fasting glucose levels (Dupuis et al., 2010). We observed an over-representation of multiple genes modestly associated with fasting glucose at different levels of significance for the different let-7 target gene sets (Table 1). The strongest enrichment was found in the genes downregulated at the mRNA level by let-7, an enrichment of 1.20 fold over expectation (p = 1*10−4). Taken together, our human genetic results support the hypothesis that genes regulated by let-7 influence human metabolic disease and glucose metabolism.
Recently it has also become clear that Lin28a/b has important let-7-independent roles in RNA metabolism, as evidenced by numerous direct mRNA targets whose translation is enhanced by LIN28A (Peng et al., 2011). Using GSEA, we found that this list of direct mRNA targets is also significantly enriched for glucose, insulin and diabetes-related genes (Table S1). Thus, Lin28a/b may regulate metabolism through direct mRNA-binding as well as let-7 targets.
DISCUSSION
Lin28 and let-7 are mutually antagonistic regulators of growth and metabolism
Our work defines a new mechanism of RNA-mediated metabolic regulation. In mice, Lin28a and LIN28B overexpression results in insulin sensitivity, enhanced glucose tolerance, and resistance to diabetes. Our analysis of iLet-7 Tg mice shows that let-7 upregulation is also sufficient to inhibit normal glucose metabolism, supporting the idea that gain of Lin28a/b exerts effects on whole animal glucose metabolism at least in part through let-7 suppression. Previously, we showed that transgenic overexpression of Lin28a causes enhanced growth and delayed puberty, phenotypes that mimicked human traits linked to genetic variation in the Lin28/let-7 pathway in GWAS (Zhu et al., 2010). Given that Lin28a/b is downregulated in most tissues after embryogenesis, while let-7 increases in adult tissues, lingering questions from our earlier report were first, whether let-7 was sufficient to influence organismal growth, and second, what function does let-7 have in adult physiology? Our observation that the iLin28a, iLIN28B, and iLet-7 Tg gain of function mice, as well as muscle-specific Lin28a loss of function mice manifest complementary phenotypes supports the notion that Lin28a/b and let-7 are both regulators of growth and developmental maturation. We propose that different developmental time-points demand distinct metabolic needs, and that global regulators such as Lin28 temporally coordinate growth with metabolism. The dynamic relationship between Lin28, let-7 and metabolic states during major growth milestones in mammals is reminiscent of the heterochronic mutant phenotypes originally defined in C. elegans (Ambros and Horvitz, 1984; Moss et al., 1997; Boehm and Slack, 2005), and suggests that metabolism, like differentiation, is temporally controlled.
Lin28a/b and let-7 influence glucose metabolism through the insulin-PI3K-mTOR pathway
We have shown that Lin28a/b and let-7 regulates insulin-PI3K-mTOR signaling, a highly conserved pathway that regulates growth and glucose metabolism throughout evolution. PI3K/Akt signaling is known to promote Glut4 translocation to upregulate glucose uptake, while mTOR signaling can promote glucose uptake and glycolysis by changing gene expression independently of Glut4 translocation (Brugarolas et al., 2003; Buller et al., 2008; Duvel et al., 2010). Previous studies have shown that Lin28a directly promotes Igf2 (Polesskaya et al., 2007) and HMGA1 translation (Peng et al., 2011), and that let-7 suppresses IGF1R translation in hepatocellular carcinoma cells (Wang et al., 2010). Consistent with these findings, our results define a model whereby Lin28a/b and let-7 coordinately regulate the insulin-PI3K-mTOR pathway at multiple points (Figure 6E), a concept that is consistent with the hypotheses that miRNAs and RNA binding proteins regulate signaling pathways by tuning the production of a broad array of proteins rather than switching single components on or off (Kennell et al., 2008; Hatley et al., 2010; Small and Olson, 2011). Coordinated regulation is important because negative feedback loops exist within the insulin-PI3K-mTOR pathway. Loss-of-function and pharmacological inhibition studies have shown that the mTOR target S6K1, for instance, inhibits and desensitizes insulin-PI3K signaling by phosphorylating IRS1 protein and suppressing IRS1 gene transcription (Harrington et al., 2004; Shah et al., 2004; Tremblay et al., 2007; Um et al., 2004). Conversely, TSC1–2 promotes insulin-PI3K signaling by suppressing mTOR signaling (Harrington et al., 2004; Shah et al., 2004). Although the effects of let-7 and Lin28a/b on the expression of individual genes are modest, simultaneous regulation of multiple components such as IGF2, IGF1R, INSR, IRS2, PIK3IP1, AKT2, TSC1, RICTOR in the insulin-PI3K-mTOR signaling pathway could explain how this RNA processing pathway coordinately regulates insulin sensitivity and glucose metabolism by effectively bypassing these negative feedback loops.
Whereas our work has implicated let-7 as a regulator of insulin-PI3K-mTOR signaling, we do not exclude a parallel role for direct mRNA targets of Lin28a/b in glucose metabolism, a hypothesis supported by the recent findings that HMGA1 is translationally regulated by LIN28A and mutated in 5–10% of T2D patients (Peng et al., 2011; Chiefari et al., 2011). Such non-let-7 functions are also suggested by the fact that muscle-specific loss of Lin28a results in glucose derangement without significant let-7 changes. Nevertheless, it remains likely that during other developmental stages or in other tissues, let-7 suppression by Lin28a or Lin28b is required for normal glucose homeostasis. The effects of the Lin28/let-7 pathway on glucose metabolism in our murine models, together with our observation that genes regulated by let-7 are associated with T2D risk in humans, indicates important functional roles for both Lin28a/b and let-7 in human metabolism.
let-7 targets are relevant to disparate human diseases: cancer and T2D
Metabolic reprogramming in malignancy is thought to promote a tumor’s ability to produce biomass and tolerate stress in the face of uncertain nutrient supplies (Vander Heiden et al., 2009). During their rapid growth phase early in development, embryos may utilize similar programs to maintain a growth-permissive metabolism. Dissecting the genetic underpinnings of embryonic metabolism would likely provide important insights into the nutrient uptake programs that are co-opted in cancer. While loss of function studies in the early embryo would help define the metabolic roles of oncofetal genes in their physiologic context, classical in vivo metabolic assays are difficult to perform in embryos. Lin28a and Lin28b are oncofetal genes, and thus highly expressed in early embryogenesis and then silenced in most adult tissues, but reactivated in cancer (Yang and Moss, 2003; Viswanathan et al., 2009). Cancer cells may utilize the embryonic function of Lin28a/b to drive a metabolic shift towards increased glucose uptake and glycolysis – a phenomenon termed the “Warburg effect.” Previously, we showed that Lin28a expression promotes glycolytic metabolism in muscle in vivo and in C2C12 myoblasts in vitro (Zhu et al., 2010). Though we cannot yet readily determine the metabolic effects of shutting off Lin28a/b within the embryo, we have dissected the potent effects of reactivating and inactivating this oncofetal program in adults. Conversely, in normal adult tissues that do not express high levels of Lin28a or Lin28b, one might ask if a role for the highly abundant let-7 is to lock cells into the metabolism of terminally differentiated cells to prevent aberrant reactivation of embryonic metabolic programs. Further studies are required to understand how this pathway may link mechanisms of tumorigenesis and diabetogenesis.
Our report implicates Lin28a/b and let-7 as important modulators of glucose metabolism through interactions with the insulin-PI3K-mTOR pathway and T2D-associated genes identified in GWAS. Although it is likely that additional mechanisms and feedback loops exist, our data suggests a model whereby Lin28a/b and let-7 coordinate the GWAS identified genes and the insulin-PI3K-mTOR pathway to regulate glucose metabolism (Figure 6E). It also suggests that enhancing Lin28 function or abrogating let-7 may be therapeutically promising for diseases like obesity and diabetes. Likewise, results from this work might shed light on the physiology of aging and, specifically, how the accumulation of let-7 in aging tissues may contribute to the systemic insulin resistance that accompanies aging.
EXPERIMENTAL PROCEDURES
Mice
All animal procedures were based on animal care guidelines approved by the Institutional Animal Care and Use Committee. Mouse lines used in this study are described in the Extended Experimental Procedures and Figure S5.
Indirect Calorimetry
The apparatus used was a set of 16 OxyMax® Metabolic Activity Monitoring chambers (Columbus Instruments; Columbus, OH, USA). Each chamber consisted of a self-contained unit capable of providing continuous measurements of an individual mouse’s total activity and feeding behavior. Monitoring occurred over a 3-day period. Each subject was placed into an individual chamber on day 1, with free access to food and water during the course of the experiment. Subjects were maintained under a normal 12:12 h light:dark cycle. All measurements were sampled periodically (at approximately 12-min intervals) and automatically recorded via the OXYMAX Windows V3.22 software. Activity measures over the final 24-h period were parceled into 2-h bins and these were used to express diurnal activity levels.
Quantitative RT-PCR
Performed with standard methods, which are described in detail in the Extended Experimental Procedures.
Histology
Tissue samples were fixed in 10% buffered formalin or Bouin’s solution and embedded in paraffin.
Glucose and insulin tolerance tests
Overnight-fasted mice were given i.p. glucose (2 mg/g body weight). For insulin tolerance test, 5 hour fasted mice were given 0.75 U insulin/kg body weight by i.p. injection (Humulin). Blood glucose was determined with a Lifescan One Touch glucometer. Insulin, GH, and Igf1 levels were measured by ELISA (Crystal Chem).
Cloning
Murine Lin28a and human LIN28B cDNA was subcloned into pBabe.Puro and pMSCV.Neo retroviral vectors. LIN28B and Control shRNA in lentiviral plasmids were purchased from Sigma-Aldrich and previously reported in Viswanathan et al., 2009. UTR cloning for luciferase reporters is described in Supplemental Table S2.
Cell culture, viral production, and transfection
Performed using standard methods as described in the Extended Experimental Procedures.
Glucose uptake assay
In vitro glucose uptake assays were performed as described in Berti and Gammeltoft, 1999.
Drug treatments
Rapamycin was injected i.p. 3 times a week for mouse experiments. For cell culture, C2C12 myotubes differentiated for 3 days were incubated with inhibitors for 1 day prior to glucose uptake assays. See Extended Experimental Procedures for further details.
Western blot assay
Performed using standard methods. Detailed methods and reagents used are described in the Extended Experimental Procedures.
Luciferase reporter assay
10 ng of each construct was co-transfected with 10 nM miRNA duplexes or into HEK-293T cells in a 96-well plate using lipofectamin-2000 (Invitrogen). After 48 hours, the cell extract was obtained; firefly and Renilla luciferase activities were measured with the Promega Dual-Luciferase® reporter system.
MAGENTA analysis
See Results, Table 1 Legend, and Extended Experimental Procedures for detailed methods.
Statistical analysis
Data is presented as mean ± SEM, and Student’s t-test (two-tailed distribution, two-sample unequal variance) was used to calculate p values. Statistical significance is displayed as p < 0.05 (one asterisk) or p < 0.01 (two asterisks). The tests were performed using Microsoft Excel where the test type is always set to two-sample equal variance.
Acknowledgments
We thank John Powers, Harith Rajagopalan, Jason Locasale, Abdel Saci, Akash Patnaik, Charles Kaufman, Christian Mosimann and Lewis Cantley for invaluable discussions and advice, Roderick Bronson and the Harvard Medical School Rodent Histopathology Core for mouse tissue pathology, and the Harvard Neurobehavior Laboratory for CLAMS experiments. This work was supported by grants from the US NIH to G.Q.D., a Graduate Training in Cancer Research Grant and a American Cancer Society Postdoctoral Fellowship to H.Z., the NSS Scholarship from the Agency for Science, Technology and Research, Singapore for N.S.C, an NIH NIDDK Diseases Career Development Award to M.G.K, and an American Diabetes Association Postdoctoral Fellowship for A.V.S. J.M.E. was supported by the National Human Genome Research Institute (NHGRI). R.I.G. was supported by US National Institute of General Medical Sciences (NIGMS) and is a Pew Research Scholar. D.A. is a Distinguished Clinical Scholar of the Doris Duke Charitable Foundation. G.Q.D. is a recipient of Clinical Scientist Awards in Translational Research from the Burroughs Wellcome Fund and the Leukemia and Lymphoma Society, and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research.
Footnotes
AUTHOR CONTRIBUTIONS
H.Z. and N.S.C designed and performed the experiments, and wrote the manuscript. A.V.S. and D.A. performed bioinformatic analysis on let-7 targets in GWAS. G.S. and S.P.S. performed expression analysis, metabolic assays and mouse husbandry. G.S., W.S.E. and A.T. generated the mouse strains. J.E.T, R.T. and R.I.G. assisted with the luciferase assays. J.P.H., R.I.G., G.S., and A.T generated the conditional knockout mice. M.G.K. helped to design the experiments. G.Q.D. designed and supervised experiments, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. [Europe PMC free article] [Abstract] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. [Abstract] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. [Abstract] [Google Scholar]
- Berti L, Gammeltoft S. Leptin stimulates glucose uptake in C2C12 muscle cells by activation of ERK2. Mol Cell Endocrinol. 1999;157:121–130. [Abstract] [Google Scholar]
- Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. [Abstract] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. [Abstract] [Google Scholar]
- Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–158. [Abstract] [Google Scholar]
- Buller CL, Loberg RD, Fan MH, Zhu Q, Park JL, Vesely E, Inoki K, Guan KL, Brosius FC., 3rd A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am J Physiol Cell Physiol. 2008;295:C836–843. [Europe PMC free article] [Abstract] [Google Scholar]
- Chiefari E, Tanyolac S, Paonessa F, Pullinger CR, Capula C, Iiritano S, Mazza T, Forlin M, Fusco A, Durlach V, et al. Functional variants of the HMGA1 gene and type 2 diabetes mellitus. JAMA. 305:903–912. [Abstract] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. [Abstract] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 42:105–116. [Europe PMC free article] [Abstract] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 39:171–183. [Europe PMC free article] [Abstract] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. [Abstract] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. [Europe PMC free article] [Abstract] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. [Europe PMC free article] [Abstract] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. [Abstract] [Google Scholar]
- Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. In Gene. 2006a:51–61. [Abstract] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. [Europe PMC free article] [Abstract] [Google Scholar]
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 18:282–293. [Europe PMC free article] [Abstract] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008:276–284. [Abstract] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. [Abstract] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. [Abstract] [Google Scholar]
- Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci U S A. 2008;105:15417–15422. [Europe PMC free article] [Abstract] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008:3903–3908. [Europe PMC free article] [Abstract] [Google Scholar]
- Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–1052. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee Y, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & Development. 2007:1025–1030. [Europe PMC free article] [Abstract] [Google Scholar]
- Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J Biol Chem. 2009;284:6605–6609. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. [Abstract] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. [Europe PMC free article] [Abstract] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 463:621–626. [Europe PMC free article] [Abstract] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. [Abstract] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008:1539–1549. [Europe PMC free article] [Abstract] [Google Scholar]
- Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. [Europe PMC free article] [Abstract] [Google Scholar]
- Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 29:496–504. [Abstract] [Google Scholar]
- Peter M. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle 2009 [Europe PMC free article] [Abstract] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. [Abstract] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes & Development. 2007:1125–1138. [Europe PMC free article] [Abstract] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. [Abstract] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. [Abstract] [Google Scholar]
- Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia-Naji N, Dupuis J, Jackson AU, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 42:142–148. [Europe PMC free article] [Abstract] [Google Scholar]
- Schwanhausser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–209. [Abstract] [Google Scholar]
- Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. :6. [Europe PMC free article] [Abstract] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. [Abstract] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. [Abstract] [Google Scholar]
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 469:336–342. [Europe PMC free article] [Abstract] [Google Scholar]
- Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J, Bouatia-Naji N, Langenberg C, Prokopenko I, Stolerman E, et al. Common variants at 10 genomic loci influence hemoglobin A(C) levels via glycemic and nonglycemic pathways. Diabetes. 59:3229–3239. [Europe PMC free article] [Abstract] [Google Scholar]
- Tremblay F, Brule S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, et al. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A. 2007;104:14056–14061. [Abstract] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. [Abstract] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. [Europe PMC free article] [Abstract] [Google Scholar]
- Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 140:445–449. [Abstract] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008:97–100. [Europe PMC free article] [Abstract] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. [Europe PMC free article] [Abstract] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 42:579–589. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang YC, Chen YL, Yuan RH, Pan HW, Yang WC, Hsu HC, Jeng YM. Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma. Carcinogenesis. 31:1516–1522. [Abstract] [Google Scholar]
- Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–4263. [Europe PMC free article] [Abstract] [Google Scholar]
- Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003:719–726. [Abstract] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. [Abstract] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 42:626–630. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2011.08.033
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867411010038/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
A comprehensive study of genetic regulation and disease associations of plasma circulatory microRNAs using population-level data.
Genome Biol, 25(1):276, 21 Oct 2024
Cited by: 0 articles | PMID: 39434104 | PMCID: PMC11492503
Nephron-Specific Lin28A Overexpression Triggers Severe Inflammatory Response and Kidney Damage.
Int J Biol Sci, 20(10):4044-4054, 22 Jul 2024
Cited by: 0 articles | PMID: 39113694 | PMCID: PMC11302891
Multi-step regulation of microRNA expression and secretion into small extracellular vesicles by insulin.
Cell Rep, 43(7):114491, 13 Jul 2024
Cited by: 0 articles | PMID: 39002127 | PMCID: PMC11363058
Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat Struct Mol Biol, 31(9):1426-1438, 25 Jul 2024
Cited by: 1 article | PMID: 39054354 | PMCID: PMC11402785
The lncRNA LINC01605 promotes the progression of pancreatic ductal adenocarcinoma by activating the mTOR signaling pathway.
Cancer Cell Int, 24(1):262, 24 Jul 2024
Cited by: 0 articles | PMID: 39048994 | PMCID: PMC11271012
Go to all (588) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Lin28a expression protects against streptozotocin-induced β-cell destruction and prevents diabetes in mice.
Cell Biochem Funct, 37(3):139-147, 18 Mar 2019
Cited by: 8 articles | PMID: 30883865
Sex-specific regulation of weight and puberty by the Lin28/let-7 axis.
J Endocrinol, 228(3):179-191, 23 Dec 2015
Cited by: 32 articles | PMID: 26698568 | PMCID: PMC4772724
Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies.
Nat Genet, 42(7):626-630, 30 May 2010
Cited by: 199 articles | PMID: 20512147 | PMCID: PMC3069638
Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer.
Mol Cancer, 14:125, 30 Jun 2015
Cited by: 101 articles | PMID: 26123544 | PMCID: PMC4512107
Review Free full text in Europe PMC
Funding
Funders who supported this work.
British Heart Foundation (1)
Psycho-social and biological factors in the occurrence of cardiovascular disease: The Whitehall II study
Professor Sir Michael Marmot, University College London
Grant ID: RG/07/008/23674
Howard Hughes Medical Institute
Medical Research Council (9)
MRC Clinical Research professorship
Professor Sir Michael Marmot, University College London
Grant ID: G19/35
Nutritional epidemiology
Professor Nita Forouhi, MRC Epidemiology Unit
Grant ID: MC_UP_A100_1003
Quantitative trait locus (QTL) identification in a Croatian isolate
Professor Alan Wright, MRC Human Genetics Unit
Grant ID: MC_U127561128
Social Determinants of Health: Psychological and Biological Pathways
Professor Sir Michael Marmot, University College London
Grant ID: G0100222
Sarcopenia, Frailty and Clinical Practice in Older People
Professor Avan Aihie Sayer, MRC Lifecourse Epidemiology Unit
Grant ID: MC_UP_A620_1015
Social influences on health
Professor Sir Michael Marmot, University College London
Grant ID: G8802774
Quantitative trait locus (QTL) identification in a Croatian isolate
Dr Chris Hayward, University of Edinburgh
Grant ID: MC_PC_U127561128
Grant ID: U1475000002
Causes of heterogeneity in ageing - the Whitehall II study
Professor Sir Michael Marmot, University College London
Grant ID: G0902037
NCI NIH HHS (2)
Grant ID: K08 CA157727
Grant ID: T32 CA009172
NIDDK NIH HHS (2)
Grant ID: P30 DK079637
Grant ID: R01 DK070055
Wellcome Trust (1)
Understanding the genetic basis of common human diseases: core funding for the Wellcome Trust Centre for Human Genetics.
Professor Peter Donnelly, University of Oxford
Grant ID: 090532





