Abstract
Free full text

Inaugural Article
Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity
Associated Data
Abstract
Staufen (STAU)1-mediated mRNA decay (SMD) is a posttranscriptional regulatory mechanism in mammals that degrades mRNAs harboring a STAU1-binding site (SBS) in their 3′-untranslated regions (3′ UTRs). We show that SMD involves not only STAU1 but also its paralog STAU2. STAU2, like STAU1, is a double-stranded RNA-binding protein that interacts directly with the ATP-dependent RNA helicase up-frameshift 1 (UPF1) to reduce the half-life of SMD targets that form an SBS by either intramolecular or intermolecular base-pairing. Compared with STAU1, STAU2 binds ~10-fold more UPF1 and ~two- to fivefold more of those SBS-containing mRNAs that were tested, and it comparably promotes UPF1 helicase activity, which is critical for SMD. STAU1- or STAU2-mediated augmentation of UPF1 helicase activity is not accompanied by enhanced ATP hydrolysis but does depend on ATP binding and a basal level of UPF1 ATPase activity. Studies of STAU2 demonstrate it changes the conformation of RNA-bound UPF1. These findings, and evidence for STAU1−STAU1, STAU2−STAU2, and STAU1−STAU2 formation in vitro and in cells, are consistent with results from tethering assays: the decrease in mRNA abundance brought about by tethering siRNA-resistant STAU2 or STAU1 to an mRNA 3′ UTR is inhibited by downregulating the abundance of cellular STAU2, STAU1, or UPF1. It follows that the efficiency of SMD in different cell types reflects the cumulative abundance of STAU1 and STAU2. We propose that STAU paralogs contribute to SMD by “greasing the wheels” of RNA-bound UPF1 so as to enhance its unwinding capacity per molecule of ATP hydrolyzed.
Mammalian cells contain two Staufen (STAU) paralogs, STAU1 and STAU2. Each derives from a separate gene that produces multiple protein isoforms via alternative pre-mRNA splicing and/or polyadenylation. Every STAU1 and STAU2 isoform contains multiple conserved dsRNA-binding domains (RBDs), of which dsRBD3 and dsRBD4 constitute the major if not sole active dsRNA-binding sites (1, 2).
Although STAU1 and probably STAU2 are expressed ubiquitously, STAU2 is most abundant in heart and brain (1, 3). Both paralogs are present in ribonucleoprotein particles (RNPs) within neuronal cell bodies and dendrites (1, 4–7). Both are also involved in the microtubule-dependent transport of RNAs to dendrites of polarized neurons (5, 8), presumably via the tubulin-binding domain (2). Moreover, both paralogs play a crucial role in the formation and maintenance of the dendritic spines of hippocampal neurons and are required for synaptic plasticity and memory (7, 9–11), although the detection of paralog-specific RNA granules in the distal dendrites of rat hippocampal neurons has been used to argue for paralog-distinct functions (1). Additional support for paralog-specific roles derives from the demonstration using rat hippocampal slice cultures that STAU1 but not STAU2 functions in the late phase of forskolin-induced long-term potentiation (10–12), whereas STAU2 but not STAU1 is involved in metabotropic glutamate receptor-mediated long-term depression (12). As another example of apparent paralog specificity, we have shown that STAU1 degrades mRNAs that harbor a STAU1-binding site (SBS) downstream of their normal termination codon in a pathway called STAU1-mediated mRNA decay or SMD (13, 14), and work published by others indicates that SMD does not involve STAU2 (3, 15).
According to our current model for SMD, when translation terminates upstream of an SBS, recruitment of the nonsense-mediated mRNA decay (NMD) factor UPF1 to SBS-bound STAU1 triggers mRNA decay. SMD influences a number of cellular processes, including the differentiation of mouse C2C12 myoblasts to myotubes (16), the motility of human HaCaT keratinocytes (17), and the differentiation of mouse 3T3-L1 preadipocytes to adipocytes (18). Thus, SMD may explain the essential role of STAU1 in embryonic stem-cell differentiation (19).
We report here that SMD is triggered not only by STAU1 but also by STAU2, each of which can self-associate as well as associate with one another. Our findings that SMD requires the ATP-dependent helicase activity of UPF1, each STAU paralog promotes UPF1 helicase activity without promoting UPF1 ATP hydrolytic activity, and STAU2 increases the RNA footprint of UPF1 allow us to expand on the existing model for SMD. We propose that STAU paralog-binding to UPF1 induces a change in UPF1 conformation that resembles its conformation during intermediate or product steps of ATP hydrolysis and enhances the amount of unwinding per ATP molecule hydrolyzed.
Results
STAU2 Functions in SMD.
Human embryonic kidney (HEK) 293T and HeLa cells express multiple isoforms of STAU2 and STAU1 (1, 13, 20) (Fig. 1A and Fig. S1A). STAU2 and STAU1 are ~50% identical (20). dsRBDs 3 and 4 have retained the ability to bind dsRNA and are 78% and 81% identical, respectively (2). dsRBD2, which is 48% identical between the two paralogs, and dsRBD5, the full-length of which is unique to STAU1 (21) (Fig. 1A), have been implicated in the formation of STAU1 homodimers and homomultimers in complex with RNA (22).

siRNA-mediated down-regulation of STAU2 inhibits SMD. (A) Diagrams of human STAU1 and STAU2 isoforms, where boxes represent functional and/or structural domains. 1, N-terminal amino acid; TBD, tubulin-binding domain (where the hatched STAU2 TBD is ~18% identical to the STAU1 TBD). (B) Partial diagrams of pcFLUC SMD-reporter plasmids. (C and D) HeLa cells (4 × 106 per 100-mm dish) were transiently transfected with 1.5 μg of each pcFLUC SMD-reporter plasmid and 1 μg of the phCMV-MUP reference plasmid. Cells were retransfected 1 d later with 100 nM of Control siRNA, STAU1 siRNA, or STAU2 siRNA and harvested 2 d later. (C) WB, where Calnexin controlled for variations in protein loading. The left-most four lanes show serial threefold dilutions of protein. Arrows pointing right denote STAU isoforms, and unidentified bands do not interfere with the analyses. (D) RT-PCR, where the level of each FLUC mRNA was normalized to the level of MUP mRNA, the level of cellular GAP43 mRNA was normalized to the level of its pre-mRNA, and normalized levels in the presence of Control siRNA were defined as 100. All results are representative of at least three independently performed experiments.
To assay for STAU2 function in SMD, STAU2 and, for comparison, STAU1 were down-regulated to the same extent in HeLa cells, where their abundance is comparable (Fig. S1 B and C). Subsequently, the levels of SMD reporters (normalized to the level of a reference mRNA to control for variations in cell transfection) and a cellular SMD target (normalized to the level of its pre-mRNA to control for effects on transcription) were quantitated. Thus, HeLa cells were transiently transfected with the phCMV-MUP [which produces major urinary protein (MUP) mRNA driven by a cytomegalovirus (CMV) promoter] reference plasmid and a mixture of the five pcFLUC SMD-reporter plasmids, which encode firefly luciferase (FLUC) mRNAs and vary in their 3′ UTRs (Fig. 1B) (13, 14, 17). The 3′ UTRs consist of the growth-associated protein 43 (GAP43) 3′ UTR, the serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (SERPINE1) 3′ UTR, the v-jun avian sarcoma virus 17 oncogene homolog (c-JUN) SBS (nucleotides 485–675 of the c-JUN 3′ UTR), the ADP-ribosylation factor 1 (ARF1) SBS (nucleotides 1–300 of the ARF1 3′UTR), or the ankyrin-repeat domain 57 (FLJ21870, also called ANKRD57) 3′ UTR. The ARF1 SBS and probably the GAP43 and c-JUN SBSs are formed by intramolecular base-pairing, whereas the SERPINE1 and FLJ21870 SBSs are formed by intermolecular base-pairing with a long-noncoding RNA (1/2-sbsRNA1) via partially complementary Alu elements (17). One day later, cells were transiently transfected with Control, STAU1, or STAU2 siRNA.
Western blotting (WB) demonstrated that STAU1 and STAU2 siRNAs specifically reduced the levels of their target isoforms to <10% the levels in Control siRNA-treated cells (Fig. 1C). RT-PCR revealed that STAU1 and STAU2 siRNAs augmented the level of each SMD reporter mRNA ~two- to threefold, indicating that each paralog functions in SMD to a comparable extent [Fig. 1D; corroborating RT–quantitative PCR (qPCR) data in Fig. S1D and data obtained using STAU2(A) or STAU2(B) siRNA in Fig. S1 E and F]. Consistent with STAU2 function in SMD, STAU2 siRNA, like STAU1 siRNA, up-regulated the level of endogenous GAP43 mRNA (Fig. 1D and Fig. S1F) and reduced the half-life of mRNA harboring a 3′ UTR SBS (Fig. S1 G–I).
STAU2, Like STAU1, Interacts Directly with UPF1.
To begin to assess STAU2 isoforms for function in SMD, extracts of HeLa cells that stably express FLAG-UPF1 at ≤twofold the level of cellular UPF1 (23) were immunoprecipitated in the presence or absence of RNase A using anti-FLAG or, as a control for nonspecific immunoprecipitation (IP), mouse (m)IgG. Approximately 10% of each isoform of STAU1 or STAU2 coimmunoprecipitated with FLAG-UPF1 (Fig. 2A). Furthermore, compared with the STAU1 isoforms (see also ref. 18), the co-IP efficiency of each STAU2 isoform was ~10-fold more resistant to RNase A (Fig. 2A). As indications that RNase A treatment was successful, the co-IP of UPF2 with FLAG-UPF1 was partially sensitive to RNase A (Fig. 2A) (24), the co-IP of poly(A)-binding protein C1 (PABPC1) was completely sensitive to RNase A (Fig. 2A) (24), and cellular suppressor with morphogenetic effect on genitalia protein 7 (SMG7) mRNA was detectable in the absence but not the presence of RNase A (Fig. 2A). IPs of cellular UPF1 using lysates from formaldehyde cross-linked human neuroblastoma SK-N-MC cells demonstrated that STAU1 and STAU2 isoforms also coimmunoprecipitate with cellular UPF1, indicating that the interactions occur within cells rather than as an artifact after cell lysis (Fig. S2A).
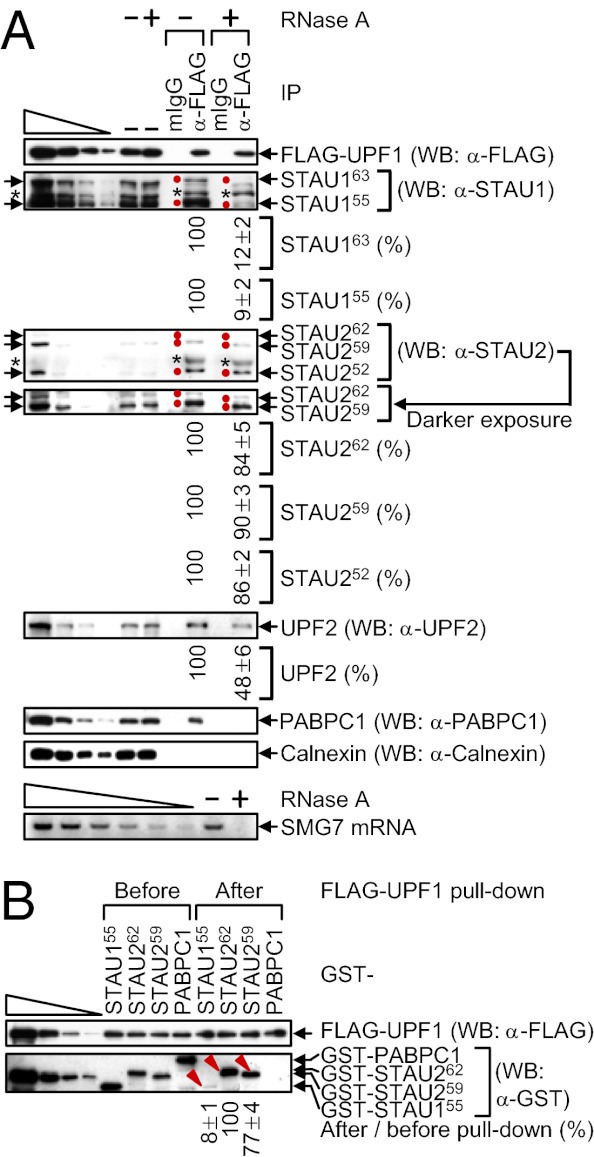
Compared with STAU1 isoforms, STAU2 isoforms interact more efficiently with UPF1. (A) HeLa cells stably expressing FLAG-UPF1 (8 × 107 per 150-mm dish) were lysed and analyzed before (−) or after IP using anti(α)-FLAG or, as a negative control, mouse (m)IgG in the absence (−) or presence (+) of RNase A. WB was performed, where the left-most four lanes show threefold dilutions of protein before IP. Arrows pointing right and red dots denote STAU isoforms, and the asterisks specify unidentified proteins. The level of each coimmunoprecipitated STAU protein in the absence of RNase A was defined as 100. (A, Lower) SMG7 mRNA was analyzed using RT-PCR before IP to demonstrate that the RNase A digestion was complete. The left-most six lanes represent twofold dilutions of RNA. (B) GST-STAU155, GST-STAU262, GST-STAU259, or GST-PABPC1 purified from E. coli (100 nM) was incubated with baculovirus-produced and purified FLAG-UPF1 (200 nM), subjected to FLAG-UPF1 pull-down, and analyzed using WB. Protein levels after pull-down were normalized to the level before pull-down, and the normalized level of GST-STAU262 was defined as 100. The left-most four lanes show threefold dilutions of protein. Red arrowheads denote the GST-STAU protein under analysis (Fig. S2B).
To examine whether the interaction of STAU2 with UPF1 is direct, and to determine whether the higher association of UPF1 with STAU2 relative to STAU1 is recapitulated in vitro, GST-STAU155 (13), GST-STAU262, GST-STAU259 or, as a negative control, GST-PABPC1 was produced and purified from Escherichia coli, mixed with baculovirus-produced and purified FLAG-UPF1 (25), and subjected to FLAG-UPF1 pull-down. Of the STAU2 isoforms, we focused on STAU262 and STAU259 because their binding to mRNA has been characterized using a genome-wide approach (20). FLAG-UPF1 pulled down ~10-fold more GST-STAU262 and GST-STAU259 compared with GST-STAU155 (Fig. 2B and Fig. S2B). As expected, FLAG-UPF1 failed to pull down GST-PABPC1 (Fig. 2B). These results are consistent with the finding that the co-IP of STAU2 with FLAG-UPF1 is less sensitive to RNase A than the co-IP of STAU1 with FLAG-UPF1.
STAU2 and STAU1 Paralogs Self-Associate and Associate with One Another.
STAU1 has been found to dimerize if not multimerize with itself in vitro and in cells (22). To better define STAU2 interactions in vitro, pull-downs using E. coli-produced proteins (Fig. S2C) were performed. GST-STAU155 pulled down not only HIS-STAU155 but also STAU262-HIS and STAU259-HIS with comparable efficiencies and, as expected, did not pull down BSA (Fig. 3A). Furthermore, STAU262-HIS and STAU259-HIS pulled down a comparable amount of GST-STAU155, STAU262-GST, or STAU259-GST and, as expected, did not pull down GST-PABPC1 (Fig. 3 B and C) (26). These findings indicate that the efficiencies with which each STAU2 isoform interacts with itself, with one another, or with STAU155 are indistinguishable in vitro (Fig. S2D).
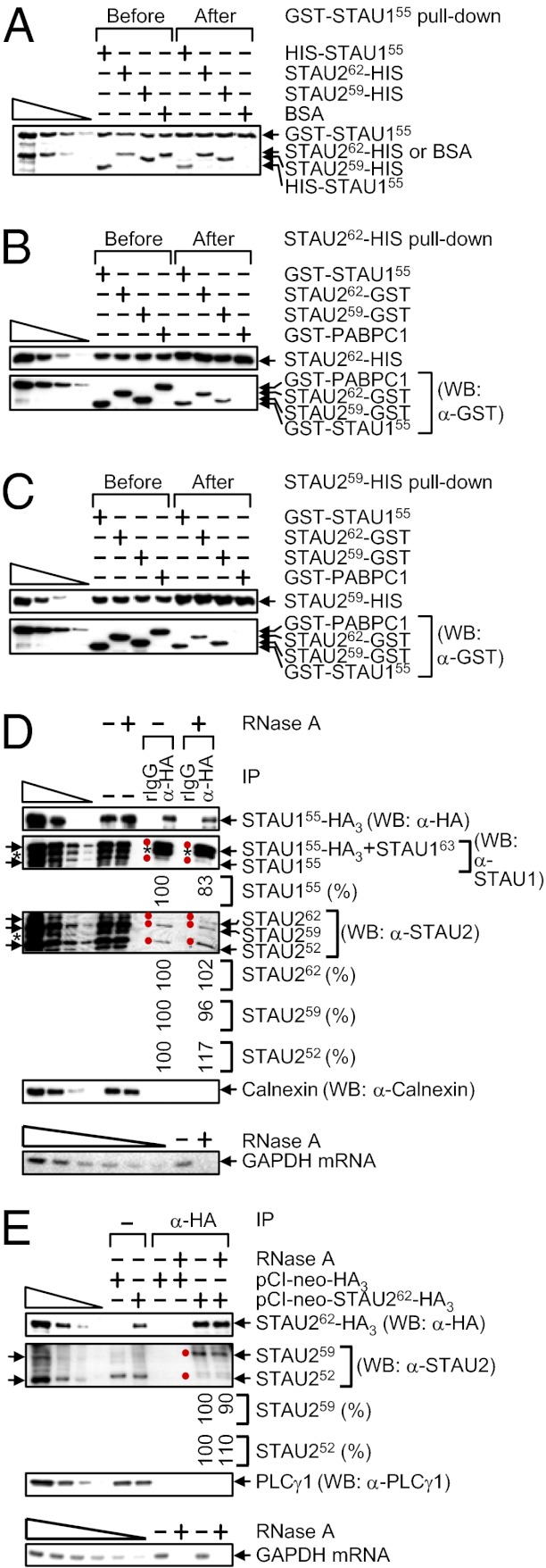
STAU262 and STAU259 self-associate and associate with one another and with STAU155. (A) GST-STAU155 (100 nM) was incubated with 100 nM of HIS-STAU155, STAU262-HIS, STAU259-HIS or, as a negative control, BSA, subjected to GST-STAU155 pull-down, resolved in SDS-polyacrylamide, and stained using SYPRO Ruby. The left-most four lanes are threefold dilutions of GST-STAU155 and BSA. (B and C) WB before and after HIS pull-down of 100 nM of STAU262-HIS (B) or STAU259-HIS (C) that was incubated with 100 nM of GST-STAU155, STAU262-GST, STAU259-GST or, as a negative control, GST-PABPC1. (D) HEK293T cells (4 × 107 per 150-mm dish) were transiently transfected with 10 μg of pSTAU155-HA3, lysed, and analyzed before (−) or after IP using anti(α)-HA or, as a negative control, rat (r)IgG in the absence (−) or presence (+) of RNase A. The level of each coimmunoprecipitated STAU protein in the absence of RNase A was defined as 100. Arrows pointing left and red dots denote STAU isoforms, and the asterisks specify unidentified proteins. Calnexin controlled for variations in protein loading before IP and demonstrates the specificity of the IP. (D, Lower) GAPDH mRNA was analyzed using RT-PCR before IP to demonstrate that the RNase A digestion was complete. The six left-most lanes represent twofold serial dilutions of RNA. (E) HEK293T cells (4 × 107 per 150-mm dish) were transiently transfected with 5 μg of pCI-neo-HA3 or pCI-neo-STAU262-HA3, lysed, and analyzed before or after IP using α-HA in the absence or presence of RNase A. The level of each coimmunoprecipitated STAU1 isoform in the absence of RNase A was defined as 100. Notations, serial dilutions, and GAPDH mRNA analyses were as in D.
To assay for STAU1 and STAU2 interactions in vivo, HEK293T cells were transiently transfected with a STAU155-HA3 expression vector (13, 14; where it was called pSTAU1-HA) to test for the ability of STAU155-HA3 to interact with cellular STAU1 and STAU2. STAU155-HA3 indeed coimmunoprecipitated with cellular STAU155, STAU262, STAU259, and STAU252 in the absence or presence of RNase A (Fig. 3D; it was not possible to assay cellular STAU163 because it comigrates with STAU155-HA3). Control experiments demonstrated that none of the STAU isoforms was immunoprecipitated using rat (r)IgG, Calnexin failed to coimmunoprecipitate under any circumstance, and cellular GAPDH mRNA was detectable in the absence but not the presence of RNase A (Fig. 3D). In related experiments, HEK293T cells were transiently transfected with a STAU262-HA3 expression vector. STAU262-HA3 also coimmunoprecipitated with cellular STAU259 and STAU252 in the absence or presence of RNase A (Fig. 3E; it was not possible to assay cellular STAU262 because it comigrates with STAU262-HA3). Control experiments showed that none of the STAU isoforms was immunoprecipitated in mock transfections involving an HA3 expression vector, PLCγ1 failed to coimmunoprecipitate under any circumstance, and cellular GAPDH mRNA was detectable in the absence but not the presence of RNase A (Fig. 3E).
STAU2, Like STAU1, Coimmunoprecipitates with SBSs.
STAU1 and STAU2 have been reported to bind distinct but overlapping subsets of mRNAs in HEK293T cells (20). Considering our findings that STAU2 and STAU1 interact directly (Fig. 3) and all STAU2 isoforms associate with polysomes (Fig. S2E), we next determined whether STAU2 coimmunoprecipitates with RNA sequences that are known to bind STAU1 and trigger SMD (13, 14, 16).
HeLa cells were transiently transfected with a pCI-neo-STAU155-HA3, pCI-neo-STAU262-HA3, or pCI-neo-STAU259-HA3 effector plasmid together with four of the pcFLUC SMD-reporter plasmids (Fig. 1B). Binding studies were performed using HeLa cells because, as noted above, they contain comparable amounts of endogenous STAU1 and STAU2 paralogs (Fig. S1 B and C), and comparable levels of STAU155-HA3, STAU262-HA3, and STAU259-HA3 can be expressed. The pcFLUC SMD-reporter plasmids served as positive controls for STAU155 binding. Two days after transfection, cells were exposed to formaldehyde, and lysates were immunoprecipitated using anti-HA.
STAU155-HA3, STAU262-HA3, and STAU259-HA3 were expressed at comparable levels before IP and immunoprecipitated with comparable efficiencies (Fig. 4A). STAU262-HA3 and STAU259-HA3 coimmunoprecipitate with FLUC SMD-reporter mRNAs ~two- to fivefold more efficiently than does STAU155-HA3 (Fig. 4 B and C). Because the efficiency of paralog self-association and association with one another is comparable, and the co-IP of STAU155-HA3 with the reporter mRNAs reflects binding to the SBSs, we conclude that STAU262 and STAU259 may bind other if not all SBSs more efficiently than does STAU155.

STAU262-HA3 and STAU259-HA3 coimmunoprecipitate with known SBS-containing targets more efficiently than does STAU155-HA3. HeLa cells (3 × 107 per 150-mm dish) were transiently transfected with 2 μg of each pcFLUC SMD-reporter plasmid, and 5 μg of pCI-neo-HA3, pCI-neo-STAU155-HA3, pCI-neo-STAU262-HA3, or pCI-neo-STAU259-HA3 effector plasmid. Two days later, lysates from formaldehyde-treated cells were immunoprecipitated using anti-HA, and cross-links were reversed. (A) WB of cell lysates after IP relative to before (−) IP using the specified antibody (α). PLCγ1 controlled for variations in protein loading before IP and demonstrates the specificity of the IP. The four left-most lanes show threefold dilutions of lysates from cells that expressed STAU259-HA3. (B) RT-qPCR of FLUC mRNAs. For samples before IP, the level of each FLUC mRNA was normalized to the level of GAPDH mRNA, and the normalized level of FLUC mRNA from cells that expressed STAU155-HA3 was defined as 100. For samples after IP, the level of each immunoprecipitated FLUC mRNA was normalized to its normalized level before IP. (C) After IP, samples were spiked with a small amount of E. coli RNA, and RT-PCR measurements of the level of E. coli LacZ mRNA confirmed comparable RNA recovery efficiencies.
Tethering siRNA-Resistant STAU2 or STAU1 Downstream of a Termination Codon Reduces mRNA Abundance in a Way That Is Inhibited When Cellular STAU1, STAU2, or UPF1 Is Down-Regulated.
Tethering human STAU1 sufficiently downstream of the termination codon of a reporter mRNA triggers SMD of that mRNA depending on UPF1 and translation (13, 25). Given our findings that down-regulating either STAU1 or STAU2 inhibits SMD (Fig. 1 and Fig. S1), and STAU262 and STAU259 bind UPF1, STAU1, and SBSs (Figs. 2–4), tethering STAU2 isoforms should also trigger SMD in a pathway that involves cellular UPF1 and both cellular STAU paralogs. To test this hypothesis because the literature indicates otherwise (3, 15), HeLa cells were transiently transfected with (i) a pcFLUC-MS2bs reporter construct that carries eight copies of the MS2 coat-protein binding sequence (MS2bs) in its 3′ UTR (13), or, as a negative control, pcFLUC (Fig. 5A), (ii) phCMV-MUP, and (iii) pMS2-HA-STAU155 (13), pMS2-HA-STAU262, or pMS2-HA-STAU259, each of which encodes a tetherable protein.
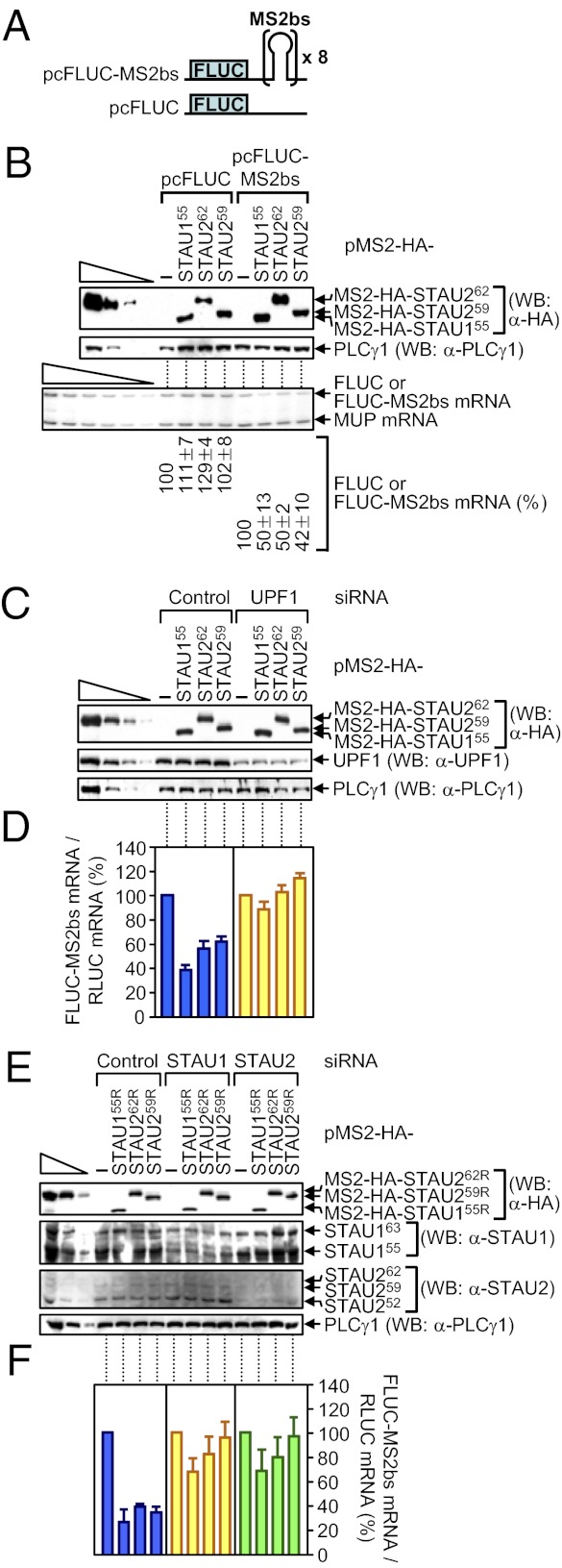
Evidence that tethering MS2-HA-STAU155, MS2-HA-STAU262, or MS2-HA-STAU259 to the FLUC-MS2bs mRNA 3′ UTR triggers FLUC-MS2bs mRNA SMD in a mechanism that depends on the cellular STAU paralogs and cellular UPF1. (A) Schematic representations of pcFLUC and pcFLUC-MS2bs, where × 8 specifies eight tandem repeats of the MS2 coat protein-binding site (MS2bs). (B) HeLa cells (2.5 × 105 per well of a six-well plate) were cotransfected with 0.2 μg of pcFLUC or pcFLUC-MS2bs, 0.1 μg of phCMV-MUP, and 1 μg of the specified pMS2-HA-STAU effector plasmid. Two days after transfection, protein and RNA were purified. WB using anti(α)-HA demonstrated comparable levels of effector protein expression. (B, Lower) Using RT-PCR, normalized levels of FLUC or FLUC-MS2bs mRNA in the presence of the specified effector protein were calculated as a percentage of the normalized level of FLUC or FLUC-MS2bs mRNA in the presence of pMS2-HA, which was defined as 100. (C and D) HeLa cells (2.5 × 105 per well) were transiently transfected with 0.1 μg of pcFLUC-MS2bs, 0.1 μg of a plasmid that produces RLUC protein (pRLUC), which encodes renilla luciferase, and 1 μg of the specified effector plasmid. One day later, cells were transfected with specified siRNA. After an additional 2 d, protein and RNA were purified. (C) WB was used to quantitate the extent of down-regulation. (D) RT-qPCR was used to quantitate the effects of tethering of the specified protein on FLUC-MS2bs mRNA abundance. (E and F) Essentially as in C and D, except the effector plasmids were siRNA resistant (R).
The expression of each tetherable protein reduced the abundance of FLUC-MS2bs reporter mRNA but not control FLUC mRNA to ~40–50% of normal (Fig. 5B). Using cells that were also transfected with Control or UPF1 siRNA (Fig. 5C), tethering not only STAU155 (13, 14) but also STAU262 or STAU259 down-regulated FLUC-MS2bs reporter mRNA abundance to ~40–60% of normal, and down-regulating UPF1 inhibited this down-regulation (Fig. 5 C and D). Thus, tethered STAU2, like tethered STAU1, triggers SMD via its interaction with UPF1. Using cells transfected with Control, STAU1, or STAU2 siRNA (Fig. 5E), tethering STAU155R, STAU262R, or STAU259R, each of which was produced from an siRNA-resistant vector, again reduced FLUC-MS2bs reporter mRNA abundance to ~20–40% of normal (Fig. 5F). STAU1 siRNA inhibited the ability of tethered STAU155R, STAU262R, or STAU259R to reduce mRNA abundance, as did STAU2 siRNA (Fig. 5F). Thus, neither STAU2 nor STAU1 can efficiently trigger SMD when the level of cellular STAU2, STAU1, or UPF1 is reduced, indicating that both paralogs contribute to SMD via their interaction with each other and UPF1.
SMD Requires ATP-Dependent UPF1 Helicase Activity, Which Is Enhanced by STAU2 and STAU1 Without a Concomitant Enhancement of UPF1 ATPase Activity.
Binding of the UPF2 NMD factor to the UPF1 CH domain promotes UPF1 ATPase and helicase activities in a way that is important for NMD (27, 28). Considering that STAU1 and STAU2 bind to a region of UPF1 that contains the CH domain (15, 16), we tested the importance of UPF1 ATPase and helicase activities to SMD by assaying UPF1(G495R,G497E) (29), which fails to bind ATP (30, 31). Human UPF1 and UPF1(G495R,G497E) were produced in and purified from E. coli (Fig. S3A). ATPase activity assays were performed in the presence of γ-[32P]ATP and, because the ATPase activity of UPF1 depends on RNA, poly(U) (30). The substrate for the helicase assays consisted of a 44-nt ssRNA annealed to an 18-nt γ-[32P]-labeled ssDNA (28) (Fig. S3C), and assays were performed in the presence of excess unlabeled 18-nt ssDNA. In contrast to UPF1, UPF1(G495R,G497E) lacks both ATPase (Fig. 6A) and helicase (Fig. 6 B and C) activities.
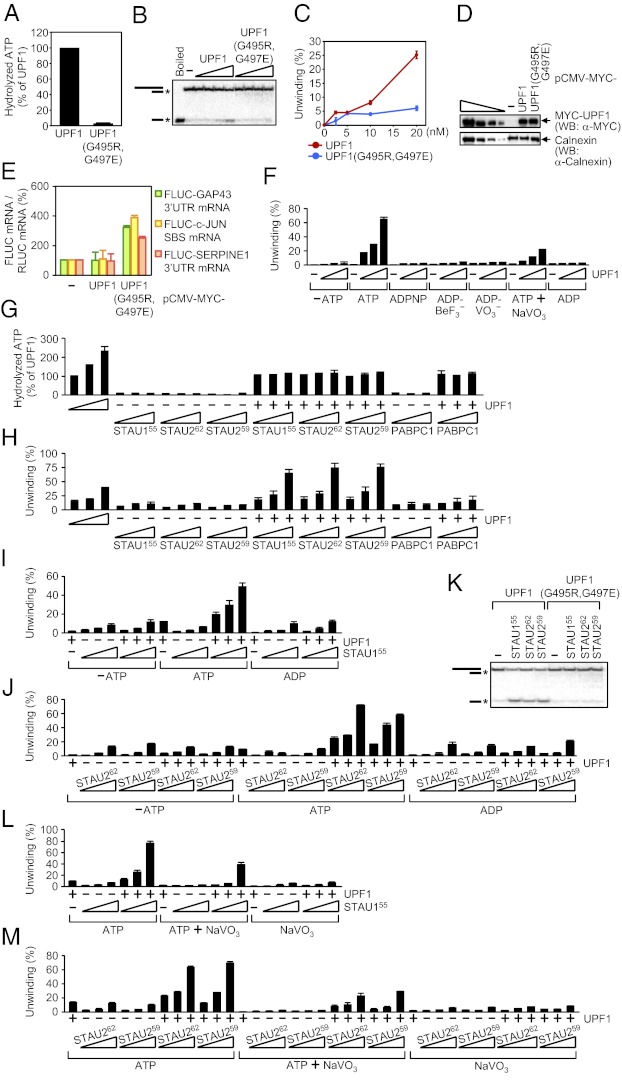
STAU155, STAU262, and STAU259 activate SMD by augmenting UPF1 helicase activity but not UPF1 ATPase activity. (A) Histogram of RNA-dependent ATPase assays using 10 nM of UPF1 or UPF1(G495R,G497E), where the activity of UPF1 was defined as 100. Assays used 1 μCi of γ-[32P]ATP and 4 μg of poly(U). (B) ATP-dependent helicase assays using 0 (−), 2.5, 5, 10, or 20 nM (wedge) of UPF1 or UPF1(G495R,G497E) in the presence of 0.25 nM of γ-[32P]-labeled RNA−DNA duplex. Mobilities of the RNA−DNA duplex and ssDNA are indicated by cartoons, where the asterisk (*) denotes γ-[32P]. (C) Plot of data in B. (D and E) HEK293T cells (4 × 107 per 150-mm dish) were transiently transfected with 1 μg of each pcFLUC SMD-reporter plasmid; 0.5 μg of the pRLUC reference plasmid; and 1 μg of pCMV-MYC(−), pCMV-MYC-UPF1, or pCMV-MYC-UPF1(G495R,G497E). Two days later, protein and RNA were purified. (D) WB using the specified antibody (α). The four left-most lanes show threefold dilutions of lysates from cells that expressed MYC-UPF1. (E) Histogram of RT-qPCR results. The level of each FLUC mRNA was normalized to the level of RLUC mRNA, and the normalized level of each in pCMV-MYC-transfected cells was defined as 100. (F) As in B, using 0 (−), 10, 20, or 40 nM (wedge) of UPF1 in the absence of nucleotide (−ATP) or in the presence of 0.5 mM of ATP, ADPNP, ADP-BeF3−, ADP-VO3−, or ADP, or 0.5 mM of ATP together with (+) 2.5 mM of NaVO3 (Fig. S3D). (G) As in A, using 0 (−), 10, 20, or 40 nM of UPF1 (wedge) or 10 nM UPF1 (+) and/or 10, 20, or 40 nM of HIS-STAU155, STAU262, STAU259-HIS, or PABPC1 (wedge). (H) Helicase assays (Fig. S3F) using samples assayed in G. (I and J) As in B, using 0 (−) or 10 nM of UPF1 (+) and/or 10, 20, or 40 nM (wedge) of (I) HIS-STAU155, (J) STAU262 or STAU259-HIS in the absence of nucleotide (−ATP) or in the presence of 2 mM of ATP or ADP (Fig. S3 G and H). (K) As in B, using 10 nM of UPF1 or UPF1(G495R,G497E) and 0 (−) or 40 nM of HIS-STAU155, STAU262, or STAU259-HIS. (L and M) Helicase assays (Fig. S3 K and L) as in I and J, in the presence of 2 mM of ATP, 2 mM of ATP+10 mM of NaVO3, or 10 mM of NaVO3.
Next, HEK293T cells were transiently transfected with a MYC-UPF1 or MYC-UPF1(G495R,G497E) expression vector (29) and a subset of the pcFLUC SMD-reporter plasmids (Fig. 1B). MYC-UPF1 proteins were each produced at a comparable level (Fig. 6D). Relative to MYC-UPF1, MYC-UPF1(G495R,G497E) inhibited SMD ~two- to fourfold (Fig. 6E). Thus, UPF1 binding to ATP and the resulting ATPase and/or helicase activities are important for SMD because of roles in SMD-target remodeling and/or indirectly because of roles in UPF1 recycling.
To characterize the dependence of UPF1 helicase activity on ATP hydrolysis, helicase assays were performed in the presence of ATP; 5′-adenylyl β,γ-imidodiphosphate (ADPNP), which is a nonhydrolyzable analog of ATP; ADP-BeF3−, which mimics the ground state of the ATP hydrolysis reaction (32); ADP-VO3−, which mimics the ADP+Pi transition state of ATP hydrolysis (33, 34); ATP together with NaVO3, where VO3− replaces Pi after ATP hydrolysis to form ADP-VO3− at the active site and inhibit further hydrolysis (35–37); ADP; or no nucleotide. UPF1 helicase activity was evident in the presence of ATP and, to a lesser extent, in the presence of ATP+NaVO3 but not in the absence of ATP or the presence of ADPNP, ADP-BeF3−, ADP-VO3−, or ADP (Fig. 6F and Fig. S3D). Thus, limiting ATP hydrolysis using ATP+NaVO3 limits UPF1 helicase activity, and helicase activity is driven by the energy released from ATP cleavage and/or a resulting cleavage-dependent change in UPF1 conformation that is not recapitulated by any of the tested nucleotide analogs.
To determine the effect of STAU1 and STAU2 on UPF1 ATPase and helicase activities, the assays were repeated in the presence of increasing amounts of HIS-STAU155, STAU262, STAU259-HIS or, as a negative control, PABPC1. In the presence of poly(U) (Fig. 6G) or the RNA−DNA duplex (Fig. S3E), none of the STAU1 or STAU2 isoforms or PABPC1 manifested appreciable ATPase activity itself or enhanced UPF1 ATPase activity. However, in the presence of ATP all STAU isoforms, unlike PABPC1, activated UPF1 helicase activity in a concentration-dependent manner more or less to the same extent (Fig. 6H and Fig. S3F; we consider the low level of helicase activity manifested by HIS-STAU155, STAU262, STAU259-HIS, or PABPC1 alone to be background activity). STAU1 or STAU2 did not enhance UPF1 helicase activity in the absence of ATP (Fig. 6 I and J and Fig. S3 G and H) and also failed to activate UPF1(G495R,G497E) helicase activity (Fig. 6K).
Results from our ATPase assays demonstrating that the STAU paralog-mediated enhancement of UPF1 unwinding is not accompanied by an increase in the number of ATP hydrolytic events predicts that limiting ATP hydrolysis using NaVO3 to “trap” UPF1 in a state that inhibits further hydrolysis would not inhibit the effect of STAU155 on UPF1 unwinding. In fact, when present in excess of ATP, NaVO3 does not inhibit the ability of STAU155, STAU262 and STAU259 to augment UPF1 helicase activity (Fig. 6 L and M and Fig. S3 I–L). We propose that when UPF1 is bound by ATP or a specific catalytic intermediate of ATP hydrolysis—one that is not recapitulated by any of the nucleotide analogs tested here—STAU1, and possibly STAU2, promotes UPF1 helicase activity by inducing and/or stabilizing a helicase-active conformation.
Evidence That STAU2 Induces a Conformational Change in RNA-Bound UPF1.
UPF2 augments UPF1 ATPase and helicase activities during NMD by binding to the UPF1 CH domain and, according to RNase protection assays, decreasing the UPF1 footprint on ssRNA (27). The decreased footprint was interpreted as a change in UPF1 conformation that loosened its grip on ssRNA, allowing it to unwind RNA more effectively.
We performed comparable RNA footprint experiments to determine whether STAU262 changes UPF1 conformation. We examined STAU262 because of its higher binding in vitro to UPF1 relative to STAU155 or STAU259 (Fig. 2B) and its greater solubility over STAU155 under footprinting conditions. STAU262 binding to UPF1 shifts the length of RNA protected by UPF1 from 11 to 12 nt to 12 to 13 nt (Fig. 7A, compare lanes 2 and 3), whereas in the absence of STAU262, ADPNP, ADP-AlF4−, or ADP-VO3− binding to UPF1 increased the length of RNA protected to 12–14 nt (Fig. 7A, compare lanes 5–7), and ADP binding further increased the length to 12–15 nt (Fig. 7A, lane 9). We propose that STAU262 alters the conformation of UPF1 so that it protects a length of RNA that is between the length protected by ATP-free UPF1 and ATP (or an ATP-hydrolysis intermediate)-bound UPF1. Thus, STAU262 binding enables UPF1 to approximate a conformation that is active in translocation and/or unwinding. In the absence of STAU262, the UPF1 footprint was not well defined when ATP or ATP+NaVO3 was present (Fig. 7A, lanes 4 and 8) as expected because either UPF1 fails to bind RNA efficiently or the position of UPF1 on RNA is not fixed (Fig. 6 F and M). Although not proven, we do not believe that the increased footprint observed when STAU262 is added to UPF1 is due to STAU262 binding to ssRNA because STAU262, unlike STAU155 (2), does not appreciably bind our RNA−DNA duplex (Fig. S4 A and B), which contains 26 nt of ssRNA. Moreover, STAU2 does not noticeably affect the ssRNA-binding activity of UPF1 (Fig. S4B). The simplest interpretation of our results is that the STAU262-mediated increase in UPF1 helicase activity is accompanied by an alteration in UPF1 conformation (Fig. 7 B–D).

STAU262 expands the UPF1 footprint on ssRNA. (A) The specified complexes were formed using 5 pmol of in vitro-transcribed α-[32P]UTP-labeled 60-nt RNA; 10 pmol of UPF1; 40 pmol of STAU262; and where specified 2 mM of the indicated nucleotide and 10 mM of the specified phosphate mimic. Notably, ADP-BeF3− was not tested because a precipitate formed at 10 mM BeF3−. Samples were then incubated with 1 μg of RNase A and 1 U of RNase T1. The sizes of the protected RNA fragments were determined using denaturing polyacrylamide gel electrophoresis followed by autoradiography. A γ-[32P]-labeled RNA ladder provided molecular weight standards (black nt), and red notations denote protected fragments. Notably, sizes assigned to the protected fragments were confirmed in gels, in which molecular weight standards were eletrophoresed in lanes residing adjacent to each experimental lane. The nature of the diffuse ~6-nt band that was evident in the presence of all tested nucleotides requires further investigation. The footprint in the presence of ADP-AlF4− was routinely faint. (B–D) Model for how STAU paralogs bind to and prolong a conformational state of UPF1 that promotes its ATP-dependent unwinding activity without promoting ATP hydrolysis. (B) Domains of UPF1, where A1 and A2 represent the two RecA-like domains, 1B is an N-terminal attachment of the A1 domain, 1C is an insertion in the A1 domain, and CH denotes the cysteine- and histidine-rich region (27, 31). (C) ATP-free UPF1 protects a smaller (11- to 12-nt) region of RNA than does ATP-bound UPF1. The footprint of ATP-bound UPF1 consists of as many as 14 nt and is maintained during the process of ATP hydrolysis and RNA unwinding. Additional rounds of ATP hydrolysis lead to more extensive duplex unwinding. (D) STAU2 association with UPF1 generates the larger UPF1 footprint that typifies UPF1 bound to ATP or a hydrolytic intermediate (e.g., ADPNP, ADP-VO3−, or ADP). Because STAU2 does not bind ssRNA, we infer that STAU2 alters the conformation of ssRNA-bound UPF1 (surrounding yellow spikes) so UPF1 is better poised to receive ATP and/or unwind RNA when ATP is already bound (surrounding green spikes). In this way, STAU2 enhances the intrinsic unwinding capability of UPF1 per ATP hydrolyzed. UPF1 helicase activity has been shown to scan and remodel mRNP during NMD (45, 46).
Discussion
STAU2 Functions in SMD.
Our results reveal that STAU2, like STAU1 (13), interacts directly with UPF1 both in vivo and in vitro, and these interactions are necessary for efficient SMD (Figs. 1, ,2,2, and and5).5). Recently, Miki et al. (15) also demonstrated that STAU2 isoforms interact directly with UPF1 in vitro. In support of STAU2 function during SMD, tethering STAU2 downstream of the termination codon of a reporter mRNA, like tethering STAU1, reduces reporter mRNA abundance in HEK293T cells in a mechanism that depends on UPF1, STAU1, and STAU2 (Fig. 5). Additionally, the efficiency of SMD in tethering assays and assays of cellular targets is determined by the abundance of both STAU1 and STAU2 (Fig. 5, Fig. S5, and SI Discussion). Our finding that the two STAU paralogs interact directly suggests that any functional differences between the two paralogs during SMD would be homogenized, but less so for tissues in which one paralog is significantly more abundant than the other.
STAU2−STAU2, STAU2−STAU1, and STAU1−STAU1 interactions are readily detected (Fig. 3) and may be required for optimal UPF1 binding and/or activation. For example, our finding that STAU1 stabilizes imperfect intermolecular RNA duplexes that range in size from ~85 to 300 bp (17) suggests that STAU association contributes to duplex stabilization. Unlike Furic et al. (20), we find no evidence for STAU1- or STAU2-specific mRNA targets (Fig. 4). Given that the paralogs interact, it is difficult to conceive of such targets unless there were conditions whereby one paralog is significantly more abundant than the other.
STAU1−STAU1 interactions have been shown using live cells to involve dsRBD2 and dsRBD5 (22). Because STAU1 dsRBD2 and parts of STAU1 dsRBD5 are conserved in all STAU2 isoforms (Fig. 1), it is possible that STAU1−STAU2 and STAU2−STAU2 interactions are also mediated by dsRBD2 and dsRBD5. Relative to STAU1, the binding of STAU2 to SBS-containing mRNAs is ~two- to fivefold higher (Fig. 4), which might be due to the additional dsRBD, dsRBD1, in STAU2 compared with STAU1 (Fig. 1). However, because dsRBD3 and dsRBD4 seem to be the only functional dsRNA-binding domains (1, 2), contributions to RNA binding by dsRBD1 or by other differences between STAU2 and STAU1 might enhance STAU2 dsRBD3 and/or dsRBD4 function. For example, individual dsRBDs that do not bind dsRNA in vitro can promote dsRNA binding of a “true” dsRBD when both reside in tandem as exemplified by Xenopus laevis RNA-binding protein A (38).
Deletion analyses indicate that STAU1 (16) and STAU2 (15) interact with a region of UPF1 that includes the CH domain. This is the same region of UPF1 that interacts with UPF2 during NMD. Relative to STAU1, the binding of STAU2 to UPF1 is ~10-fold higher (Fig. 2). Considering that residues spanning the TBD to just upstream of dsRBD5 seem to be the minimal region of STAU1 that interacts with UPF1 (16), differences in the binding of STAU2 and STAU1 to UPF1 may be attributable to sequence variations in their TBDs, which share only 18% identity (Fig. 1). Provided the paralogs bind tightly enough to overcome the activation threshold for UPF1 helicase activity, they may bind to overlapping or different regions of the CH domain with different efficiencies yet still activate UPF1 helicase activity to the same extent (Fig. 6). Alternatively, STAU2 may interact with UPF1 using an entirely different region, such as dsRBD2 or dsRBD3 (15).
STAU2 Stimulates UPF1 Unwinding Activity but Not UPF1 ATP Hydrolytic Activity and Induces a Conformational Change in UPF1.
When added to UPF1, higher concentrations of STAU2 or STAU1 increase higher levels of UPF1 RNA−DNA duplex unwinding without stimulating UPF1 ATPase activity in a mechanism that depends on the presence and/or hydrolysis of ATP (Fig. 6). There are two key differences between how STAU2 and STAU1 activate UPF1 during SMD and how UPF2 activates UPF1 during NMD (27). First, UPF2 stimulates UPF1 ATPase activity, whereas the STAU paralogs do not; they merely “grease the wheels” of the UPF1 helicase so that unwinding is more extensive per ATP hydrolyzed. Second, at least for STAU262, activation of UPF1 helicase activity is achieved by increasing rather than decreasing the UPF1 footprint on RNA (Fig. 7). It has been reported that UPF2 in the presence of ADP-BeF3 decreases the size of the UPF1 footprint (27), UPF2 in the absence of nucleotide analog loosens the grip of UPF1 on RNA (28), and UPF2 in the presence of ATP enhances UPF1 helicase activity (27). Although at pH 6.5 the footprint of UPF1 alone (28) was found to be similar in size to the footprint of UPF1 in the presence of ADPNP (27, 28), we are able to detect a difference in the size of the two footprints at the more physiological pH 7. We report here that UPF1 alone, which has a higher affinity for ssRNA than when nucleotide bound (27, 31), has a smaller footprint than when nucleotide bound. Therefore, the correlation of a smaller UPF1 footprint with activation of UPF1 helicase activity, as observed in studies using UPF2 (27), may not be universal, as evidenced from our studies using STAU2, which indicate that a larger UPF1 footprint correlates with activation of UPF1 helicase activity. Because nucleotide binding lowers the association of UPF1 with RNA (27, 31), and a lowered association correlates with enhanced helicase activity (27), STAU262 binding to UPF1 may mimic nucleotide binding. UPF1 may resemble the human coronavirus 229E helicase, which is capable of unwinding long stretches of duplexed RNA before dissociating (39), except that the processivity of UPF1 is augmented by its binding to one or both STAU cofactors.
There are other examples of cofactor-mediated helicase activation that seem to bypass coordinated ATPase stimulation. Ntr1 of the Ntr1/Ntr2 complex enhances Prp43 RNA helicase activity in an ATP-dependent manner but without an apparent enhancement of ATPase activity (40, 41). As a variation of this theme, the E. coli Rep DNA helicase uses ATP hydrolysis not for unwinding but for translocation to a dsDNA structure and, by analogy to how STAU cofactors activate the UPF1 helicase, the ØX174 gene A protein or constituents of the multi-Rep complex then promote E. coli Rep helicase activity (42, 43). Taking cues from these and other studies, we propose that STAU2 and possibly STAU1 stabilize UPF1 in a conformation that is active during unwinding and/or translocation (Fig. 7).
Summary.
We envision that STAU1 and/or STAU2 bind to an SBS either individually or, more likely, as homo- or hetero-multimers, the size of which may depend on the length of the SBS and the relative abundance of each paralog. STAU bound to an SBS recruits UPF1 either directly or through interactions with another STAU molecule, presumably after translation terminates if a comparison can be made to when UPF1 binds the stem-loop binding protein of cell cycle-regulated histone mRNAs (44). By analogy to the mechanism of NMD, after translation terminates sufficiently upstream of the SBS, STAU−UPF1 then reconfigures so that the helicase activity of UPF1 is activated by STAU.
Materials and Methods
Descriptions of plasmid constructions; cell transfection and lysis, and cell protein and RNA purification are given in SI Materials and Methods. Supporting Information also provides details for recombinant protein purification and in vitro pull-down experiments. See SI Materials and Methods for in vitro ATPase and helicase assays; and RNase protection assays.
Acknowledgments
We thank Susana de Lucas and Juan Ortín for anti-STAU1, David Bear for the GST-PABPC1 plasmid, Yalan Tang for baculovirus-produced FLAG-UPF1, Galina Pavlencheva for E. coli-produced UPF1 and UPF1(G495R,G497E), and Dmitri Ermolenko, Joseph Wedekind, Eckhard Jankowsky, Hérve Le Hir, Fabien Bonneau, and L.E.M. laboratory members for advice. This work was funded by National Institutes of Health (NIH) Grant R01 GM074593 (to L.E.M.). E.P. was supported by American Heart Association Founders Affiliate Postdoctoral Fellowship 11POST 7860051. M.L.G. was supported by NIH Grant F32 GM090479 and NIH National Cancer Institute Grant T32 CA09363.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1213508110/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1213508110
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/110/2/405.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
The role of secondary structures in the functioning of 3' untranslated regions of mRNA: A review of functions of 3' UTRs' secondary structures and hypothetical involvement of secondary structures in cytoplasmic polyadenylation in Drosophila.
Bioessays, 46(3):e2300099, 31 Dec 2023
Cited by: 0 articles | PMID: 38161240
Review
Staufen1 Represses the FOXA1-Regulated Transcriptome by Destabilizing FOXA1 mRNA in Colorectal Cancer Cells.
Mol Cell Biol, 44(2):43-56, 12 Feb 2024
Cited by: 0 articles | PMID: 38347726
Translation-coupled mRNA quality control mechanisms.
EMBO J, 42(19):e114378, 22 Aug 2023
Cited by: 14 articles | PMID: 37605642 | PMCID: PMC10548175
Review Free full text in Europe PMC
The nexus between RNA-binding proteins and their effectors.
Nat Rev Genet, 24(5):276-294, 23 Nov 2022
Cited by: 20 articles | PMID: 36418462 | PMCID: PMC10714665
Review Free full text in Europe PMC
Integrative omics indicate FMRP sequesters mRNA from translation and deadenylation in human neuronal cells.
Mol Cell, 82(23):4564-4581.e11, 09 Nov 2022
Cited by: 6 articles | PMID: 36356584 | PMCID: PMC9753132
Go to all (62) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Staufen-mediated mRNA decay.
Wiley Interdiscip Rev RNA, 4(4):423-435, 16 May 2013
Cited by: 137 articles | PMID: 23681777 | PMCID: PMC3711692
Review Free full text in Europe PMC
SMG1 regulates adipogenesis via targeting of staufen1-mediated mRNA decay.
Biochim Biophys Acta, 1829(12):1276-1287, 01 Nov 2013
Cited by: 17 articles | PMID: 24185201
Staufen1 regulates diverse classes of mammalian transcripts.
EMBO J, 26(11):2670-2681, 17 May 2007
Cited by: 140 articles | PMID: 17510634 | PMCID: PMC1888674
lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements.
Nature, 470(7333):284-288, 01 Feb 2011
Cited by: 810 articles | PMID: 21307942 | PMCID: PMC3073508
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: T32 CA009363
Grant ID: T32 CA09363
NIGMS NIH HHS (2)
Grant ID: F32 GM090479
Grant ID: R01 GM074593




