Abstract
Free full text

Activity of the thiopeptide antibiotic nosiheptide against contemporary strains of methicillin-resistant Staphylococcus aureus
Abstract
The rapid rise in antimicrobial resistance in bacteria has generated an increased demand for the development of novel therapies to treat contemporary infections, especially those caused by methicillin-resistant Staphylococcus aureus (MRSA). However, antimicrobial development has been largely abandoned by the pharmaceutical industry. We recently isolated the previously described thiopeptide antibiotic nosiheptide from a marine actinomycete strain and evaluated its activity against contemporary clinically relevant bacterial pathogens. Nosiheptide exhibited extremely potent activity against all contemporary MRSA strains tested including multiple drug-resistant clinical isolates, with MIC values ≤ 0.25 mg/L. Nosiheptide was also highly active against Enterococcus spp and the contemporary hypervirulent BI strain of Clostridium difficile but was inactive against most Gram-negative strains tested. Time-kill analysis revealed nosiheptide to be rapidly bactericidal against MRSA in a concentration- and time-dependent manner, with a nearly 2-log kill noted at 6 hours at 10X MIC. Furthermore, nosiheptide was found to be non-cytotoxic against mammalian cells at >> 100X MIC, and its anti-MRSA activity was not inhibited by 20% human serum. Notably, nosiheptide exhibited a significantly prolonged post-antibiotic effect (PAE) against both healthcare- and community-associated MRSA compared to vancomycin. Nosiheptide also demonstrated in vivo activity in a murine model of MRSA infection, and therefore represents a promising antibiotic for the treatment of serious infections caused by contemporary strains of MRSA.
Introduction
Multidrug resistant hospital-associated (HA-) and highly virulent community-associated (CA-) methicillin-resistant Staphylococcus aureus (MRSA) have shown increased tolerance or resistance in recent years to vancomycin, linezolid, and daptomycin with accompanying reduction in clinical antimicrobial efficacy. The development of novel agents without cross-resistance to current antimicrobials against MRSA infections is desperately needed.1
In our screen of marine-derived actinomycete extract libraries for anti-MRSA activity, we identified a potent fraction derived from strain CNT-373, a Streptomycetes species isolated from a marine sediment collected in Fiji. The 1221.16 mol wt active component was purified and identified by NMR as the thiopeptide antibiotic nosiheptide. Thiopeptide antibiotics encompass a large family of compounds that include thiostrepton and nocathiacin and are comprised of sulfur- and nitrogen-rich heterocycles linked to non-natural amino acids.2,3 Nosiheptide, also referred to historically as multhiomycin, was originally isolated in 1970 and shown to be structurally similar to thiostrepton.4,5 Several thiopeptide antibiotics including nosiheptide block protein synthesis by the inhibition of elongation factors Tu and G.6 Historically, nosiheptide has been used as a growth-promoting additive in animal feed7 but was never developed further as a human therapeutic.
While the discovery of new chemical scaffolds provides the potential for novel drugs with new mechanisms of action, the re-investigation of previously discovered antibacterial scaffolds also provides a valuable source of compounds with therapeutic potential. Surprisingly, despite the structural identification and description of Gram-positive antibacterial activity, no detailed characterization of nosiheptide activity against contemporary drug resistant strains such as MRSA has been undertaken. Here, we investigate nosiheptide activity in vitro against a panel of contemporary MRSA and other Gram-positive clinical isolates. MRSA killing kinetics and post-antibiotic effects are characterized, and nosiheptide in vivo activity is demonstrated. Our results indicate that “re-discovered” natural product antibiotics may harbor meaningful therapeutic activity against contemporary multidrug resistant pathogens, and therefore warrant further consideration and preclinical development in hopes of expanding our current limited pharmacological arsenal.
Materials and Methods
Strain isolation and identification
Strain CNT-373 was isolated from a marine sediment sample collected at a depth of 5 m off Nacula Island, Fiji. The sediment was dried overnight in a laminar flow hood and then stamped on agar plates containing medium A1 (10 g starch, 4 g yeast extract, 2 g peptone, 16 g agar, 1L seawater) supplemented with 100 mg/L cyclohexamide to reduce fungal growth. The strain was identified as a Streptomyces sp. based on 16S rRNA gene sequence analysis (http://eztaxon-e.ezbiocloud.net/ezt_identify) and shares greatest similarity (98.3%) with the type strain Streptomyces althioticus. This low level of sequence identity suggests it may be a new species. The 16S rRNA sequence has been deposited in GenBank under accession number JQ946086.
Fermentation and extraction
A 2 mL frozen glycerol stock of strain CNT-373 was used to inoculate 25 mL of liquid A1 medium and shaken at 230 rpm and 27°C. After 5 days, the seed culture was used to inoculate a 1 L culture in growth medium A1bfe+C (A1 medium with the addition of 1 g CaCO3, 100 mg KBr and 40 mg Fe2(SO4)3•4H2O). After 3 days of shaking, 25 mL aliquots were used to inoculate 18 Fernbach flasks (2.8 L) each containing 1 L medium A1bfe+C. After 7 days of cultivation, sterilized XAD-16 resin (20 g per L) was added, shaken for 6 hours, and collected by filtration through cheesecloth. The resin was then washed with deionized water and eluted with acetone. The acetone solvent was removed under reduced pressure and the resulting aqueous layer extracted with ethyl acetate (3 × 500 mL). The combined ethyl acetate extracts were dried over anhydrous sodium sulfate, decanted, and concentrated to dryness to yield 2.4 g of crude material.
Isolation of nosiheptide
The crude extract was dissolved in methanol and dichloromethane, adsorbed onto silica gel (2.5 g), and fractionated on a short column of silica gel (6 × 2.5 cm, h × w) using a 10% step gradient from 100% isooctane to 100% ethyl acetate as eluents). Fractions with antibacterial activity were eluted with 90% and 100% ethyl acetate. The active fractions were pooled and subjected to a size exclusion column chromatography with Sephadex LH-20 using methanol as eluent. The final purification step was achieved with silicia gel, using ethyl acetate with 10% dimethylformamide (DMF) to give nosiheptide (210 mg).
Spectroscopic analysis of nosiheptide
1H, 13C and 2D NMR spectroscopic data were obtained on a Varian Inova 500 MHz spectrometer in a solvent mixture of methanol-d4 and CDCl3 (3:1) to facilitate solubility. Offline processing was conducted using topspin NMR software by Bruker BioSpin 2011 (iNMR, http://www.inmr.net). The NMR data (Table 1) is in good agreement with previously published NMR data for nosiheptide,8 with the resultant nosiheptide structure shown (Figure 1). High resolution ESI-TOF mass spectra were provided by the mass spectrometry facility at the Department of Chemistry and Biochemistry at the University of California San Diego, CA. HR-ESI-TOF-MS [M+H]+ m/z 1222.1565 (calcd for C51H44N13O12S6 1222.1551, Δ1.13 ppm). Low resolution LC-MS data were measured using a Hewlett-Packard HP1100 integrated LC/MS system with a reversed-phase C18 column (Phenomenex Luna, 4.6 mm × 100 mm, 5 mm) at a flow rate of 0.7 mL/min.
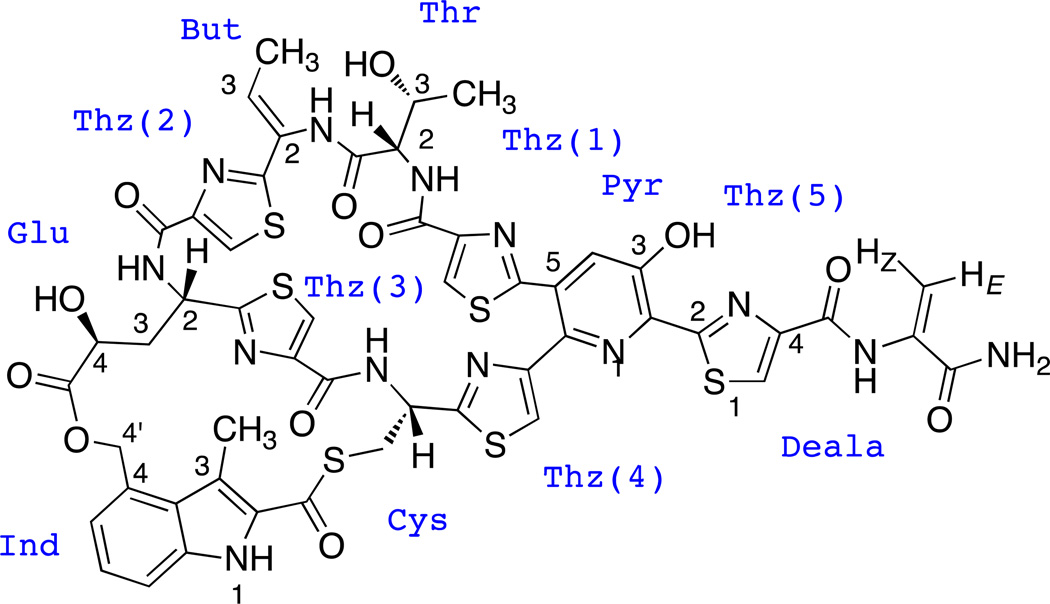
Structure of marine-derived nosiheptide.
NMR data in good agreement with:
Mocek, U., Chen, L.-C., Keller, P. J., Houck, D. R., Beale, J. M. & Floss, H. G. 1H and 13C NMR assignment of the thiopeptide antibiotic nosiheptide. J. Antibiot. 11, 1643-16-48 (1989).
Table 1
1H and 13C NMR spectral data of nosiheptide(δ in ppm, J in Hz).
| Ind CO | 180.69 | But 3 | 128.14 | 6.32 (q, J = 6.72) |
| Glu CO | 173.52 | Pyr 4 | 127.17 | 7.64 (s) |
| Thz(3) 2 | 170.18 | Thz(5) 5 | 126.52 | 8.28 (s) |
| Thz(4) 2 | 169.64 | Thz(1) 5 | 125.82 | 8.47 (s) |
| Thr CO | 169.44 | Thz(3) 5 | 125.67 | 8.18 (s) |
| Thz(5) 2 | 166.83 | Ind 6 | 125.28 | 7.41 (dd, J = 7.13, 7.33) |
| Thz(2) 2 | 166.41 | Ind 3a | 124.36 | |
| Deala CO | 165.99 | Thz(2) 5 | 123.91 | 7.91 (s) |
| Thz(1) 2 | 164.85 | Ind 5 | 122.24 | 7.12 (d, J = 7.13) |
| Thz(3) CO | 161.43 | Thz(4) 5 | 120.38 | 7.66 (s) |
| Thz(2) CO | 160.84 | Ind 3 | 118.90 | |
| Thz(1) CO | 160.57 | Ind 7 | 115.31 | 7.79 (d, J = 7.33) |
| Thz(5) CO | 158.59 | Deala 3 | 104.03 | 6.60 E (d, J = 1.83), |
| 5.58 Z (d, J = 1.73) | ||||
| Thz(4) 4 | 155.00 | Thr 3 | 67.45 | 4.04 (dd, J = 3.46, 13,65) |
| Pyr 3 | 150.30 | Glu 4 | 67.18 | 4.14 (d, J= 12.02) |
| Thz(1) 4 | 149.81 | Ind 4’ | 66.65 | 5.85 (d, J = 11.25), |
| 5.02 (d, br, J = 10.79) | ||||
| Thz(5) 4 | 149.76 | Thr2 | 56.75 | 4.36 (d, J = 7.79) |
| Thz(3) 4 | 148.45 | Cys 2 | 50.17 | 5.98 (d, br, J = 9.00) |
| Thz(2) 4 | 147.86 | Glu2 | 45.85 | 5.76 (t, J = 10.19) |
| Pyr 6 | 143.99 | Glu 3 | 36.72 | 2.35 S (m), |
| 1.80 R (m) | ||||
| Ind 7a | 137.43 | Cys 3 | 29.71 | 3.79 (dd, J = 3.66, 12.21), |
| 3.71 (dd, J = 4.07, 11.81) | ||||
| Pyr 2 | 134.46 | Thr CH3 | 17.52 | 0.95 (s) |
| Deala 2 | 132.93 | But CH3 | 14.58 | 1.68 (d, J = 6.92) |
| Ind 2 | 130.64 | Ind CH3 | 11.96 | 2.54 (s) |
| Pyr 5 | 130.41 | Ind NH | 10.83 (s, br) | |
| But 2 | 129.41 | Deala NH | 9.99 (s) | |
| Ind 4 | 128.73 | But NH | 9.75 (s) | |
| Glu NH | 7.97 (d, br, J = 7.29) | |||
| Glu OH | ||||
| Cys NH | 7.82 (s) | |||
| Thr NH | 7.50 (s) |
Bacterial Strains and Susceptibility Testing
Details on the strains used in this study are in Table 2. MRSA Sanger 252 USA 200 strain was obtained through the Network of Antimicrobial Resistance in S. aureus (NARSA) program supported under NIAID/NIH contract # HHSN272200700055C. Susceptibility testing was performed in duplicate using cation-adjusted Mueller-Hinton broth (CA-MHB) and Mueller-Hinton agar (MHA) according to CLSI methods.9 The MIC for Clostridium difficile was determined by the agar dilution reference method according to CLSI guidelines.10,11 Susceptibility of nosiheptide against MRSA strain TCH1516 was also determined in the presence of 20% activated-pooled human serum (freshly collected from consenting healthy donors) and 80% MHB by adding resazurin and assessing the conversion to resorufin exactly as previously described.12 This method provides a visual readout of color change from the blue indicator resazurin to the pink resorufin, a sign of bacterial growth.
Table 2
Nosiheptide MIC
| Strain | Details | MIC (mg/L) |
|---|---|---|
| TCH1516 | USA 300 CA-MRSA (ATCC) | 0.06 |
| (0.06)* | ||
| Sanger 252 | USA 200 HA-MRSA (NARSA) | 0.03 |
| ATCC 33591 | HA-MRSA | 0.06 |
| A593720 | Bloodstream MRSA, progenitor to A5940 | 0.125 |
| A5940 VISA20 | in vivo VISA | 0.125 |
| A630020 | MRSA, progenitor to A6298 | 0.125 |
| A6298 VISA20 | VISA | 0.125 |
| SA85312 | Progenitor to SI853b | 0.125 |
| SA853b12 | in vitro VISA | 0.125 |
| HIP5836 NJ VISA12,20 | VISA - NJ | 0.125 |
| VRSA PA12 | vanA vancomycin-resistant S. aureus | 0.06 |
| VRSA MI12 | vanA vancomycin-resistant S. aureus | 0.125 |
| A781712 | Progenitor to A7819, A7819erm | 0.06 |
| A7819LinR 12 | LinezolidR isolate | 0.06 |
| A7819ermR 12 | Erythromycin-resistant variant (plasmid) | 0.06 |
| SA35412 | Progenitor to SA355 | 0.125 |
| SA35512 | LinezolidR isolate | 0.125 |
| 061612 (DapS) | Endocarditis MSSA, progenitor to 0701 | 0.125 |
| 070112 (DapR) | Daptomycin-resistant MSSA | 0.06 |
| MSSA | ATCC 29213 | 0.125 |
| RN912020 | MSSA, agr knockout, progenitor to RN9120b | 0.06 |
| RN9120V20 | VISA selected in vitro | 0.25 |
| S. epidermidis | ATCC 12228 | 0.5 |
| VRE-CUS | VRE bloodstream left-sided endocarditis | 0.125 |
| VRE-WMC | VRE boodstream infection | 0.125 |
| BI/NAP1/027 Hypervirulent Contemporary | ||
| Clostridium difficile BI | Clostridium difficile | 0.008 |
| Moraxella catarrhalis | ATCC 25238 | 0.5 |
| E. coli 103521 | Clinical respiratory tract isolate | >4 |
| Pseudomonas aeruginosa | ATCC 27853 | >4 |
| Acinetobacter baumannii | ATCC 19606 | >4 |
| Enterobacter cloacae | ATCC 13047 | >4 |
Time Kill Studies
Time kill studies were done essentially as previously described in this laboratory.13 Briefly, nosiheptide was added at multiples of the MIC in CA-MHB. MRSA strains or vancomycin-resistant Enterococcus faecium (VRE-CUS) were added at a starting inoculum of 5 × 105 cfu/mL, and the cultures were incubated in a 37°C shaking incubator (New Brunswick Scientific, Enfield, CT). Samples were taken at specified timepoints for enumeration of viable bacteria by plating serial dilutions on Todd-Hewitt agar plates.
Post-Antibiotic Effect
Post-antibiotic effect was assessed using the viable plate count method as previously described.13 For these studies, either nosiheptide or vancomycin was added at 10X their respective MICs to 14 mL Falcon tubes containing CA-MHB. CA-MRSA strain TCH1516 or HA-MRSA strain Sanger 252 was added at 5 × 105 cfu/mL, and the tubes were incubated under shaking conditions at 37°C for 1 hour. The bacteria were then pelleted and washed in 4 mL of antibiotic-free CA-MHB. The pellets were resuspended in 4 mL CA-MHB and incubated at 37°C in a shaking incubator. Surviving bacteria were assessed at specified timepoints as for the time-kill assay. The PAE was determined as previously described.13,14
Mammalian Cell Cytotoxicity
Mammalian cell cytotoxicity was assessed as previously described.13 Briefly, 2 × 104 HeLa cells (American Type Culture Collection # CCL-2, ATCC, Manassas, VA) were seeded per well of sterile 96-well tissue culture-treated plates (Falcon; Becton Dickinson, Franklin Lakes, NJ). After 24 hours, the medium (RPMI containing 10% heat-inactivated fetal bovine serum) was replaced with fresh medium containing increasing concentrations of nosiheptide up to 128 mg/L, and the plates were incubated at 37°C in 5% CO2. Cell viability was assayed at 72 hours by measuring the reduction of MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] using the CellTiter 96 Aqueous nonradioactive cell proliferation assay according to the manufacturer's instructions (Promega, Madison, WI).
In Vivo Testing
A murine model of intraperitoneal (ip) infection was used to test in vivo efficacy of nosiheptide.15 Eight week old female CD1 mice (Charles River, Wilmington, MA, USA) were injected ip with 1 – 2 × 109 cfu of the HA-MRSA strain Sanger 252 in 4% hog gastric mucin. The mice were treated with nosiheptide (20 mg/kg) ip at 1 and 8 h after bacterial inoculation and were monitored for mortality and clinical status twice daily thereafter for five days. Moribund mice were humanely euthanized as were mice at the end of the study. These experiments were reviewed and approved by the Animal Subjects Committee of UCSD.
Statistical Analysis
The data from the survival studies were analyzed with GraphPad Prism (GraphPad Software, Incorporated, La Jolla, CA) using log-rank analysis (Mantel-Cox test). Data were considered significant at p < 0.05.
Results
In vitro antimicrobial activity
Nosiheptide exhibited strong activity against all contemporary MRSA and MSSA strains tested including multidrug-resistant clinical isolates (Table 2), with MIC values ≤ 0.25 mg/L. Notably, clinical MRSA isolates previously demonstrated to have developed resistance to front-line antibiotics including daptomycin, linezolid, and vancomycin remained highly sensitive to nosiheptide. This activity against MRSA was also completely unaffected by the presence of 20% human serum (Table 2). Nosiheptide was also highly active against Enterococcus spp and the Gram-negative pathogen Moraxella catarrhalis, but did not possess activity at ≤ 4 mg/L against the other Gram-negative strains tested including clinical isolates of E. coli and P. aeruginosa. Addressing another critical antibiotic resistance challenge, nosiheptide showed extremely potent activity against the contemporary hypervirulent BI strain of C. difficile (also known as the NAP1 or ribotype 027 strain).
Time-kill kinetics
In vitro time-kill analysis was used to assess nosiheptide killing kinetics. Nosiheptide was rapidly bactericidal against MRSA in a concentration- and time-dependent manner (Figure 2A), with a nearly 2-log kill noted within 6 h at both 10X and 20X MIC. A 2-log kill of MRSA was also noted for nosiheptide against MRSA resistant to other antibiotics such as linezolid or erythromycin (Figure 2B). While nosiheptide demonstrated bactericidal activity against MRSA at 10X and 20X the MIC, it was bacteriostatic against a clinical bloodstream isolate of vancomycin-resistant Enterococcus faecium up to 64X MIC (Figure 2C). The MRSA killing kinetics of nosiheptide also compared favorably with vancomycin kinetics at 10X and 20X their respective MICs (Figure 2D).
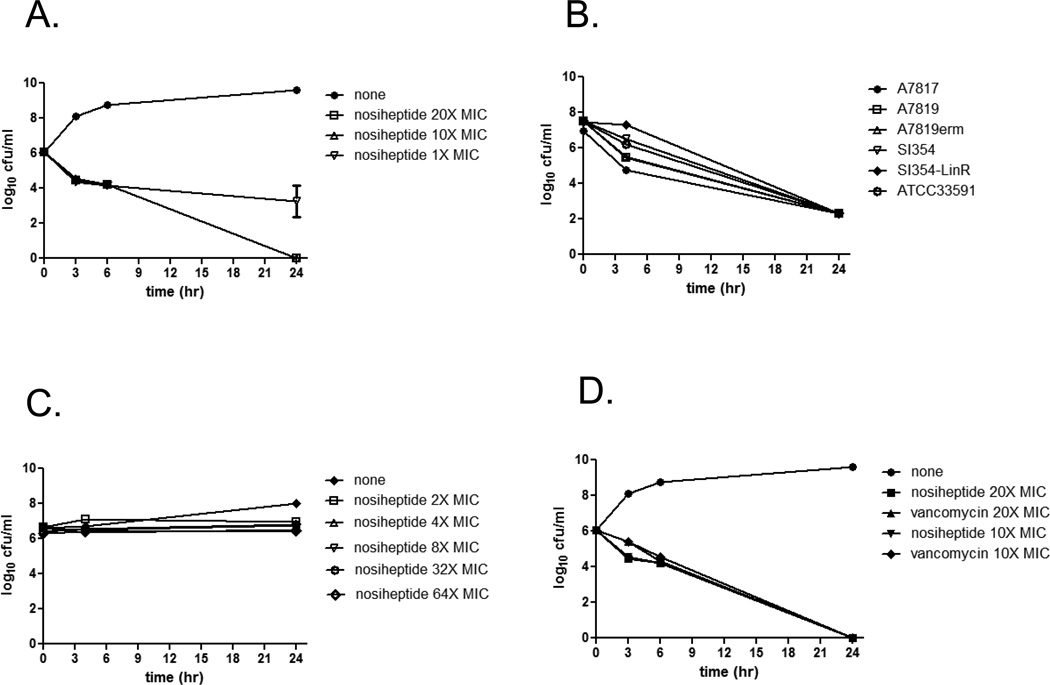
Time-kill kinetics of nosiheptide against MRSA and VRE. Nosiheptide killing kinetics was studied against MRSA and VRE at multiples of their MICs (MIC = 0.06 mg/L for nosiheptide). A.) Killing kinetics of nosiheptide at 1X, 10X, and 20X MIC against MRSA USA300 strain TCH1516 (MIC = 0.06 mg/L). B.) Nosiheptide killing kinetics against various multi-drug resistant MRSA at 10X their respective MICs. C.) Nosiheptide activity over time against VRE-CUS at multiples of the MIC (MIC = 0.125 mg/L). D.) Nosiheptide time-kill kinetics against MRSA USA300 strain TCH1516 compared to vancomycin at 10X and 20X their respective MICs (MIC = 0.06 mg/L for nosiheptide; MIC = 1.56 mg/L for vancomycin). Each assay was repeated in duplicate (for VRE) or triplicate (for MRSA), and the data shown are the mean +/− SD from one representative experiment.
Prolonged post-antibiotic effect and lack of cytotoxicity
Given its favorable MRSA killing kinetics, nosiheptide was assessed for its post-antibiotic effects (PAE). Notably, nosiheptide exhibited prolonged PAEs against both HA- and CA-MRSA compared to the most commonly used systemic antibiotic, vancomycin, at 10X their respective MICs (Figure 3A and B). The PAE for nosiheptide was calculated to exceed 9 h for both HA- and CA-MRSA. We also tested nosiheptide for in vitro evidence of mammalian cell cytotoxicity as a prelude to our in vivo experiments. No evidence for nosiheptide cytotoxicity was observed when incubated for 72 hours with the cervical carcinoma HeLa cell line at up to 128 mg/L, which is approximately 1000-fold above the MIC against MRSA. We also found that nosiheptide activity against USA300 MRSA was not inhibited in 20% human serum (Table 2).
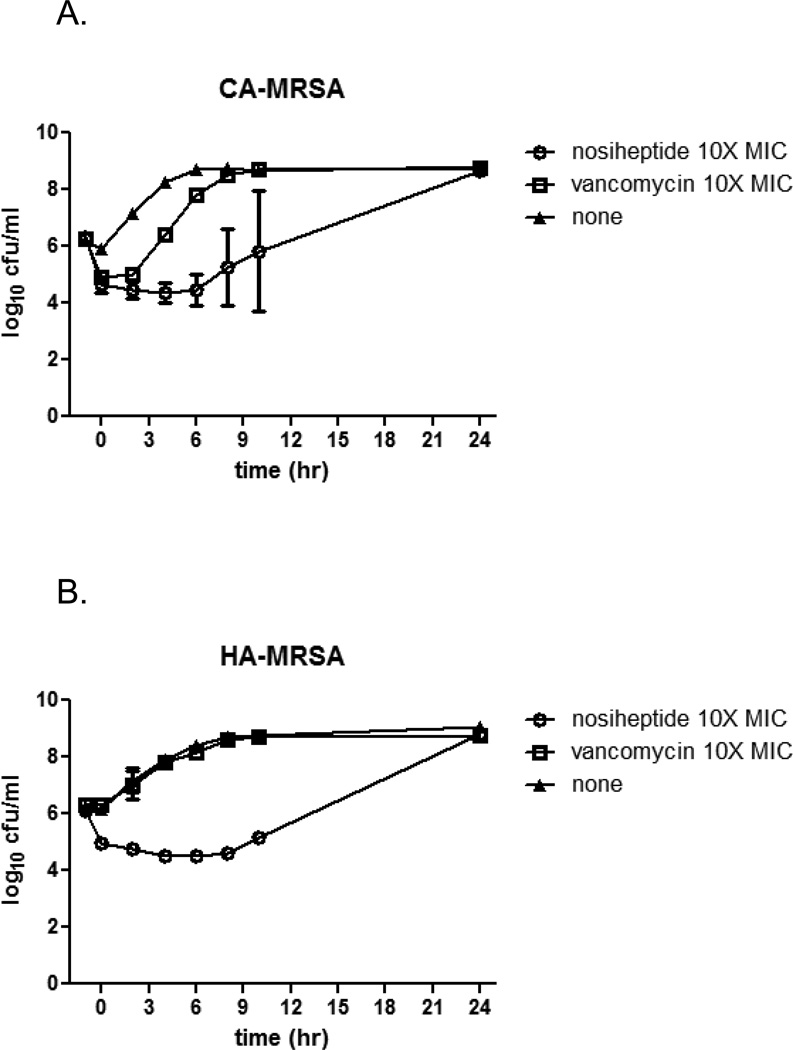
Post-antibiotic effect of nosiheptide on contemporary MRSA strains. CA-MRSA strain TCH-1516 (Panel A) or HA-MRSA strain Sanger 252 (Panel B) were incubated for one hour with nosiheptide or vancomycin at 10X MIC. The antibiotics were removed, and the bacteria were washed and allowed to recover in antibiotic-free media. The curves show the rates of growth following antibiotic treatment. The data represent the mean +/− SD of one representative assay repeated in duplicate.
In Vivo Activity
Due to its potent anti-MRSA activities in vitro, nosiheptide was tested in a murine model of intraperitoneal MRSA infection. Mice were infected with HA-MRSA strain Sanger 252, followed by nosiheptide treatment (20 mg/kg, ip) at 1 and 8 h post-infection. Although all mice became lethargic and exhibited piloerection within approximately 4 h of infection, nosiheptide provided significant (p < 0.03) protection against mortality (Figure 4). Ten out of 10 of the nosiheptide-treated mice remained alive on day 3, while 6/10 of the controls died on day 1. By the end of the study, only one mouse in the nosiheptide group had died. These results provide evidence of significant in vivo activity for nosiheptide.
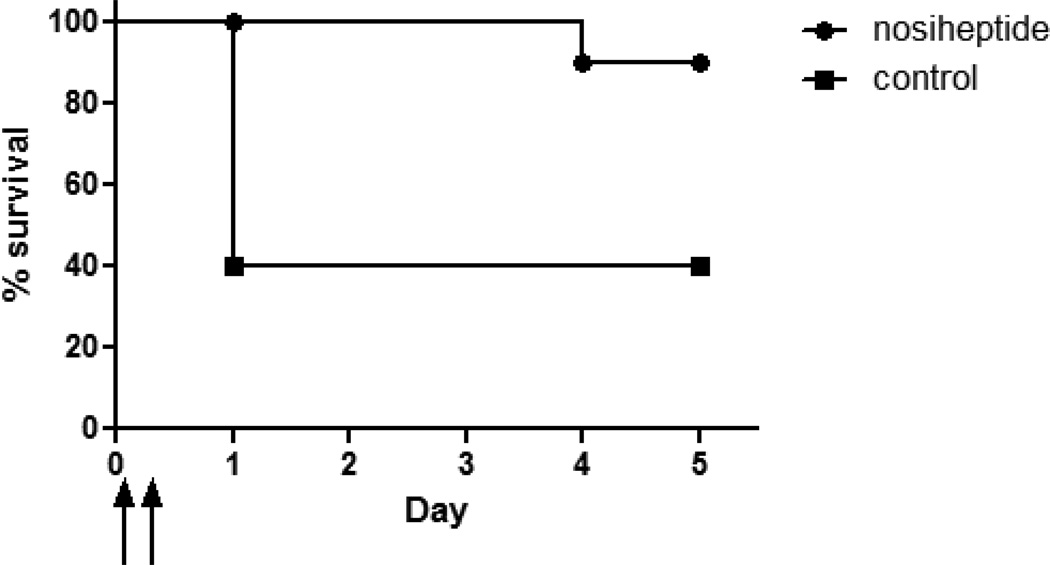
Nosiheptide protection in a murine model of intraperitoneal MRSA infection. Plot showing survival of mice (n = 10 per group) infected ip with HA-MRSA strain Sanger 252, followed by ip treatment with either nosiheptide (20 mg/kg, filled circles) or vehicle control (filled squares) at 1 hour or 8 hours after infection (represented by upward arrows on the x-axis). N = 10 mice per treatment group (p < 0.03 by log-rank analysis).
Discussion
In our search for new anti-MRSA antibiotics from marine-derived microorganisms, we identified an actinomycete strain that produced a metabolite shown to be nosiheptide. Although the structure of this compound was originally elucidated several decades ago and activity against Gram-positive activity was noted, a paucity of data exist on the therapeutic potential for this compound given contemporary concerns of multi-drug resistant bacterial pathogens. With growing antibiotic resistance exhibited by MRSA and other Gram-positive pathogens driving the need for new antimicrobials, we sought to further characterize this compound for its activity against HA- and CA-MRSA. Nosiheptide was exquisitely active against all contemporary MRSA strains tested regardless of their resistance to other front-line therapeutics including vancomycin, linezolid, and daptomycin. Importantly, high level activity was also noted against the increasingly problematic pathogen, Clostridium difficile, especially significant since the FDA has approved only one new drug (Fidaxomicin) to treat C. difficile in the last two decades. Although the activity of some thiazole antibiotics may be reduced in the presence of serum, we found that nosiheptide activity against USA300 MRSA was unaffected in the presence of 20% human serum, suggesting that serum inhibition may not necessarily be a characteristic of all thiazole-containing antibiotics.
Nosiheptide exhibited favorable MRSA killing kinetics, a very prolonged post-antibiotic effect, and was active in a murine model of intraperitoneal infection. Although in vivo evidence of nosiheptide activity has not been published, Benazet et al16 report that nosiheptide protected mice from S. aureus mortality only when the compound was administered at the site of infection. However, no data were presented to support this claim. We report in the current paper nosiheptide activity when the compound is administered one and eight hours after MRSA injection at the site of infection as well (Figure 4). Our data are consistent with the claims of Benazet et al16 and point to the fact that nosiheptide is efficacious in vivo but may either be metabolized or not distributed (due to localized precipitation or lack of absorption) when injected. A closely related compound, glycothiohexide α, was once shown to be quite active against MRSA and vancomycin-resistant E. faecium.17–19 However, despite its in vitro potency, this compound had minimal activity in a S. aureus model of intraperitoneal infection when administered subcutaneously. It is unknown whether in the case of glycothiohexide α pharmacokinetic and/or possible metabolic factors abrogated potential efficacy. In pilot studies, we also found that nosiheptide exhibited significantly reduced activity when injected subcutaneously (data not shown) following intraperitoneal MRSA inoculation and are currently embarking upon collaborative studies of liposomal and other lipophilic drug formulations to optimize nosiheptide delivery in vivo.
In sum, nosiheptide represents a promising antibiotic scaffold for further development due to its potent anti-MRSA and other Gram-positive activity, lack of inhibition by human serum, lack of mammalian cell cytotoxicity, and demonstrated tolerability when delivered orally to animals.7 Many of the synthesis/manufacturing issues have already been addressed, since nosiheptide is produced in mass quantities by manufacturers worldwide. Cumulatively, these properties provide nosiheptide an advantage over other early-stage antibacterials, many of which are later found to be cytotoxic or serum-inhibited, or are fraught with difficulties in scale-up of production.
Acknowledgements
N.M.H. was supported by the National Institutes of Health (NIH) Training Program in Marine Biotechnology (T32 GM067550) and a Ruth L. Kirschstein National Research Service Award (NRSA) from National Institutes of Health grants (5 F31 GM090658-02). S.L. was supported by the German Research Foundation (DFG). W.F., V.N., P.R.J., and M.E.H. were supported by National Institutes of Health grant GM084350. PRJ and WF acknowledge financial support from the NIH Fogerty Center International Cooperative Biodiversity Groups program (grant U01-TW00007-401). We thank K. Freel and C. Kauffman for assistance with fieldwork and W. Aalbersberg (University of the South Pacific) for providing laboratory space and facilitating the field collections. We gratefully acknowledge the people of Fiji for their hospitality and permission to collect samples from their local waters. We are also grateful to Diane M. Citron and Ellie Goldstein MD of R.M. Alden Research Laboratories (Culver City, CA) for performing susceptibility testing against Clostridium difficile.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ja.2012.77
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/ja201277.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Cell-Free Systems: Ideal Platforms for Accelerating the Discovery and Production of Peptide-Based Antibiotics.
Int J Mol Sci, 25(16):9109, 22 Aug 2024
Cited by: 0 articles | PMID: 39201795 | PMCID: PMC11354240
Review Free full text in Europe PMC
Discovery and Biosynthesis of Persiathiacins: Unusual Polyglycosylated Thiopeptides Active Against Multidrug Resistant Tuberculosis.
ACS Infect Dis, 10(9):3378-3391, 27 Aug 2024
Cited by: 1 article | PMID: 39189814 | PMCID: PMC11406533
Rule-based omics mining reveals antimicrobial macrocyclic peptides against drug-resistant clinical isolates.
Nat Commun, 15(1):4901, 08 Jun 2024
Cited by: 3 articles | PMID: 38851779
Selective biosynthesis of a rhamnosyl nosiheptide by a novel bacterial rhamnosyltransferase.
Microb Biotechnol, 17(1):e14412, 24 Jan 2024
Cited by: 2 articles | PMID: 38265165 | PMCID: PMC10832541
Design of a chimeric glycosyltransferase OleD for the site-specific O-monoglycosylation of 3-hydroxypyridine in nosiheptide.
Microb Biotechnol, 16(10):1971-1984, 22 Aug 2023
Cited by: 3 articles | PMID: 37606280 | PMCID: PMC10527214
Go to all (35) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - JQ946086
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Anthracimycin activity against contemporary methicillin-resistant Staphylococcus aureus.
J Antibiot (Tokyo), 67(8):549-553, 16 Apr 2014
Cited by: 19 articles | PMID: 24736856 | PMCID: PMC4146678
Comparative time-kill study of doxycycline, tigecycline, cefazolin and vancomycin against several clones of Staphylococcus aureus.
Curr Clin Pharmacol, 8(4):332-339, 01 Nov 2013
Cited by: 1 article | PMID: 23590512
Pharmacodynamic activity of ceftobiprole compared with vancomycin versus methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-intermediate Staphylococcus aureus (VISA) and vancomycin-resistant Staphylococcus aureus (VRSA) using an in vitro model.
J Antimicrob Chemother, 64(2):364-369, 19 May 2009
Cited by: 15 articles | PMID: 19454524
In-vitro profile of a new beta-lactam, ceftobiprole, with activity against methicillin-resistant Staphylococcus aureus.
Clin Microbiol Infect, 13 Suppl 2:17-24, 01 Jun 2007
Cited by: 21 articles | PMID: 17488372
Review
Funding
Funders who supported this work.
FIC NIH HHS (3)
Grant ID: U01 TW007401
Grant ID: U01-TW00007-401
Grant ID: U19 TW007401
NIGMS NIH HHS (4)
Grant ID: 5 F31 GM090658-02
Grant ID: T32 GM067550
Grant ID: F31 GM090658
Grant ID: R01 GM084350





