Abstract
Free full text

Phylogeography and Population Dynamics of Dengue Viruses in the Americas
Abstract
Changes in Dengue virus (DENV) disease patterns in the Americas over recent decades have been attributed, at least in part, to repeated introduction of DENV strains from other regions, resulting in a shift from hypoendemicity to hyperendemicity. Using newly sequenced DENV-1 and DENV-3 envelope (E) gene isolates from 11 Caribbean countries, along with sequences available on GenBank, we sought to document the population genetic and spatiotemporal transmission histories of the four main invading DENV genotypes within the Americas and investigate factors that influence the rate and intensity of DENV transmission. For all genotypes, there was an initial invasion phase characterized by rapid increases in genetic diversity, which coincided with the first confirmed cases of each genotype in the region. Rapid geographic dispersal occurred upon each genotype's introduction, after which individual lineages were locally maintained, and gene flow was primarily observed among neighboring and nearby countries. There were, however, centers of viral diversity (Barbados, Puerto Rico, Colombia, Suriname, Venezuela, and Brazil) that were repeatedly involved in gene flow with more distant locations. For DENV-1 and DENV-2, we found that a “distance-informed” model, which posits that the intensity of virus movement between locations is inversely proportional to the distance between them, provided a better fit than a model assuming equal rates of movement between all pairs of countries. However, for DENV-3 and DENV-4, the more stochastic “equal rates” model was preferred.
Introduction
Dengue virus (DENV) is the most widely distributed flavivirus species, found in almost all tropical and subtropical regions where the mosquito vectors Aedes aegypti and Aedes albopictus thrive (Halstead 1992). This distribution places approximately 2.5 billion people at risk of infection (Guzman et al. 2010). DENV exists as four antigenically distinct serotypes (DENV-1 to DENV-4) that are further subdivided into numerous phylogenetically distinct genotypes. All four serotypes are known to be the causative agents of dengue fever and of the more severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Dengue is perhaps the most important emerging vector-borne disease of the 21st century. Recent figures estimate that there are 50 million infections annually, including 500,000 hospitalizations due to DHF alone (Guzman et al. 2010), and since the 1980s, there has been a 4.5-fold increase in the number of reported cases Americas alone (San Martin et al. 2010).
Phylogenetic analyses suggest that currently circulating serotypes diverged from their sylvatic ancestors approximately 1,000 years ago (Twiddy et al. 2003); however, the geographic origins of DENV remain unclear and have been alternatively attributed to Africa or Asia (Smith 1956; Christophers 1960; Powell et al. 1980; Wang et al. 2000; Diallo et al. 2005). Currently, the greatest genetic diversity is found in Southeast Asia where all four serotypes have been cocirculating since World War II (Gubler 2006). This region has been identified as a source population for the rapid and dramatic reemergence of DENV globally over the last half-century, initially facilitated by the movement of ships and goods during the Second World War (Gubler 1997) and more recently by rapid increases in human population size, uncontrolled urbanization, and the advent of rapid mass human movement (Gubler 1998; Kroeger and Nathan 2006).
In the Americas, patients with dengue-like clinical symptoms were recorded as early as 1780 in Philadelphia (Rush 1789) but such outbreaks were rare. Before 1963, only DENV-2 American genotype was reported in the Americas, but since then the region has been subject to repeated introductions (fig. 1). This shift from hypo- to hyperendemicity was associated with a rise in the size and frequency of DENV outbreaks (Gubler 1997). In addition, there was a rise in DHF and DSS cases after the introduction of DENV-2 Asian–American genotype in 1981. Today, all four serotypes exist in the Americas, and since 1963, the region has experienced outbreaks caused by at least one DENV-1 genotype, two DENV-2 genotypes, three DENV-3 genotypes, and one DENV-4 genotype (fig. 1).
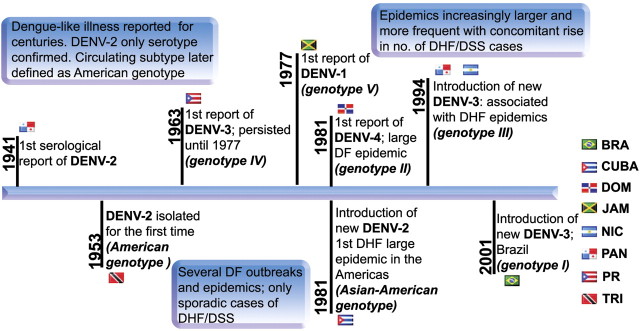
Timeline showing the introductions of the various genotypes of the four serotypes of DENV into the Americas. The flags of the countries associated with the first reports are displayed. BRA, Brazil; CUBA, Cuba; DOM, Dominica; JAM, Jamaica; NIC, Nicaragua; PAN, Panama; PR, Puerto Rico; and TRI, Trinidad and Tobago.
There is a clear seasonality to DENV outbreaks in the Americas, with peak incidence occurring when rainfall and mosquito population densities are highest (Focks and Barrera 2006; Halstead 2008). Although multiple serotypes, and even multiple genotypes of a serotype, may be detected in a given season, typically, a single serotype will predominate regionally and to an even greater extent within individual countries. At small epidemiological scales, movement of DENV between locales has resulted in the establishment of certain lineages within naive populations, accompanied by the extinction of local circulating genotypes (Zhang et al. 2005). Adams et al. (2006) investigated the complex epidemic dynamics of all four serotypes in Bangkok, Thailand, using an epidemic model with stochastic seasonal forcing and found them to be characterized by 8- to 10-year epidemic oscillations. This dynamic was reflected in DENV-1, DENV-2, and DENV-3 gene sequences collected monthly over almost three decades. It was suggested that moderate cross-protective immunity gave rise to persistent out-of-phase epidemic oscillations among serotypes but that strong or weak cross-protection/cross-enhancement only produced in-phase patterns (Adams et al. 2006). Zhang et al. (2005) proposed that the genetic diversity of a given serotype increases at times of relative abundance of the particular serotype and that hyperendemicity may lead to complex patterns of selective competition, influenced by differential susceptibility to cross-reactive host immunological responses to cocirculating serotypes, genotypes, or lineages within genotypes (Zhang et al. 2005).
Work done in the Caribbean, specifically in Puerto Rico by Bennett et al. (2003, 2006), used epidemiological data from passive surveillance to reveal similar DENV evolutionary dynamics among genotypes. In Puerto Rico, viral diversity tends to be maintained in situ, driven by genetic drift with notable clade extinction and replacement events over time. Occasionally, new lineages are exchanged with South and Central America and other regions in the Caribbean.
In the past, efforts have been made to infer the dispersal of DENV and to elucidate the diffusion patterns of the virus over decades (Carrington et al. 2005; Araujo et al. 2009). However, these studies have been restricted to low-resolution heuristic approaches that provide few insights into the timescale of DENV spatial dynamics. Recently, Lemey et al. (2009) introduced a Bayesian phylogeographic inference framework, which incorporates the spatial and temporal dynamics of viral lineages for inference, visualization, and phylogeographic hypothesis testing. In this framework, different scenarios and models of spatial spread can be investigated and compared by specifying different prior distributions for the diffusion rates among sampling locations (Auguste et al. 2010; Talbi et al. 2010).
In this study, we have inferred the population dynamics of DENV in the Americas and compared these with previously published results for DENV-2 and DENV-4 (Carrington et al. 2005). We also undertook a thorough analysis of the phylogeography of DENV in the Americas, which allowed us to reconstruct the spatiotemporal spread of the virus and to provide statistical assessment of the geographic locations of ancestral infections. With these results, we also sought to uncover the factors influencing the unique patterns of population dynamics and spatial diffusion of DENV in the Americas (Minin et al. 2008; Lemey et al. 2009).
Materials and Methods
Virus Isolation, Reverse Transcription Polymerase Chain Reaction Amplification, and Nucleotide Sequencing of Envelope (E) Gene
A total of 18 DENV-1 and 25 DENV-3 isolates from 10 Caribbean countries (see table 1) were obtained from the Caribbean Epidemiology Centre (CAREC), Trinidad and Tobago, where the viruses were isolated from sera by culturing in C6/36 cells according to Tesh (1979). Viral RNA was extracted from 140 μl of cell culture supernatant using QIAamp Viral RNA Mini Kits (Qiagen, Germany). Envelope (E) genes were then amplified in a two-step process: Reverse transcription was performed using SuperScript III Reverse Transcriptase (Invitrogen) and amplified by polymerase chain reaction (PCR) using PfuUltra II Fusion HS DNA polymerase (Stratagene), using primers designed for 2× coverage of the entire open reading frame (see supplementary table S2, Supplementary Material online). Amplicons were visualized on a 2% agarose gel and subsequently excised and purified using QIAquick Gel Extraction kit (Qiagen). Purified PCR amplicons were sequenced at the Greenwood Molecular Biology Facility, University of Hawaii, HI. Sequences were assembled using Sequencher 4.5 (Gene Codes, Michigan) and were submitted to GenBank under the accession numbers shown in table 1.
Table 1.
DENV-1 and DENV-3 Sequences Derived from This Study.
| Year | Isolate Name | Location | Serotype | Accession Number |
| 1977 | CAREC777882 | Bahamas | 1 | JN379475 |
| 1977 | CAREC778136 | Grenada | 1 | JN379476 |
| 1977 | CAREC778140 | Grenada | 1 | JN379477 |
| 1977 | CAREC778164 | Grenada | 1 | JN379478 |
| 1977 | CAREC778169 | Grenada | 1 | JN379479 |
| 1977 | CAREC778776 | Grenada | 1 | JN379480 |
| 1978 | CAREC780174 | Grenada | 1 | JN379481 |
| 1978 | CAREC780184 | Grenada | 1 | JN379482 |
| 1981 | CAREC817486 | Trinidad and Tobago | 1 | JN379483 |
| 1981 | CAREC817593 | Grenada | 1 | JN379484 |
| 1981 | CAREC817838 | Suriname | 1 | JN379485 |
| 1995 | CAREC9510153 | Barbados | 1 | JN379486 |
| 1999 | CAREC9911056 | Barbados | 1 | JN379487 |
| 2001 | CAREC0101764 | Barbados | 1 | JN379470 |
| 2001 | CAREC0101765 | Barbados | 1 | JN379471 |
| 2003 | CAREC0308216 | Barbados | 1 | JN379472 |
| 2004 | CAREC0403888 | Aruba | 1 | JN379472 |
| 2005 | CAREC0511227 | Belize | 1 | JN379474 |
| 1998 | CAREC 9810082 | Jamaica | 3 | JN379502 |
| 1998 | CAREC 9810095 | Jamaica | 3 | JN379503 |
| 1998 | CAREC 9811479 | Belize | 3 | JN379504 |
| 2001 | CAREC0106464 | Barbados | 3 | JN379488 |
| 2001 | CAREC0201394 | Antigua | 3 | JN379489 |
| 2002 | CAREC0201402 | St Vincent and the Grenadines | 3 | JN379490 |
| 2002 | CAREC0201710 | St Vincent and the Grenadines | 3 | JN379491 |
| 2002 | CAREC0106464 | Barbados | 3 | JN379492 |
| 2002 | CAREC0202137 | Trinidad and Tobago | 3 | JN379512 |
| 2002 | CAREC020213 | Trinidad and Tobago | 3 | JN379496 |
| 2002 | CAREC0202139 | Trinidad and Tobago | 3 | JN379493 |
| 2002 | CAREC0202259 | Trinidad and Tobago | 3 | JN379494 |
| 2002 | CAREC0202260 | Trinidad and Tobago | 3 | JN379505 |
| 2002 | CAREC0202290 | Grenada | 3 | JN379495 |
| 2002 | CAREC0202314 | Grenada | 3 | JN379506 |
| 2002 | CAREC0202329 | Grenada | 3 | JN379507 |
| 2003 | CAREC0301734 | Barbados | 3 | JN379497 |
| 2003 | CAREC0302290 | Grenada | 3 | JN379498 |
| 2003 | CAREC0306149 | Trinidad and Tobago | 3 | JN379508 |
| 2003 | CAREC0310367 | Trinidad and Tobago | 3 | JN379499 |
| 2004 | CAREC0400408 | Barbados | 3 | JN379500 |
| 2004 | CAREC0400409 | Barbados | 3 | JN379509 |
| 2004 | CAREC0414035 | Belize | 3 | JN379510 |
| 2004 | CAREC0414066 | Belize | 3 | JN379511 |
| 2005 | CAREC0508129 | Suriname | 3 | JN379501 |
Data Sets
Derived DENV-1 and DENV-3 nucleotide sequences were manually aligned together with previously published E gene sequences from GenBank belonging to DENV-1 genotype V and DENV-3 genotype III then trimmed to a common length (see supplementary table S1, Supplementary Material online). DENV-2 Asian-American genotype and DENV-4 genotype II sequences (the genotypes currently circulating in the Americas) were also obtained from GenBank and similarly prepared. In order to prevent overrepresentation of come countries, data sets were downsampled to no more than five sequences per country per year. Sequence genotypes were confirmed through maximum likelihood tree estimation using a genotype reference alignment (data not shown). All sequences were confirmed as nonrecombinant using the Recombination Detection Program (RDP) v3 (Martin et al. 2005) and Genetic algorithms for recombination detection program (Pond and Frost 2005). Additionally, using the programs Datamonkey and HyPhy v0.99 (Pond et al. 2005), we confirmed that there was no evidence for site-specific positive selection in the data sets (at the P = 0.05 significance level). Together, the four data sets represent 35 countries and a time period covering 1977–2009 (available from the authors on request).
Evolutionary Rates, Population Dynamics, and Spatial Diffusion
Nucleotide substitution rates, divergence times, spatial diffusion rates (i.e., rate at which viral lineages move among sampled locations), and demographic histories were jointly estimated from the date- and location-stamped DENV sequences using a Bayesian approach implemented in the Markov chain Monte Carlo (MCMC) inference framework of the BEAST v1.6.1 software package (Drummond and Rambaut 2007; Lemey et al. 2009), This program uses Bayesian MCMC methods to integrate across all plausible tree topologies and thereby estimate evolutionary parameters while taking into account statistical uncertainty arising from both phylogenetic inference and the spatial diffusion process. Analyses were carried out using a general time reversible (GTR) model with a discretized gamma-distributed across-site rate variation (GTR + Γ4) substitution model, a Bayesian skyride plot (BSP) coalescent model (Drummond et al. 2005) and both a strict molecular clock model and a relaxed (uncorrelated lognormal) molecular clock model (Drummond et al. 2006).
Demographic histories were illustrated using BSPs (Minin et al. 2008), which show temporal changes in the mean estimate for relative genetic diversity (Neτ, where Ne is the effective population size and τ is the generation time).
To determine the viral transmission network within the Americas, a nonreversible discrete phylogeography model was applied to all the data sets individually, with each country used as discrete states. To ensure statistical efficiency, we employed a Bayesian stochastic search variable selection procedure (Lemey et al. 2009), which imposes a prior distribution on the spatial model that favors a limited number of nonzero movement rates among pairs of locations, thus reducing the number of estimated parameters and identifying only those rates that are required to explain the observed data. A truncated Poisson prior with a mean of log 2 and an offset of K − 1 (where K = number of discretized location states) was used. In order to ensure adequate sampling of the parameters of the model (with effective sample sizes for all parameters > 200), MCMC chain was run for 100 million states, with a 10% burn-in period (to remove nonstationarity samples), and data were then subsampled every 10,000 states. Model comparisons were performed using Bayes factors (BFs) (Suchard et al. 2001), which are estimated using harmonic mean estimates of the model marginal likelihood (Newton and Raftery 1994; Suchard et al. 2003; Redelings and Suchard 2005).
Hypothesis Testing
The Bayesian phylogeographic inference framework incorporated into BEAST allows for testing of scenarios about the evolutionary and phylogeographic history. We create pairwise location matrices representing the different diffusion predictors, which are used as priors for the relative rate parameters (Talbi et al. 2010). The different diffusion predictors explored were
Equal rates: spatial diffusion model that assumes equal rates of virus movement between all countries a priori.
Distance-informed rates: formalizes a model of more intense virus movement between nearby locations (≈rates of movement being inversely proportional to the distance between locations). Two different measures of distance were used: GeoDistance (log of the great-circle distance between centroids of each pair of countries) and PopDistance (log of the great-circle distance between the largest and closest population centres between each pair of countries).
Air traffic informed rates: formalizes a model of more intense virus movement between countries with more air traffic between them (~rates of movement being proportional to the number of airline seats available between each pair of countries). Data for this model were obtained from OAG Max database and represent the airline seat capacity in 2007.
The fit of the scenarios were assessed using BF comparison, which allows for the comparison of the predictors on each data set.
Results
Population Genetic History and Serotype Prevalence
Figure 2 depicts the estimates of relative genetic diversity for each genotype on the same timescale as a histogram showing the percentage of infections confirmed by virus isolation for each serotype in each year. The latter was derived from records of virus isolations performed by the CAREC and is used as a proxy for relative serotype prevalence in the absence of reliable data from all of the countries surveyed. The census data indicate that DENV-1, which was first reported in 1977 (fig. 1), predominated until 1979. It was then quickly replaced in 1981 by DENV-4, which was first reported in the region in that same year. DENV-2 Asian–American genotype, which was also first reported in 1981, was slower to become established (Carrington et al. 2005) and did not dominate the virus isolations until 1986. Subsequently, there was a reversion to mainly DENV-4 isolations between 1992 and 1994, then DENV-1 for 2 years, and DENV-2 for a year. Between 1997 and 1999, DENV-2 was most prevalent, but there were also significant numbers of DENV-1 and DENV-4 isolations and a gradual increase in DENV-3, which became more prevalent from 1996. The latter predominated from 2000 to 2003, followed by an abrupt decline in 2004.
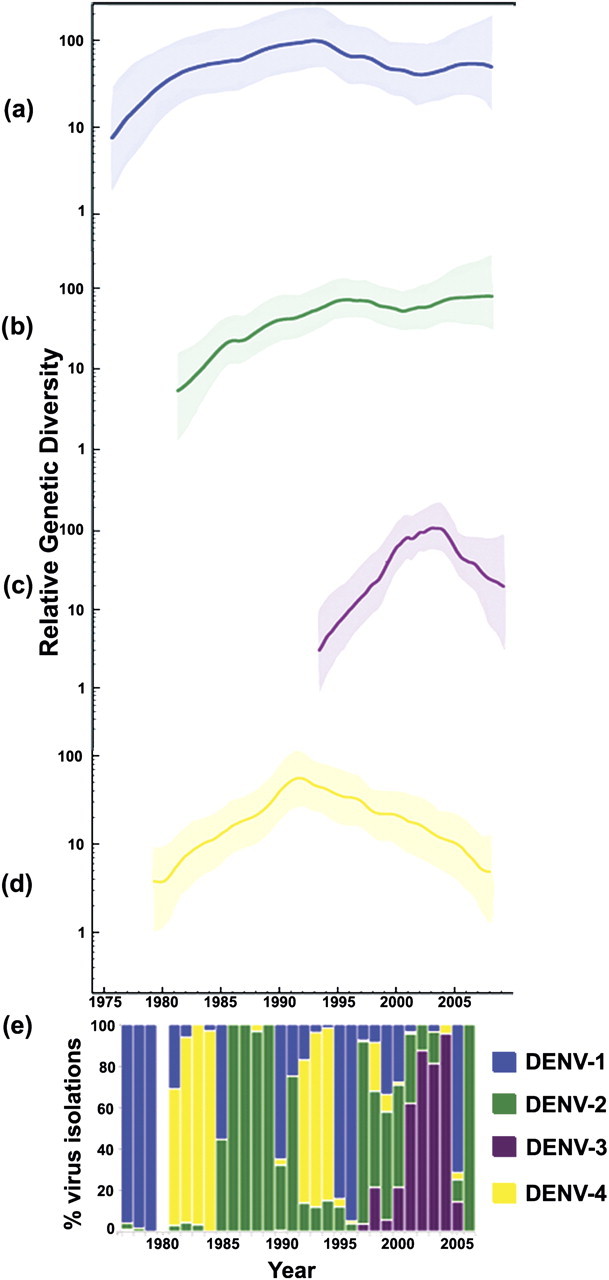
Demographic history of DENV-1 to DENV-4 (a–d) circulating in the Americas using Bayesian skyride model. The y axes of the skyline plots represent relative genetic diversity (Neτ). The x axis represents the number of years prior to the date of the most recent isolate in each analysis (i.e., 2007). The thick colored line is the mean estimate, and the 95% HPD are indicated by the shaded area. Below is a histogram (e) showing the percentage of infections confirmed by virus isolation for each serotype in each year.
In all cases, the initial increase in genetic diversity for each serotype (fig. 2) began prior to the date when that serotype was first reported. The date of first detection of each genotype also corresponded to the times when these viruses first accounted for the majority of isolations. The plots suggest that the period when a genotype's genetic diversity is highest coincided with the period that serotype is most prevalent in the Caribbean. Also, the introduction and sharp increase of the genetic diversity of DENV-3 coincided with a decline in DENV-4.
Inference of Substitution Rates and Dates of Divergence
The posterior mean substitution rates inferred for DENV-1 to DENV-4 under the relaxed clock model are shown in table 2, along with the inferred date for the most recent common ancestor (MRCA) of each genotype in the Americas. The mean substitution rates for the four genotypes were very similar, ranging from 7.8 × 10−4 subs/site/year to 9.9 × 10−4 subs/site/year, with overlaps among the four pairs of the 95% highest posterior density (HPD) intervals. The date that the MRCA of each genotype was estimated to have existed was prior to the year that genotype was first reported in the Americas. Only in the case of DENV-2 did the 95% HPD intervals for the time of the MRCA include the year of first detection.
Table 2.
Mean Substitution Rates and the Ages of the MRCA of Main Genotypes of DENV-1 to DENV-4 Circulating in the Americas.
| Serotype | MRCA (HPD) | Year of First Report | Root State Location | Country of First Report | Rates (HPD) (subs/site/year) |
| DENV-1 | 1973 | 1977 | Grenada | Jamaica | 7.773 × 10−4 |
| (1970–1975) | (6.528 × 10−4, 9.081 × 10−4) | ||||
| DENV-2 | 1979 | 1981 | Jamaica | Cuba | 8.600 × 10−4 |
| (1978–1981) | (7.408 × 10−4, 9.836 × 10−4) | ||||
| DENV-3 | 1992 | 1994 | Nicaragua | Nicaragua and Panama | 9.851 × 10−4 |
| (1991–1993) | (7.337 × 10−4, 10.396 × 10−4) | ||||
| DENV-4 | 1979 | 1981 | Dominica | St Barthelemy, St Martin, and Dominica | 9.712 × 10−4 |
| (1978–1980) | (7.944 × 10−4, 10.627 × 10−4) |
Note.—HPD—95% highest posterior density interval.
Bayesian Phylogeography
We summarized the results of the Bayesian phylogeographic analyses using maximum clade credibility (MCC) trees (supplementary fig. S1, Supplementary Material online), branches of which have been colored according to the most probable location of the nodes subtending them. As has been previously reported for DENV in this region (Foster et al. 2004; Carrington et al. 2005; Araujo et al. 2009), there is a strong signal of phylogenetic clustering according to sample location, reflected by clades comprising of isolates from either the mainland (South or Central America) or island (Caribbean) locations, with few exceptions. In all cases, the data are consistent with a single introduction of each genotype followed by rapid spatial dispersion within the Americas. From the estimated root state probability distribution, we infer that these introductions were mostly likely to Grenada (88%), Jamaica (66%), Nicaragua (67%), and Dominica (39%) for DENV-1 to DENV-4, respectively (data not shown). The countries that first reported each of these viruses were Jamaica (Pan American Health Organization 1979), Cuba (Rico-Hesse 1990), Nicaragua and Panama (Centers for Disease Control and Prevention 1995), and St Barthelemy, St Martin, and Dominica (Centers for Disease Control and Prevention 1981), respectively (table 2).
Interactive virtual globe animations that demonstrate the diffusion dynamics of DENV-1 to DENV-4 through time after each introduction are available online at http://www.phylogeography.org. These dispersal histories are summarized in figure 3, which shows snapshots of dispersal patterns among countries from 1985 to 2008. The results suggest that after DENV-1 genotype V was introduced into Grenada in the Lesser Antilles, the virus rapidly spread to South and Central America and then to the Greater Antilles. The Lesser Antilles remained the only source population from 1973 until 1983, after which movement was also observed from the Greater Antilles to other islands and to nearby mainland regions. From 1985, Brazil, Venezuela, Argentina, and then Colombia served as source populations to other mainland regions. Figure 4 shows only those links that are statistically well supported (BF ≥ 6). The strongest epidemiological links were between Argentina and Paraguay, Venezuela and Colombia, and Nicaragua and both Mexico and Belize. Grenada, the most probable root state location for DENV-1, is involved in eight well-supported links, mostly with other Caribbean islands. The only other country with as many links is Trinidad and to a lesser extent Brazil.
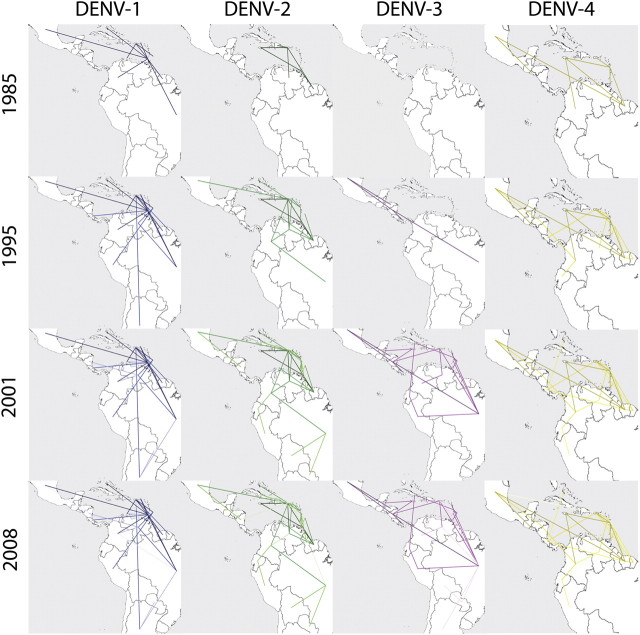
Spatiotemporal dynamics of four DENV serotypes circulating in the Americas represented with snapshots of the dispersal pattern for January 1985, 1995, 2001, and 2008. Lines represent MCC phylogeny branches projected on the surface. The intensity of the color indicates the relative age of the dispersal pattern (darker = older).
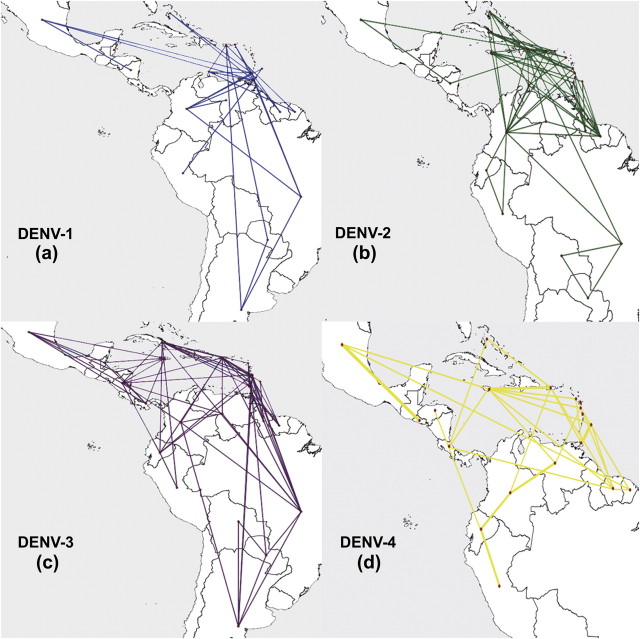
Significant nonzero rates of movement for DENV-1 to DENV-4 (labeled a–d, respectively) as identified by Bayesian stochastic search variable selection using a BF test. Only links with BF > 6 are shown.
In the case of DENV-2 Asian–American genotype, the results suggest that upon introduction to the Greater Antilles around 1979, this genotype spreads southward along the island chain to the Lesser Antilles and northeastern coast of South America. From 1984 onward, there was gene flow back and forth along the island chain and between the islands and South America. Dissemination within South America is estimated to have begun around 1992, and after 1995, Central America became involved as a result of gene flow from the islands. More statistically supported links were noted for DENV-2 than for any other serotype, and the countries with the most links were Colombia, Dominican Republic, and Suriname and to a lesser extent Barbados and Dominica.
The MCC phylogeny for DENV-3 genotype III suggests that there was a single introduction into the Americas through Nicaragua. Initially gene flow occurred northward and southward along the mainland. From 1994, Brazil operated as the major source population (evidenced by the existence of Brazilian isolates scattered throughout the tree). However, Central America (Nicaragua) was the source of initial gene flow to the islands (Greater Antilles), beginning in 1996. As was the case for DENV-2, the links with the strongest statistical support were observed within South America, Central America, and between mainland regions and the islands, where the strongest links were seen between Colombia and Venezuela and Paraguay with both Brazil and Bolivia. The highest number of links was seen from Martinique, Nicaragua, Brazil, Cuba, and Puerto Rico.
The first reports of DENV-4 genotype II in the Americas were from St Barthelemy and St Martin, and Dominica in 1981 (Centers for Disease Control and Prevention 1981), the latter of which was also the root location inferred by our analysis. The phylogeographic reconstruction suggests that following introduction DENV-4 was rapidly disseminated to both South and Central America. The BSP for DENV-4 (fig. 2) describes a second period of exponential growth beginning in 1989, and the phylogeographic reconstruction (fig. 3) suggests that Puerto Rico and Suriname the major source population for this second wave. The most significant links seen were between Barbados and Trinidad, French Guiana and Martinique, and Ecuador and Peru. The countries with the most links are Suriname, Jamaica, and Barbados.
Testing Spatial Diffusion Scenarios
In order to investigate the factors influencing rates of DENV gene flow in the Americas, we explored four different scenarios for rates of movement among the countries represented in the data sets. These included 1) equal rates of virus movement between all countries (serving as a reference model for comparison), 2) rates of virus movement that were inversely proportional to the distance between locations (cities and countries), and 3) a model which expects rates of movement between pairs of countries to be proportional to the number of airline seats available between them. Surprisingly, the factors dictating the diffusion of all the circulating genotypes appear to be different, as reflected by table 3. For the serotypes DENV-1 and DENV-2, the models with the distance-based rates performed better (had a higher marginal likelihood) than the airline-based model or the equal rates model in all cases. For these serotypes above, the model utilizing airline seat data did not perform as well as the distance models but did perform significantly better than the equal rates model. For the serotypes DENV-3 and DENV-4, the equal rates seemed to best describe diffusion in the region and significantly so for DENV-4. Definitive conclusions could not be made about DENV-1 and DENV-3, for when the different rates were compared for each serotype, the marginal likelihoods did not significantly differ from each other.
Table 3.
Marginal Likelihood Values of the Four Different Data-Informed Models of Virus Diffusion for Each DENV Serotype.
| Model (lnL) | ||||
| Serotype | Equal | GeoDistance | PopDistance | Air |
| DENV-1 | −8051.366 | −8050.789 | −8049.084 | −8049.143 |
| DENV-2 | −8830.731 | −8788.436 | −8790.572 | −8792.343 |
| DENV-3 | −7652.318 | −7652.818 | −7652.45 | −7653.351 |
| DENV-4 | −6391.16 | −6394.209 | −6404.984 | −6404.449 |
Discussion
In this study, we sought to elucidate the spatiotemporal dynamics of the four main circulating serotypes of DENV in the Americas. Our Bayesian analyses incorporated a more realistic relaxed clock model and utilized larger and more temporally and geographically diverse data sets than those used the previous study (Carrington et al. 2005). The inferred posterior mean substitution rates for the four serotypes ranged from 7.8 × 10−4 subs/site/year to 9.9 × 10−4 subs/site/year, which were comparable to those obtained in previous studies (Twiddy et al. 2003; Carrington et al. 2005; Araujo et al. 2009; Bennett et al. 2010).
Bayesian phylogeographic analyses of all four serotypes indicate that each arose from a single introduction prior to the first epidemiological reports of the virus in the region. For all DENV serotypes, initial growth in viral genetic diversity occurred shortly before the period when the virus isolation data suggest an increase in incidence of that genotype, an outcome observed previously on a more confined spatial scale (Bennett et al. 2010). Overall, the population genetic histories of DENV-1, DENV-2, and DENV-4 were similar, with an increase in genetic diversity upon introduction into the region, followed a maintenance phase where genetic diversity remains more of less constant with only gradual increases or decreases depending on the serotype. The maintenance of genetic diversity through time—despite significant epidemic troughs and shifts in relative prevalence—most likely results from the spatial population structure that is established when each genotype invades the region (Carrington et al. 2005). DENV-3 genotype III, the last genotype to be introduced into the region, had a different population genetic history, with no obvious maintenance phase. As this was the second genotype of DENV-3 to have circulated in the region, herd immunity may have limited the rate at which this genotype could expand in the population.
The epidemiological data used in this study were obtained from virus isolations from the English-speaking Caribbean and include a selection of symptomatic cases only. However, the pattern of fluctuating virus activity and changes in predominant serotypes described by these data concur with other census data from the Americas (San Martín et al. 2010), which indicate that DENV-1 and DENV-2 were the most prevalent circulating serotypes prior to 2002, after which there was a switch to predominantly DENV-2 and DENV-3, with DENV-4 detected in a low percentage of countries during this period. These results continue to emphasize the ability of coalescent-based models to recover information from genetic data in the absence of sufficient epidemiological data.
With respect to spatial epidemiology, our most probable estimates of ancestral locations matched the countries in which serotypes were first reported in two cases (DENV-3 and DENV-4) and identified a neighboring country in one case (DENV-2; first reported in Cuba and estimated to have been first introduced into Jamaica). In the case of DENV-1, Grenada was identified as the origin, although Jamaica was the country of first report. Although the latter are relatively far apart, they do share linguistic, historical, and thus strong socioeconomic links. In the Americas today, levels of surveillance vary considerably among countries and are very poor in less-developed countries. Furthermore, historical epidemiological data for some countries are nonexistent and incidence measures collected by some countries in the region are rough estimates, limited by the accuracy of diagnosis, number of suspected cases confirmed, and reporting rates to public health agencies. For DENV-1 to DENV-4, the MRCAs were estimated to have existed 2–4 years, depending on the serotype, before the first outbreaks of these serotypes were reported. This result is consistent with a scenario in which the virus remains undetected until number of infections or disease incidence reaches a detection threshold, which may be quite high due to poor surveillance in most countries in the Americas. Also, given the length of time between the first reports of outbreaks and the estimated MRCA, there is the possibility that virus could have moved to another country undetected.
The cocirculation of multiple serotypes and/or genotypes in a particular region may result in complex patterns of competition, leading to fluctuations in genotype genetic diversity. Bennett et al. (2010) investigated the epidemic history of DENV-4 in Puerto Rico from its introduction in 1981–1998. Estimates of DENV-4 relative genetic diversity in this study exhibited cyclical dynamics that closely followed epidemiological estimates of prevalence, reflecting a periodicity of transmission within the population (Bennett et al. 2010). This periodicity has also been noted in Thailand (Nisalak et al. 2003; Cummings et al. 2004) and Singapore (Ooi et al. 2006). However, within-country variation in relative genetic diversity (Mondini et al. 2009; Bennett et al. 2010) was not observed in our analysis, which is a broader region-wide study. Instead, as noted above, genetic diversity in our study is maintained despite apparent fluctuations in epidemic activity (as measured by the percentage of virus isolations by CAREC), a pattern previously described for DENV-2 and DENV-4 in the Americas (Carrington et al. 2005) by population subdivision among locations. Clearly, the phylodynamic behavior of DENV—as with other viruses—depends on the scale at which it is being studied (Pybus and Rambaut 2009). With additional sampling among the locations and over time, more fine-scale patterns may emerge.
One would generalize that the strongest supported migration events occur mainly between neighboring countries, but several countries (i.e., Barbados, Puerto Rico, Colombia, Suriname, Venezuela, and Brazil) had significantly more linkages than the others and as such possibly play the role of hubs for the dissemination of this virus on a seasonal basis. The possibility of other confounding factors dictating the diffusion of DENV (e.g., linguistic and/or socioeconomic relations) has been previously suggested (Carrington et al. 2005).
The ability to integrate useful geographic and epidemiological information with viral gene sequences offers new opportunities for hypothesis testing (Lemey et al. 2009). In our analyses, we compared four different scenarios in an attempt to explain the level of DENV gene flow within the Americas. Given the nature of the anthropophilic nature of the urban-dwelling vector species Ae. aegypti, the assumption one would make is that gene flow between adjacent or nearby countries is more likely than between distant locales, and as a result, the distance models would perform better. More specifically, that the PopDistance model (which considered distances between the closest and largest population centers) would be a better fit than the GeoDistance model. However, the distance models were preferred only for DENV-1 and DENV-2 and without statistical support in the case of DENV-1.
DENV can be disseminated by infected mosquitoes or infected humans. Ae. aegypti typically travel relatively short distances of about 100 m, although there are reports of individuals moving up to 800 m (Reiter et al. 1995; Harrington et al. 2005). In an attempt to distinguish between human and mosquito movement, we formalized an air-traffic model that incorporates airline seat capacity between each pair of countries. This model did not perform as well as the distance-informed models but was better than the equal rates model for DENV-1 and DENV-2. However, a major limitation here was the fact that the seat capacity data were from a single year, that is, 2007, whereas the genetic data spanned 32 years. With increasing air traffic and globalization, flight patterns will have changed considerably over this period. Also this analysis does not consider the role of other methods of long-distance travel in the dissemination of infected mosquitoes and humans between countries (e.g., shipping or travel by road on the mainland). Given the historical importance of shipping in the introduction of Ae. aegypti, DENV, and other arboviruses to the Americas (Soper 1967; Halstead 1992), it would be useful to investigate the role of shipping traffic in DENV dissemination on a hemispheric scale. However, for most of the Caribbean islands in our data set, sufficient shipping data were unavailable.
The marginal likelihood estimation for the majority of the serotypes indicated either that the equal rates model was the best fit or that no other model was a significantly better fit than the equal rates model. The harmonic mean approach was used for estimating the marginal likelihoods. Although this approach is currently one of the more common approaches, there are some major limitations, where it can lead to a strong overestimation of the marginal likelihood when more complex models are analyzed. To this end, we are investigating more effective methods of Bayesian model comparison, along with the development of models that allow estimating and testing different contributions of these predictors simultaneously.
Airline traffic is an important parameter that deserves further investigation, as the air transport network has played a key role in the global dissemination of other acute infections, such as influenza and severe acute respiratory syndrome (SARS) (Colizza et al. 2006; Baker et al. 2010). Although it would not be surprising to find that air traffic is important in the case of DENV, this approach warrants development as it could be applied to viruses that are less well understood in terms of the nature of their transmission cycles. Further exploration of the historical role of human movement versus mosquito movement within the Americas, and differences between the Americas and Southeast Asia, is essential in gaining better insights into DENV transmission patterns and evolutionary dynamics.
Supplementary Material
Supplementary tables S1 and S2 and figure S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grants from the Caribbean Health Research Council to C.V.F.C. O.M.A. is a member of the Tropical Medicine Cluster (University of the West Indies) and was supported by grants and scholarships from the University of the West Indies, St Augustine Campus Research and Publications Fund, and the Commonwealth Scholarship Commission. P.L., M.A.S., and A.R. acknowledge support from the European Research Council (ERC) under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement no. 260864. S.B. and B.M. acknowledge support from NIH-RR018727; COBRE Bioinformatics Facility, NIH-AI065359;, NIH-RR003061, DOD-06187000;, and NSF-IGERT 0549514. A.J.T. acknowledges support from the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. The authors would like to acknowledge the CAREC for providing the viral isolates included in the study, and the National Institute of Allergy and Infectious Diseases-funded Genomic Sequencing Center at the Broad Institute, for permission to use sequences generated under Dengue Virus Portal.
References
- Adams B, Holmes EC, Zhang C, Mammen MP, Jr, Nimmannitya S, Kalayanarooj S, Boots M. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc Natl Acad Sci U S A. 2006;103(38):14234–14239. [Europe PMC free article] [Abstract] [Google Scholar]
- Araujo JM, Nogueira RM, Schatzmayr HG, Zanotto PM, Bello G. Phylogeography and evolutionary history of dengue virus type 3. Infect Genet Evol. 2009;9(4):716–725. [Abstract] [Google Scholar]
- Auguste AJ, Lemey P, Pybus OG, Suchard MA, Salas RA, Adesiyun AA, Barrett AD, Tesh RB, Weaver SC, Carrington CVF. Yellow fever virus maintenance in Trinidad and its dispersal throughout the Americas. J Virol. 2010;84(19):9967. [Europe PMC free article] [Abstract] [Google Scholar]
- Baker MG, Thornley CN, Mills C, Roberts S, Perera S, Peters J, Kelso A, Barr I, Wilson N. Transmission of pandemic A/H1N1 2009 influenza on passenger aircraft: retrospective cohort study. BMJ. 2010;340:c2424. [Europe PMC free article] [Abstract] [Google Scholar]
- Bennett SN, Drummond AJ, Kapan DD, Suchard MA, Munoz-Jordan JL, Pybus OG, Holmes EC, Gubler DJ. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2010;27(4):811–818. [Europe PMC free article] [Abstract] [Google Scholar]
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20(10):1650–1658. [Abstract] [Google Scholar]
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol. 2006;87(Pt 4):885–893. [Abstract] [Google Scholar]
- Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79(23):14680–14687. [Europe PMC free article] [Abstract] [Google Scholar]
- Centers for Disease Control and Prevention. Dengue type 4 infections in U.S. travelers to the Caribbean. MMWR Morb Mortal Wkly Rep. 1981;30(21):249–250. [Abstract] [Google Scholar]
- Centers for Disease Control and Prevention. Dengue type 3 infection—Nicaragua and Panama, October-November 1994. MMWR Morb Mortal Wkly Rep. 1995;44(2):21–24. [Abstract] [Google Scholar]
- Christophers S. Aedes aegypti (L.) the yellow fever mosquito: its life history, bionomics and structure. London: Cambridge University Press; 1960. [Google Scholar]
- Colizza V, Barrat A, Barthelemy M, Vespignani A. The role of the airline transportation network in the prediction and predictability of global epidemics. Proc Natl Acad Sci U S A. 2006;103(7):2015–2020. [Europe PMC free article] [Abstract] [Google Scholar]
- Cummings DAT, Irizarry RA, Huang NE, Endy TP, Nisalak A, Ungchusak K, Burke DS. Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature. 2004;427(6972):344–347. [Abstract] [Google Scholar]
- Diallo M, Sall AA, Moncayo AC, Ba Y, Fernandez Z, Ortiz D, Coffey LL, Mathiot C, Tesh RB, Weaver SC. Potential role of sylvatic and domestic African mosquito species in dengue emergence. Am J Trop Med Hyg. 2005;73(2):445–449. [Abstract] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. [Europe PMC free article] [Abstract] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. [Europe PMC free article] [Abstract] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22(5):1185–1192. [Abstract] [Google Scholar]
- Focks DA, Barrera R. Report of the Scientific Working Group Meeting on Dengue. Geneva (Switzerland): WHO; Dengue transmission dynamics: assessment and implications for control; pp. 92–108. [Google Scholar]
- Foster JE, Bennett SN, Carrington CV, Vaughan H, McMillan WO. Phylogeography and molecular evolution of dengue 2 in the Caribbean basin, 1981-2000. Virology. 2004;324(1):48–59. [Abstract] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. In: Gubler DJ, Kuno G, editors. Dengue and dengue hemorrhagic fever. London: CAB International; 1997. pp. 1–22. [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3–16. ; discussion 16–22, 71–3, 251–3. [Abstract] [Google Scholar]
- Guzman MG, Halstead SB, Artsob H, et al. (11 co-authors) Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8(Suppl 12):S7–S16. [Abstract] [Google Scholar]
- Halstead SB. The XXth century dengue pandemic: need for surveillance and research. World Health Stat Q. 1992;45(2–3):292. [Abstract] [Google Scholar]
- Halstead SB. Dengue virus-mosquito interactions. Annu Rev Entomol. 2008;53:273–291. [Abstract] [Google Scholar]
- Harrington LC, Scott TW, Lerdthusnee K, et al. (11 co-authors) Dispersal of the dengue vector Aedes aegypti within and between rural communities. Am J Trop Med Hyg. 2005;72(2):209–220. [Abstract] [Google Scholar]
- Kroeger A, Nathan MB. Dengue: setting the global research agenda. Lancet. 2006;368(9554):2193. [Abstract] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5(9):e1000520. [Europe PMC free article] [Abstract] [Google Scholar]
- Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21(2):260. [Abstract] [Google Scholar]
- Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 2008;25(7):1459. [Europe PMC free article] [Abstract] [Google Scholar]
- Mondini A, de Moraes Bronzoni RV, Nunes SHP, Chiaravalloti Neto F, Massad E. Spatio-temporal tracking and phylodynamics of an urban dengue 3 outbreak in Sao Paulo, Brazil. PLoS Negl Trop Dis. 2009;3:e448. [Europe PMC free article] [Abstract] [Google Scholar]
- Newton MA, Raftery AE. Approximate Bayesian inference with the weighted likelihood bootstrap. J R Stat Soc B Met. 1994;56(1):3–48. [Google Scholar]
- Nisalak A, Endy TP, Nimmannitya S, Kalayanarooj S, Thisayakorn U, Scott RM, Burke DS, Hoke CH, Innis BL, Vaughn DW. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am J Trop Med Hyg. 2003;68(2):191–202. [Abstract] [Google Scholar]
- Ooi EE, Goh KT, Gubler DJ. Dengue prevention and 35 years of vector control in Singapore. Emerg Infect Dis. 2006;12(6):887–893. [Europe PMC free article] [Abstract] [Google Scholar]
- Pan American Health Organization. Proceedings of a workshop held in Montego Bay, Jamaica, 8–11 May, 1978. PAHO scientific publication 375. Washington (DC): Pan American Health Organization; 1979. Dengue in the Caribbean, 1977. [Google Scholar]
- Pond K, Sergei L, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208. [Abstract] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21(10):2531–2533. [Abstract] [Google Scholar]
- Powell JR, Tabachnick WJ, Arnold J. Genetics and the origin of a vector population: Aedes aegypti, a case study. Science. 1980;208(4450):1385–1387. [Abstract] [Google Scholar]
- Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10(8):540–550. [Abstract] [Google Scholar]
- Redelings BD, Suchard MA. Joint Bayesian estimation of alignment and phylogeny. Syst Biol. 2005;54(3):401. [Abstract] [Google Scholar]
- Reiter P, Amador MA, Anderson RA, Clark GG. Short report: dispersal of Aedes aegypti in an urban area after blood feeding as demonstrated by rubidium-marked eggs. Am J Trop Med Hyg. 1995;52(2):177–179. [Abstract] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174(2):479–493. [Abstract] [Google Scholar]
- Rush AB. Medical inquiries and observations. Philadelphia (PA): Prichard and Hall; 1789. An account of the bilious remitting fever, as it appeared in Philadelphia in the summer and autumn of the year 1780; pp. 89–100. [Google Scholar]
- San Martín JL, Brathwaite O, Zambrano B, Solorzano JO, Bouckenooghe A, Dayan GH, Guzman MG. The epidemiology of dengue in the americas over the last three decades: a worrisome reality. Am J Trop Med Hyg. 2010;82(1):128–135. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith CE. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. J Trop Med Hyg. 1956;59(10):243–251. [Abstract] [Google Scholar]
- Soper FL. Aedes aegypti and yellow fever. Bull World Health Organ. 1967;36(4):521–527. [Europe PMC free article] [Abstract] [Google Scholar]
- Suchard MA, Kitchen CMR, Sinsheimer JS, Weiss RE. Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol. 2003;52(5):649. [Abstract] [Google Scholar]
- Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18(6):1001. [Abstract] [Google Scholar]
- Talbi C, Lemey P, Suchard MA, et al. (11 co-authors) Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog. 2010;6(10):e1001166. [Europe PMC free article] [Abstract] [Google Scholar]
- Tesh RB. A method for the isolation and identification of dengue viruses, using mosquito cell cultures. Am J Trop Med Hyg. 1979;28(6):1053–1059. [Abstract] [Google Scholar]
- Twiddy SS, Holmes EC, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol. 2003;20(1):122–129. [Abstract] [Google Scholar]
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Weaver SC. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74(7):3227–3234. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang C, Mammen MP, Jr, Chinnawirotpisan P, Klungthong C, Rodpradit P, Monkongdee P, Nimmannitya S, Kalayanarooj S, Holmes EC. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol. 2005;79(24):15123–15130. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Molecular Biology and Evolution are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/molbev/msr320
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/mbe/article-pdf/29/6/1533/13646693/msr320.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Dengue Outbreak Caused by Multiple Virus Serotypes and Lineages, Colombia, 2023-2024.
Emerg Infect Dis, 30(11):2391-2395, 08 Oct 2024
Cited by: 0 articles | PMID: 39378873 | PMCID: PMC11521178
Re-Emergence of DENV-3 in French Guiana: Retrospective Analysis of Cases That Circulated in the French Territories of the Americas from the 2000s to the 2023-2024 Outbreak.
Viruses, 16(8):1298, 14 Aug 2024
Cited by: 0 articles | PMID: 39205272 | PMCID: PMC11360160
Age-specific case data reveal varying dengue transmission intensity in US states and territories.
PLoS Negl Trop Dis, 18(3):e0011143, 01 Mar 2024
Cited by: 0 articles | PMID: 38427702
Detection of a Multiple Circulation Event of Dengue Virus 2 Strains in the Northern Region of Brazil.
Trop Med Infect Dis, 9(1):17, 09 Jan 2024
Cited by: 0 articles | PMID: 38251214 | PMCID: PMC10818346
Computational Advancement towards the Identification of Natural Inhibitors for Dengue Virus: A Brief Review.
Comb Chem High Throughput Screen, 27(17):2464-2484, 01 Jan 2024
Cited by: 0 articles | PMID: 37859315
Review
Go to all (82) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 42 of 42)
- (2 citations) ENA - JN379472
- (1 citation) ENA - JN379479
- (1 citation) ENA - JN379512
- (1 citation) ENA - JN379478
- (1 citation) ENA - JN379511
- (1 citation) ENA - JN379475
- (1 citation) ENA - JN379497
- (1 citation) ENA - JN379474
- (1 citation) ENA - JN379496
- (1 citation) ENA - JN379477
- (1 citation) ENA - JN379499
- (1 citation) ENA - JN379510
- (1 citation) ENA - JN379476
- (1 citation) ENA - JN379498
- (1 citation) ENA - JN379482
- (1 citation) ENA - JN379481
- (1 citation) ENA - JN379484
- (1 citation) ENA - JN379483
- (1 citation) ENA - JN379480
- (1 citation) ENA - JN379501
- (1 citation) ENA - JN379489
- (1 citation) ENA - JN379500
- (1 citation) ENA - JN379503
- (1 citation) ENA - JN379502
- (1 citation) ENA - JN379486
- (1 citation) ENA - JN379485
- (1 citation) ENA - JN379488
- (1 citation) ENA - JN379487
- (1 citation) ENA - JN379509
- (1 citation) ENA - JN379508
- (1 citation) ENA - JN379505
- (1 citation) ENA - JN379504
- (1 citation) ENA - JN379507
- (1 citation) ENA - JN379506
- (1 citation) ENA - JN379471
- (1 citation) ENA - JN379493
- (1 citation) ENA - JN379470
- (1 citation) ENA - JN379492
- (1 citation) ENA - JN379495
- (1 citation) ENA - JN379494
- (1 citation) ENA - JN379491
- (1 citation) ENA - JN379490
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evolutionary history and spatiotemporal dynamics of DENV-1 genotype V in the Americas.
Infect Genet Evol, 45:454-460, 03 Oct 2016
Cited by: 23 articles | PMID: 27713055
Worldwide spread of Dengue virus type 1.
PLoS One, 8(5):e62649, 13 May 2013
Cited by: 54 articles | PMID: 23675416 | PMCID: PMC3652851
Spatiotemporal dynamics of DENV-2 Asian-American genotype lineages in the Americas.
PLoS One, 9(6):e98519, 04 Jun 2014
Cited by: 21 articles | PMID: 24897118 | PMCID: PMC4045713
Origin, tempo, and mode of the spread of DENV-4 Genotype IIB across the state of São Paulo, Brazil during the 2012-2013 outbreak.
Mem Inst Oswaldo Cruz, 114:e180251, 07 Jan 2019
Cited by: 9 articles | PMID: 30624458 | PMCID: PMC6333047
Review Free full text in Europe PMC
Funding
Funders who supported this work.
European Commission FP7 (1)
Grant ID: FP7_260864
European Research Council (1)
Evolutionary reconstruction of viral spread in time and space (ViralPhylogeography)
Prof Philippe LEMEY, University of Leuven
Grant ID: 260864
NCRR NIH HHS (4)
Grant ID: RR018727
Grant ID: P20 RR018727
Grant ID: G12 RR003061
Grant ID: RR003061
NIAID NIH HHS (2)
Grant ID: AI065359
Grant ID: U54 AI065359
NIGMS NIH HHS (1)
Grant ID: P20 GM103516
Wellcome Trust (1)
Centre for Immunity, Infection and Evolution.
Prof Keith Matthews, University of Edinburgh
Grant ID: 095831





