Abstract
Free full text

Translation‐coupled mRNA quality control mechanisms
Abstract
mRNA surveillance pathways are essential for accurate gene expression and to maintain translation homeostasis, ensuring the production of fully functional proteins. Future insights into mRNA quality control pathways will enable us to understand how cellular mRNA levels are controlled, how defective or unwanted mRNAs can be eliminated, and how dysregulation of these can contribute to human disease. Here we review translation‐coupled mRNA quality control mechanisms, including the non‐stop and no‐go mRNA decay pathways, describing their mechanisms, shared trans‐acting factors, and differences. We also describe advances in our understanding of the nonsense‐mediated mRNA decay (NMD) pathway, highlighting recent mechanistic findings, the discovery of novel factors, as well as the role of NMD in cellular physiology and its impact on human disease.
Abstract
This review discusses mRNA quality control mechanisms including non‐stop and no‐go mRNA decay pathways and the nonsense‐mediated mRNA decay pathway.
Introduction
Translation‐coupled RNA quality control mechanisms sense ribosome stalling, or premature translation stops and elicit mRNA degradation and ribosome recycling. In eukaryotic cells, there is an intricate relationship between mRNA turnover and active translation (Fig 1A). This was recently confirmed in mammalian cells in culture, with the use of single‐molecule imaging approaches that revealed translation‐dependent destabilization of mRNA (Dave et al, 2023). In addition, the presence of a series of non‐optimal codons can negatively influence protein production by decreasing ribosome translocation rates leading to ribosome collisions that have the potential to trigger RNA quality control pathways and lead to mRNA decay (Hanson & Coller, 2018; Wu & Bazzini, 2023). This process, known as codon optimality‐mediated mRNA decay (COMD), allows the cell to distinguish between variation in normal translation speeds and terminal ribosome stalling, which triggers alternative RNA quality control pathways (Fig 1B) (Wu et al, 2019; D’Orazio & Green, 2021).
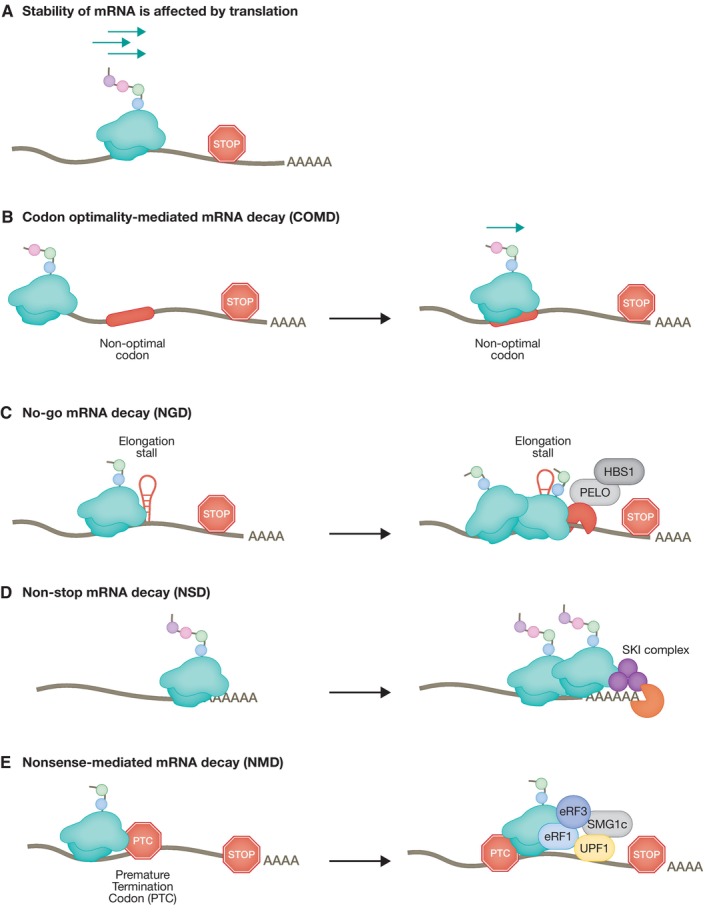
(A) Stability of mRNAs is affected by translation. Several layers of regulation monitor the efficiency of mRNA translation, including the translation rate, amino acid composition, and mRNA secondary structures. (B) The mRNA translation rate is slowed down when the ribosome encounters sub‐optimal codons leading to a decrease in mRNA stability. (C) No‐go mRNA decay (NGD) is triggered by the presence of mRNA secondary structures leading to ribosome stalling. (D) The absence of a stop codon results in slowing of the ribosome reading through the poly (A) tail triggering the non‐stop mRNA decay (NSD) pathway. (E) Recognition of a PTC sets in motion a cascade of event involving the UPF family of proteins, resulting in mRNA degradation by the nonsense‐mediated mRNA decay (NMD) pathway.
Two major pathways of co‐translational mRNA surveillance, no‐go mRNA decay (NGD) and non‐stop mRNA decay (NSD), sense aberrant translation by monitoring the stalling of ribosomes and/or ribosomes that fail to encounter a stop codon, respectively (Fig 1C and D). A common feature of both NGD and NSD is that upon sensing defective ribosome translocation, these pathways activate mechanisms leading to mRNA and nascent peptide degradation, and ribosome recycling (Brandman & Hegde, 2016; Powers et al, 2020). NGD and NSD use similar factors, but differ mainly in their substrate mRNAs and initial triggering mechanism (Simms et al, 2016). In both pathways, ribosome stalling is followed by recruitment of nucleases leading to degradation of mRNA (Powers et al, 2020). By contrast, the nonsense‐mediated mRNA decay (NMD) pathway senses inappropriate translation termination (Kervestin & Jacobson, 2012; Hug et al, 2016) (Fig 1E). NMD eliminates mRNAs that harbor premature termination codons (PTCs), thus, preventing the synthesis of truncated proteins. Significantly, NMD has a more global role in post‐transcriptional regulation of gene expression and also regulates the stability of many cellular non‐mutated transcripts, which do not harbor PTCs. These non‐canonical functions of the NMD pathway, which do not represent RNA quality control per se, are important for the regulation of many cellular pathways, including differentiation, neurogenesis, synaptic control, as well as the response to viral infections and stress (Jaffrey & Wilkinson, 2018; Kurosaki et al, 2019).
In this review article, we will focus primarily on three translation‐coupled RNA quality control mechanisms: NGD, NSD, and NMD. We will review similarities and differences among these pathways and cover recent advances related to their mechanisms, targets, and trans‐acting factors that ensure a tight control of aberrant RNAs that fail to be properly translated.
No‐go mRNA decay
Initially discovered in S. cerevisiae, the no‐go mRNA decay (NGD) pathway is triggered when ribosomes move slowly or stall during translation elongation, leading to ribosome collisions with trailing ribosomes, forming disomes (Doma & Parker, 2006; Harigaya & Parker, 2010). Ribosome stalling can be triggered by a number of stimuli, such as the presence of stable stem loops, pseudoknots, GC‐rich sequences, or damaged RNA bases (Shoemaker & Green, 2012). Persistent collisions trigger a two‐pronged cellular response that aims to inhibit translation re‐initiation on the problematic mRNA, and simultaneously remove faulty mRNAs and nascent peptides, and recycle the ribosomes (Fig 2).
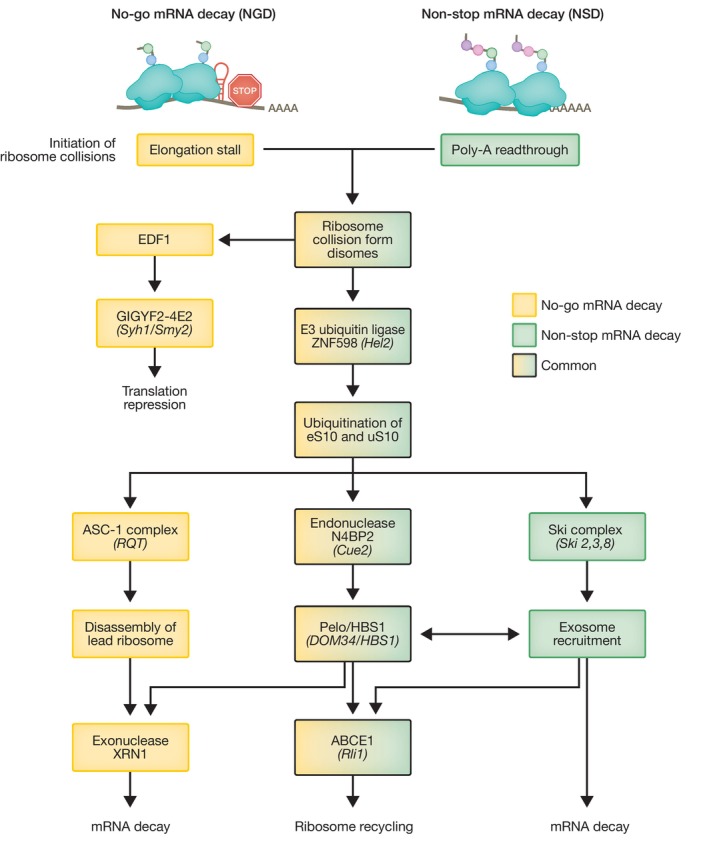
Flow chart depicting steps and trans‐acting factors shared or unique to these two RNA quality control pathways from initial ribosome collision, ubiquitination and endonucleolytic cleavage to ultimate mRNA degradation and ribosome recycling. S. cerevisiae homologs are indicated in brackets.
Initially, translational repression is mediated by EDF1, which binds to collided ribosomes and recruits the translational repressor complex GIGYF2‐4E2 (Syh1‐Smy2 in S. cerevisiae) (Hickey et al, 2020; Sinha et al, 2020; Veltri et al, 2022). Upon ribosome collision, leading and trailing ribosomes form a ‘rotated’ interface of 40S subunits recognized by the E3 ubiquitin ligase ZNF598 (Hel2 in yeast) that ubiquitinates ribosomal proteins eS10 and uS10 and acts independently of the GIGYF2‐4E2 branch (Juszkiewicz et al, 2018; Ikeuchi et al, 2019) to initiate a degradation pathway. Ubiquitination of the 40S subunit is critical for the recruitment of an endonuclease, which cleaves mRNAs in the middle of a stalled disome, exposing fragmented mRNA to further degradation by XRN1 and potentially the exosome. A genetic screen in yeast recently identified the endonuclease Cue2 as a component of the NGD pathway (D’Orazio et al, 2019). Cue2 is a low‐abundance protein that cleaves mRNA at the A site of the rotated collided ribosome in a Hel2‐dependent manner and has homologues in C. elegans (NONU1) and in mammalian cells (N4BP2; D’Orazio et al, 2019; Glover et al, 2020; Monem et al, 2023). As a consequence of mRNA cleavage, stalled ribosomes appear near the 3′ end of mRNA, which favors the binding of an evolutionarily conserved complex formed by PELOTA (PELO) and its GTPase cofactor HBS1 (Dom34/Hbs1 in yeast; Doma & Parker, 2006; Pisareva et al, 2011). Despite structural similarities with release factors eRF1/eRF3, instead of detecting a stop codon, the PELO/HBS1 complex preferentially binds an empty ribosomal A site. This induces ribosome splitting into 40S bound to the mRNA, and 60S with the attached nascent peptide, and is mediated by the ribosome recycling factor ABCE1 (Rli1 in yeast; Shoemaker et al, 2010). The nascent peptide is subsequently targeted for Listerin‐mediated ubiquitination and degraded by the proteasome as part of the ribosome quality control (RQC) pathway (Shao et al, 2013). However, despite widespread conservation of the Cue2‐Dom34/Hbs1 pathway, recent evidence suggests that this represents a secondary decay method in NGD and may only be required when the capacity of the XRN1‐mediated degradation pathway is saturated (Juszkiewicz et al, 2020). In the majority of mRNAs with ribosome stalls within the open reading frame (ORF), Hel2‐mediated 40S ubiquitination of disomes recruits the ribosome quality control trigger (RQT) complex containing the RNA helicase Slh1 in yeast, with the homologous complex ASC‐1 performing the same role in mammals (Matsuo et al, 2017). Upon 40S ubiquitination of collided ribosomes, the RNA helicase ASCC3 (part of the ASC‐1 complex) selectively disassembles the lead ribosome in an ATP‐dependent manner, in a process not requiring PELO/HBS1 activity (Juszkiewicz et al, 2020; Matsuo et al, 2020). After removing the roadblock of the lead stalled ribosome, normal elongation can resume, or in case of persistent stalls, subsequent ribosome collisions induce further quality control and mRNA is subjected to XRN1‐mediated degradation, likely with a contribution of the PELO/HBS1 branch of NGD.
Non‐stop mRNA decay
The non‐stop mRNA decay pathway (NSD) degrades mRNAs that lack a stop codon, ensuring the degradation of mRNAs that will fail to be properly translated (Frischmeyer et al, 2002; van Hoof et al, 2002). It can be initiated by a number of aberrant translation events which promote readthrough into the poly(A) tail resulting from lack of an in‐frame stop codon or polyadenylation within the ORF. Poly(A) translation within the coding region slows down when the nascent poly(lysine) peptide (encoded by poly(A)) interferes with the ribosomal exit tunnel. This attenuation of translation elongation allows stretches of poly(A) to adopt a conformation that impedes further mRNA decoding at the ribosome A site resulting in stalling of the translating ribosome (Ito‐Harashima et al, 2007; Chandrasekaran et al, 2019; Tesina et al, 2020). Interestingly, hindered elongation can induce ribosome sliding on poly(A) sequences causing frameshifting that may lead to an out‐of‐frame PTC and subsequent triggering of NMD‐mediated mRNA degradation (Koutmou et al, 2015). Importantly, elongation stalling causes collisions with trailing ribosomes and the resulting disomes are recognized and ubiquitinated by the ZNF598 E3 ubiquitin ligase with subsequent Cue2‐mediated endonucleolytic cleavage, steps that are also common to NGD, as described above (Juszkiewicz & Hegde, 2017; Powers et al, 2020).
In yeast, 80S ribosomes stalled at the 3′end of an mRNA lacking a stop codon are bound by the SKI complex, leading to recruitment of the exosome by the exosome‐associated factor SKI7, resulting in degradation of the aberrant mRNA in a 3′ to 5′ direction. SKI7 binds the empty A site of the stalled ribosome at the 3′ end of mRNA and directs mRNA degradation through an eRF3‐like domain (Saito et al, 2013). This process also requires the RNA helicase SKI2 (SKIV2L in mammals) to guide RNA molecules to the exosome complex, triggering mRNA degradation upon ribosome stalling on A‐rich sequences (Tuck et al, 2020). In mammalian cells where SKI7 is not present, bridging the SKI complex to the exosome is carried out by HBS1, its closest homologue. This triggers the interaction with Dom34/Pelota‐Hbs1 complex shared with NGD and acts to dissociate the stalled ribosome with the help of ribosome recycling factors Rli1 (yeast)/ABCE1 (mammals; Tsuboi et al, 2012; Saito et al, 2013).
Fundamentally, NGD and NSD are parallel mRNA quality control pathways that share many similarities, such as ubiquitination of crucial ribosome sites, Cue2‐mediated endonucleolytic cleavage, activation of mRNA decay, and ribosome recycling; however, they have some key differences (Fig 2). The NSD helicase SKI2 is highly related to the NGD helicase Slh1 in yeast/ASCC3 in mammals; however, the Ski complex directly recruits the exosome, thus directly linking mRNA degradation and ribosome recycling. Conversely, in NGD, the Slh1/ASCC3 complex removes stalled ribosomes first, resulting in mRNA degradation mediated by XRN1. NSD has been implicated in the development of several human diseases, highlighting the physiological importance of this RNA quality control pathway. For example, non‐stop mutations in the Dysferlin gene (DYSF) lead to degradation of its mRNA and a subsequent reduction in Dysferlin expression, contributing to the progression of muscular dystrophy (Cacciottolo et al, 2011). Insufficient NSD activity can also lead to disease, such as mutations which eliminate the stop codon in the skeletal muscle alpha actin (ACTA1) gene. Here, incomplete NSD fails to remove mRNAs encoding 47 additional amino acids that are translated within the 3′UTR, leading to large protein aggregates that are responsible for the development of severe skeletal myopathy (Wallefeld et al, 2006).
Nonsense‐mediated mRNA decay
NMD is an RNA quality control mechanism that targets mutated mRNAs harboring PTCs for degradation, but also regulates the stability of many cellular transcripts. As such, NMD modulates the phenotypic outcome of genetic disorders caused by frameshift or nonsense mutations that generate PTCs (Bhuvanagiri et al, 2010; Karousis & Mühlemann, 2022). In contrast to NGD and NSD, which are triggered by the collision of elongating ribosomes, NMD initiates mRNA degradation in response to faulty translation termination events. Similar to NGD and NSD, NMD degrades mRNAs co‐translationally, leading to production of truncated nascent peptides. In the case of NGD and NSD, these potentially toxic peptides are rapidly degraded by the RQC pathway. By contrast, peptides produced from PTC‐containing transcripts are targeted by an ubiquitin proteasome system, which specifically targets peptides remaining tethered to the ribosome following decay of a nonsense mRNA (Inglis et al, 2023).
The function of NMD in preventing the accumulation of truncated proteins can have a positive or negative cellular effect, depending on whether the truncated protein has a deleterious dominant negative function, or whether it retains at least partial function, elimination of which by NMD could be detrimental. For example, inhibition of NMD by antisense oligonucleotides (ASOs) targeting the W1282X mutation in the CFTR gene increases production of a partially functional protein that enhances the CFTR‐mediated chloride current in human bronchial cells (Kim et al, 2022). On the other hand, failure of NMD to degrade β‐globin transcripts harboring a PTC in the last exon leads to expression of truncated, dominant‐negative protein causing severe beta‐thalassemia (Hall & Thein, 1994). The effect of NMD must be carefully considered in the development of any therapeutic approach for diseases caused by PTC mutations. Importantly, NMD also has a role in many physiological processes, for example in the regulation of the stress response and as a modulator of neural development (Jaffrey & Wilkinson, 2018; Kurosaki et al, 2019).
In mammals, NMD is linked to the process of pre‐mRNA splicing via the exon junction complex (EJC), a multi‐subunit protein complex that is deposited 20‐24 nucleotides upstream of most exon‐exon junctions (Le Hir et al, 2000a, 2000b). The splicing factor CWC22 interacts with the core EJC factor eIF4AIII and links splicing with EJC deposition, leading to NMD activation (Alexandrov et al, 2012; Barbosa et al, 2012; Steckelberg et al, 2012). Mechanistically, the NMD response is coupled to mRNA translation since EJCs remain bound to mRNAs until they are displaced by the translation machinery. Initially, it was suggested that NMD activation occurs as a consequence of ribosome stalling at the termination codon (TC). However, recent data challenge this model and revealed that NMD activation in humans is not necessarily linked to stable stalling of ribosomes at TCs (Karousis et al, 2020). A ribosome terminating prematurely at a PTC located ≥ 50–55 nucleotides upstream of the final exon–exon junction will not remove an EJC (Nagy & Maquat, 1998; Thermann et al, 1998) and this initiates the NMD response (Metze et al, 2013; Kurosaki et al, 2019).
50–55 nucleotides upstream of the final exon–exon junction will not remove an EJC (Nagy & Maquat, 1998; Thermann et al, 1998) and this initiates the NMD response (Metze et al, 2013; Kurosaki et al, 2019).
Core NMD factors
A central player in the NMD pathway is the ATP‐dependent RNA helicase of the SF1 superfamily, Upstream Frameshift 1 (UPF1), which harbors an amino‐terminal cysteine‐and histidine‐rich (CH) domain and a carboxy‐terminal RNA helicase domain (Fig 3; Kim & Maquat, 2019).
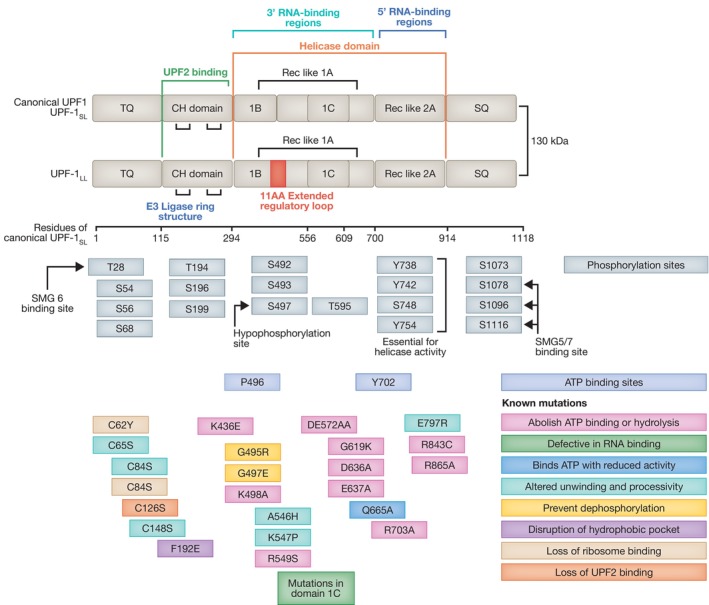
Diagram depicting the domain structure of the canonical UPF1SL isoform, highlighting key residues for its phosphorylation and ATP binding capacity. An isoform generated via alternative splicing, UPFLL, includes an extra 11 amino acid extended regulatory region, while all other structural elements remain the same. Mutations and their effect on UPF1 function are indicated by the colored boxes. Some mutations in UPF1 have the capacity to eliminate NMD function, whilst some mutations sustain functional NMD. All residue numbers relate to the canonical UPF1SL isoform.
UPF1 is activated by phosphorylation carried out by the SMG1c complex, comprised of the phosphoinositide 3‐kinase (PI3K)‐like kinase, SMG1, and two additional subunits, SMG8 and SMG9, that negatively regulates its activity (Yamashita et al, 2001, 2009; Fernández et al, 2011; Langer et al, 2021). SMG1 phosphorylates UPF1 at multiple SQ and TQ motifs located in the amino‐ and carboxy‐terminal domains (Yamashita et al, 2001). Until recent years it was widely accepted that initially, the association of UPF1 with SMG1 and the eukaryotic release factors eRF1 and eRF3 form the surveillance complex (SURF) in the vicinity of the PTC (Kashima et al, 2006) (Fig 4). Subsequently, interaction of the SURF complex with UPF2 and UPF3B and an EJC downstream of the PTC leads to the assembly of a decay‐inducing complex (DECID), where UPF1 is phosphorylated and eRF1 and eRF3 are released (Kashima et al, 2006; Chamieh et al, 2008; López‐Perrote et al, 2016). In addition to this, a central role for UPF3B in translation termination has been highlighted. A fully reconstituted in vitro translation system showed the predominance of the interaction of UPF3B with ribosome release factors, to delay translation termination and dissociate post‐termination ribosomal complexes that are devoid of the nascent peptide. UPF1 was shown to interact transiently with the termination factors and UPF3B to initiate the subsequent mRNA decay (Neu‐Yilik et al, 2017). Despite the lack of mammalian in vivo data, a recent in vivo study in yeast re‐established the central role of the UPF1:80S interaction for translation termination and NMD initiation (Ganesan et al, 2022).
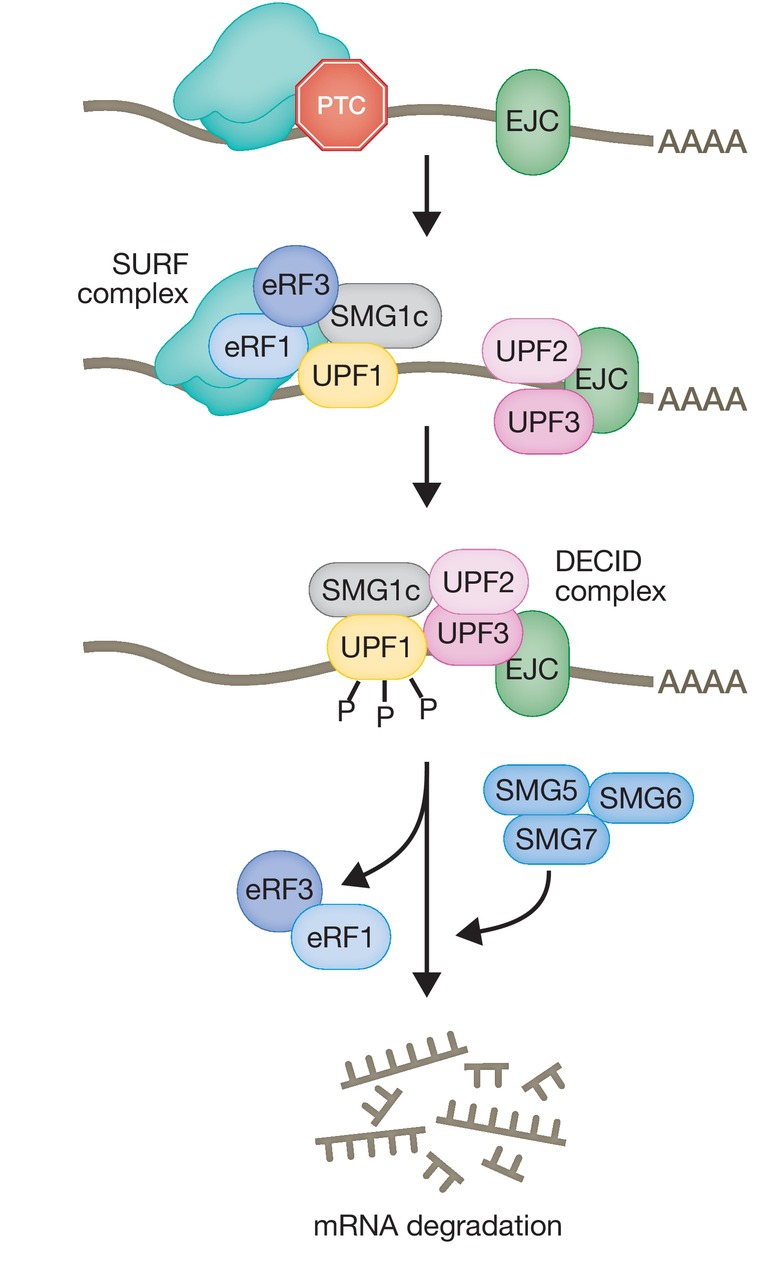
Schematic depicting the widely accepted molecular events leading to the assembly of the surveillance complex (SURF), its transition to the decay‐inducing complex (DECID) leading to UPF1 phosphorylation and recruitment of SMG6 and/or SMG5/SMG7 that elicit mRNA degradation.
Under normal conditions UPF1 has low basal helicase activity; however, upon recognition of a PTC and subsequent binding of UPF1 to UPF2, this activity is greatly increased. Alongside this, a large conformational change of the CH inhibitory domain modifies the RNA‐binding properties and the catalytic activity of UPF1, causing a switch from an RNA‐clamping mode to an RNA‐unwinding mode (Chakrabarti et al, 2011). The active UPF1 helicase functions as an RNPase translocating along the mRNA with a 5′ to 3′ polarity, acting to resolve secondary structures, remove proteins from mRNA, and provide access to nucleases (Franks et al, 2010; Fiorini et al, 2015). Importantly, NMD can also be elicited on mRNAs that do not have a downstream EJC, although the mechanism of non‐EJC dependent NMD is less well defined (Bühler et al, 2006; He & Jacobson, 2015). Interestingly, alternative branches of the NMD pathway that act independently of UPF2, UPF3B, or the EJC have also been described (Gehring et al, 2005; Chan et al, 2007; Ivanov et al, 2008). In mammals, there are two highly related UPF3 paralogs, UPF3A and UPF3B (also called UPF3X due to its location in chromosome X). Recent studies have shown that UPF3A and UPF3B perform redundant functions and can activate NMD without EJC binding, suggesting that UPF3 paralogs play a more active role in NMD than simply bridging the EJC and the UPF complex. UPF1 almost exclusively associates with UPF3B and only minimally with UPF3A; however, when UPF3B is mutated or removed, the association of UPF1 with UPF3A is enhanced 4‐6 times, independent of RNA. Thus, UPF3A seems almost dispensable for NMD; however, it performs a compensatory role and can maintain NMD in the absence of UPF3B (Wallmeroth et al, 2022; Yi et al, 2022; Chen et al, 2023). In cells lacking both UPF3 paralogs, although NMD is not completely abrogated, its activity is reduced significantly (Bufton et al, 2022; Yi et al, 2022).
A current model postulates that UPF1 binding to mRNAs does not inevitably mark mRNAs for NMD‐mediated degradation. Use of CLIP (cross‐linking and immunoprecipitation) revealed that UPF1 binds target RNAs prior to mRNA translation. Once translating ribosomes are engaged, they displace UPF1 from coding sequences, leading to UPF1 enrichment at 3′UTRs (Hurt et al, 2013; Zünd et al, 2013). By contrast, binding of phosphorylated UPF1 (P‐UPF1) marks mRNAs for NMD‐mediated degradation, since P‐UPF1 is enriched on endogenous transcripts degraded by NMD, whereas unphosphorylated UPF1 is released from non‐targeted transcripts in an ATP‐dependent manner (Kurosaki et al, 2014; Lee et al, 2015). Interestingly, UPF1 mutants with substantially impaired processing and slower unwinding rates are still functional in NMD and still have the capacity to restore NMD functionality upon loss of WT UPF1 (Fig 3) (Chapman et al, 2022).
In addition to a rapidly increasing number of NMD regulators, it was recently shown that UPF1LL, an alternative mammalian‐specific isoform of the core NMD factor UPF1 (UPF1SL), harbors a regulatory loop that is 11‐residues longer and preferentially binds and down‐regulates a different subset of NMD targets through a reduction in the RNA dissociation potential (Fig 3) (Gowravaram et al, 2018; Fritz et al, 2022). Canonical NMD is mediated by UPF1SL that represents >75% of the total UPF1 pool and is inhibited by moderate translational repression. By contrast, the UPF1LL isoform triggers NMD in response to activation of the integrated stress response (ISR), and repression of translation, targeting novel mRNA, including stress response genes (Fritz et al, 2022). Interestingly, UPF1LL requires translation termination events; however, due to its improved RNA‐binding capacity in the presence of ATP compared to UPF1SL, its sustained RNA interaction favors a reduced frequency of termination events in conditions of attenuated translation. Therefore, under conditions of moderate translation inhibition where NMD is inhibited, UPF1LL activity is enhanced, changing the specificity of NMD in response to stress conditions (Fritz et al, 2022).
UPF1 phosphorylation represents a non‐reversible point in NMD progression during which the transcript is committed for degradation, leading to repression of further translation initiation, a key step in the NMD pathway (Isken et al, 2008). This phosphorylation event leads to the recruitment of additional NMD factors, namely the phospho‐binding proteins SMG6 and/or SMG5/SMG7, which function in two independent, yet overlapping pathways, and lead to further recruitment of nucleases to elicit mRNA degradation. The SMG6 endonuclease cleaves NMD targets in the vicinity of the PTC (Huntzinger et al, 2008; Eberle et al, 2009) and generates a 5′ cleavage product that is most likely degraded 3′ to 5′ by the exosome and/or DIS3L2 (DIS3‐like exonuclease 2; Kurosaki et al, 2018). The resulting 3′ cleavage product is cleared of protein components by UPF1 to provide access to the exoribonuclease XRN1 (Kurosaki et al, 2019). Alternatively, the heterodimer SMG5/SMG7 binds to P‐UPF1 and recruits the CCR4/NOT complex to promote deadenylation, leading to 3′ to 5′ decay (Loh et al, 2013) and decapping, resulting in XRN1‐catalyzed 5′ to 3′ degradation (Unterholzner & Izaurralde, 2004). Transcriptome profiling revealed that SMG6 and SMG7 act on essentially the same transcripts, suggesting extensive redundancy between the endo‐ and exonucleolytic decay pathways (Colombo et al, 2017). There is a tight regulation between these two RNA degrading pathways, since inactivation of the SMG5‐SMG7 pathway also abrogates the SMG6‐dependent mechanism, supporting a mechanism that involves UPF1 phosphorylation and SMG5‐SMG7 recruitment to access SMG6 activity (Boehm et al, 2021).
Additional NMD factors
Core NMD factors SMG1‐7 were initially discovered using genetic screens in C. elegans and in S. cerevisiae and were later found by homology searches in other species, including Arabidopsis, Drosophila, and mammals. Since then, a variety of experimental approaches have been employed to identify novel regulators of this pathway, most of which are still being investigated and therefore do not feature in models of NMD (Table 1) (Hug et al, 2016).
Table 1
Factors involved in nonsense‐mediated mRNA decay.
| Protein | Role in nonsense‐mediated mRNA decay | References |
|---|---|---|
| SMG1 | A phosphoinositide 3‐kinase (PI3K)‐like kinase that phosphorylates UPF1 | Yamashita et al (2001), Kashima et al (2006) |
| SMG8/9 | Two subunits of the SMG‐1 complex which negatively regulate SMG‐1 kinase activity | Yamashita et al (2009) |
| UPF1 | An ATP‐dependent RNA helicase of the SF1 superfamily, which undergoes cycles of phosphorylation and dephosphorylation | Sun et al (1998), Bhattacharya et al (2000), Kim and Maquat (2019) |
| UPF2 | Recruited following recognition of a PTC leading to conformational changes in UPF1 and formation of the DECID complex | Serin et al (2001), Chamieh et al (2008) |
| UPF3A/B | UPF3 paralogs that bridge the EJC and the surveillance complex | Serin et al (2001), Neu‐Yilik et al (2017) |
| SMG5/7 | A heterodimer that binds to phosphorylated UPF1 and recruits the exonucleolytic RNA degrading machinery | Unterholzner and Izaurralde (2004), Loh et al (2013) |
| SMG6 | An endonuclease that cleaves NMD targets in the vicinity of the PTC | Huntzinger et al (2008), Eberle et al (2009) |
| DHX34 | A DExH/D box helicase that activates NMD by promoting a transition from the SURF to DECID complex | Hug and Cáceres (2014) |
| NBAS | NMD factor localized at the membrane of the ER that recruits UPF1 to activate a local NMD response | Longman et al (2007, 2020) |
| MOV10 | A 5′ to 3′ RNA helicase that contributes to UPF1‐mediated mRNA target degradation | Gregersen et al (2014) |
| RUVBL1/2 | AAA+ adenosine triphosphatases involved in the early stages of NMD that interact with SMG‐1 and promote the formation of the SURF complex | Izumi et al (2010) |
| ICE1 | An EJC‐associated protein that promotes UPF3B recruitment to the EJC and provides a link between splicing and NMD | Baird et al (2018) |
| GIGYF2/EIF4E2 | Mediate translational repression following recognition of a PTC | Zinshteyn et al (2021) |
| AKT | AKT signaling leads to formation of alternative EJCs where AKT replaces UPF2. AKT‐mediated phosphorylation of UPF1 activates NMD. | Palma et al (2021), Cho et al (2022) |
| CWC22 | Essential splicing factor which interacts with eIFA3 to activate NMD | Alexandrov et al (2012), Barbosa et al (2012), Steckelberg et al (2012) |
| ABCE1 | Responsible for ribosome recycling necessary to initiate translation termination and initiation of NMD via SMG6‐mediated endonucleolytic pathway | Annibaldis et al (2020), Zhu et al (2020) |
We developed RNAi screens in C. elegans that resulted in the identification of novel NMD factors, including smgl‐2/DHX34 and smgl‐1/NBAS that are conserved throughout evolution and function in NMD in nematodes, zebrafish, and human cells (Longman et al, 2007, 2013; Anastasaki et al, 2011). DHX34, a DExH/D box RNA helicase, forms a complex with SMG1 and UPF1 and activates NMD by promoting the transition from the surveillance to the decay‐inducing complex (Hug & Cáceres, 2014; Melero et al, 2016). Heterozygous mutations in DHX34 were identified in four families affected with inherited acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). These mutations map to different domains of DHX34, and all germline variants identified in these families abrogated NMD activity (Rio‐Machin et al, 2020). DHX34 is also associated with the human spliceosomal catalytic C complex and regulates a large number of alternative splicing (AS) events in mammalian cells in culture, establishing a dual role for DHX34 in both NMD and pre‐mRNA splicing (Hug et al, 2022).
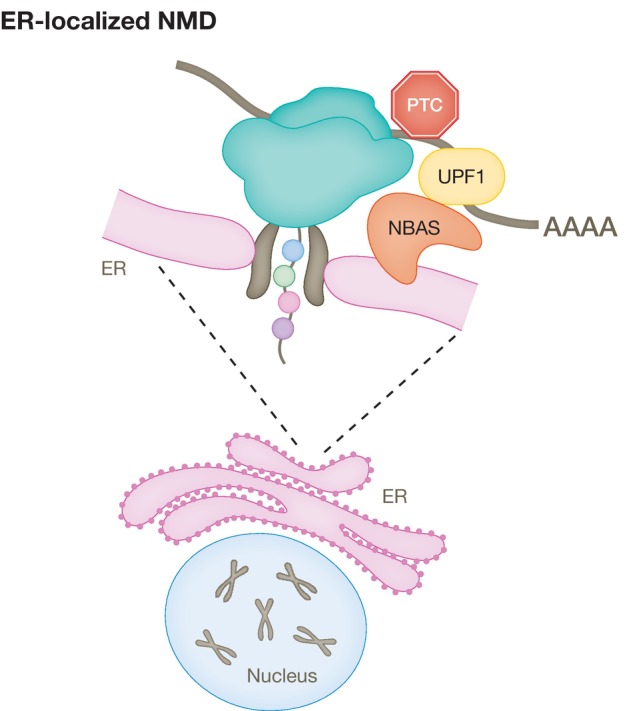
Neuroblastoma amplified sequence (NBAS) encodes a protein localized to the endoplasmic reticulum (ER) that is a component of the Syntaxin 18 complex and fulfils a role in Golgi‐to ER retrograde transport, which is independent of its function in NMD (Aoki et al, 2009; Longman et al, 2020; discussed below in the “Localized NMD and the stress response” section).
Interactome studies, RNAi screens, and newer CRISPR screens in mammalian cells in culture have led to the identification of novel NMD regulators, which are still being characterized (Table 1). The RNA helicase MOV10, a member of the UPF1‐like group helicase superfamily 1 (SF1), interacts with UPF1 and promotes degradation of UPF1‐regulated mRNA transcripts (Gregersen et al, 2014). An interactome of the SMG1 protein kinase identified the AAA ATPases, RuVB‐like 1 (RUVBL1) and RuVB‐like 2 (RUVBL2). These proteins are involved in a variety of cellular functions, including transcription and DNA repair, and were previously shown to have a role in the early stages of NMD (Izumi et al, 2010). We recently showed that RUVBL1/2 also interact with DHX34, coupling their ATPase activity to the assembly of factors required to initiate the NMD response (López‐Perrote et al, 2020).
A genome‐wide RNAi screen in human cells identified ICE1, an EJC‐associated factor that promotes the interaction of UPF3B with the EJC and activates NMD (Baird et al, 2018). CRISPR screens identified several candidate NMD genes (Alexandrov et al, 2017) and highlighted a role for the ribosome recycling factor ABCE1 in NMD (Annibaldis et al, 2020; Zhu et al, 2020). A CRISPR screen in K562 cells identified the translational repressors GIGYF2 and EIF4E2, suggesting a model wherein recognition of a stop codon as premature leads to its translational repression mediated by GIGYF2 and EIF4E2, a process shared with the NGD pathway (Zinshteyn et al, 2021). Finally, a haploid‐cell genetic screen for NMD effectors identified several components of the AKT signaling pathway. It was shown that AKT‐mediated phosphorylation of the UPF1 CH domain at T151D overcomes auto‐inhibition of UPF1 helicase activity, which is critical for NMD and decreases the dependence of helicase activity on ATP. AKT also promotes formation of EJCs that contain AKT at the expense of UPF2, potentially facilitating a UPF2 independent branch of NMD (Cho et al, 2022). Interestingly, AKT1 had been independently shown to phosphorylate UPF1 and activate NMD (Palma et al, 2021; Table 1).
NMD factors have also been shown to display additional cellular functions. This is particularly prominent in the case of UPF1 that not only functions in genome stability (Azzalin & Lingner, 2006), but also contributes to several RNA decay pathways (Kim & Maquat, 2019). These include the Staufen‐mediated decay pathway (SMD), which involves UPF1 recruitment to stem‐loops in the 3′UTRs of mRNAs bound by the RNA‐binding proteins STAUFEN1 and its paralog STAUFEN 2 (Kim et al, 2005; Park et al, 2013), and in the degradation of histone mRNAs, where UPF1 is recruited to target mRNAs via the stem‐loop binding protein (SLBP) (Kaygun & Marzluff, 2005). Interestingly, UPF1 has also been proposed to act as an E3‐ubiquitin ligase and promote degradation of the truncated polypeptide produced by translation of a PTC‐containing transcript (Takahashi et al, 2008; Feng et al, 2017), establishing a link between RNA degradation and protein decay (Kim & Maquat, 2019; Inglis et al, 2023). In S. cerevisiae, UPF1 facilitates proteasome degradation of truncated polypeptides in a ubiquitin dependent manner. The truncated protein product gets released from the ribosome following NMD‐mediated mRNA degradation, but remains associated with UPF1 which directs it to the proteasome for removal, thus leading to a protein and mRNA turnover coupled process (Kuroha et al, 2013).
NMD targets
Despite decades of research, it is still not entirely clear what constitutes a bona fide NMD target. The introduction of a mutation, such as a single nucleotide variant, which gives rise to a PTC, represents the most obvious target for NMD. However, transcriptome profiling to identify NMD targets in cells of different species revealed that the majority of NMD‐sensitive transcripts do not contain PTCs but are rather mRNAs coding for full‐length proteins. This led to the hypothesis that a combination of NMD‐inducing and NMD‐antagonizing features will contribute to determine NMD susceptibility for any given mRNA. Some of the common features that render mRNAs susceptible to NMD include the presence of a PTC located at least 50‐55 nucleotides upstream of an EJC (Colombo et al, 2017), mRNAs with upstream ORFs (uORFs; Calvo et al, 2009), and the presence of a long 3′UTR (Hogg & Goff, 2010). However, recent use of cDNA Nanopore sequencing combined with short RNA‐seq allowed the detection of full‐length NMD substrates that are highly unstable and only display an increase in RNA levels when NMD is inhibited. This analysis identified NMD target mRNAs derived mainly from alternative exon usage, yet it did not identify long 3′UTRs as a common feature for NMD regulated mRNAs (Karousis et al, 2021). RNA‐seq allows for the analysis of steady‐state changes, which are influenced by stability, degradation, and transcription rates. Recently, SH‐linked alkylation for the metabolic sequencing of RNA (SLAM‐seq) using 4‐thiouracil pulse‐chase labeling (Herzog et al, 2017) was used to accurately measure changes in RNA half‐lives and to identify new targets of the NMD pathway in S. cerevisiae (Alalam et al, 2022). SLAM seq analysis of Smg5‐7 genetic knockouts in mouse ESCs revealed that NMD controls expression levels of the translation initiation factor Eif4a2 and its alternative splicing isoform that harbors a PTC‐encoding isoform (Eif4a2 PTC ). Upon NMD inhibition, aberrant expression of the eIF4A2PTC elicits increased mTORC1 activity and translation rates and causes differentiation delays, highlighting a role of RNA stability regulation in development (Huth et al, 2022).
NMD regulation
The NMD pathway is dynamic and subject to regulation and impacts several physiological processes, such as the stress and immune responses. A correct magnitude of NMD activity is particularly important for proper brain function and indeed, NMD activity is extensively regulated during neural development (Jaffrey & Wilkinson, 2018). It was shown that NMD activity in human neuroblastoma cells is attenuated by fragile X protein FMRP, which is recruited to NMD targets by UPF1. FMRP acts as an NMD repressor in neural cells and in its absence, NMD is hyperactivated, leading to widespread transcriptome changes which contributes to intellectual disability and autism (Kurosaki et al, 2021). Recently, a conditional Smg6 mutant mouse model revealed that the NMD pathway has a role in controlling circadian clock regulation (Katsioudi et al, 2023). The NMD response varies within and across cell lines (Sato & Singer, 2021), in different cell tissues (Zetoune et al, 2008), and even among individuals (Nguyen et al, 2014; Rivas et al, 2015). NMD is tightly regulated by a negative feedback network that leads to a large proportion of core NMD factors being regulated by NMD itself in several organisms, including mammalian cells, nematodes, zebrafish, and plants (Huang et al, 2011; Yepiskoposyan et al, 2011; Longman et al, 2013). NMD can occur with equal probability during each round of translation of an mRNA molecule; however, this probability is variable and linked to sequence features, including the exon sequence downstream of the PTC, the PTC‐to‐intron distance, and the number of introns both upstream and downstream of the PTC. Furthermore, a subpopulation of mRNAs can escape NMD, further contributing to variation in NMD efficiency (Hoek et al, 2019). Use of a single‐cell approach comprising a bi‐directional NMD reporter expressing two β‐globin genes with or without a PTC in the same cell, allowed the characterization NMD efficiency in individual cells. This revealed a broad range of NMD efficiencies in the population (where some cells degraded essentially all mRNAs and others escaped NMD almost completely) and was correlated to the differential level of SMG1 expression and P‐UPF1. Mechanistically, this escape occurred either by translational read‐through at the PTC or by inefficient mRNA degradation following translation termination at the PTC (Sato & Singer, 2021).
Localized NMD and the stress response
The decay of NMD reporters in mammalian cells occurs in the cytoplasm (Trcek et al, 2013) and is closely linked to mRNA translation (Kervestin & Jacobson, 2012). We previously identified two novel NMD factors, NBAS and SEC13, which localize to the ER, raising the possibility that they could be involved in a localized NMD pathway (Longman et al, 2007; Casadio et al, 2015). There are precedents for a localized NMD response in neurons that regulate the expression of dendritic and axonal mRNAs upon the activation of their localized mRNA translation (Colak et al, 2013). It has been shown that mRNAs coding for secreted or transmembrane proteins are translated only when they encounter the ER (Wu et al, 2016). However, it remained largely unknown how NMD regulates the stability of RNAs translated at the ER, which due to their intrinsic localized translation, will not have sufficient exposure to cytoplasmic NMD surveillance. NBAS is a member of the Syntaxin 18 complex as part of the COPI vesicle and is involved in Golgi‐to‐ER retrograde transport (Aoki et al, 2009). We showed that NBAS fulfils a second, independent function and recruits the core NMD factor UPF1 to the membrane of the ER and activates a local NMD response that regulates RNAs associated with cellular stress and membrane trafficking (Fig 5; Longman et al, 2020). Loss‐of‐function mutations in NBAS have been found in diseases affecting bone, connective tissue, and acute liver failure (Hug et al, 2016; Staufner et al, 2020). We identified compound heterozygous variants in NBAS as a cause of atypical osteogenesis imperfecta (Balasubramanian et al, 2017). It remains to be seen whether the phenotype of NBAS mutations is due to faulty NMD response and/or defective Golgi‐to‐ER retrograde transport (Haack et al, 2015).
The Unfolded Protein Response (UPR) senses and responds to excessive amounts of misfolded proteins in the ER, that cause ER stress (Walter & Ron, 2011). Inappropriate UPR activation contributes to many human pathologies, most notably related to neurodegeneration (Hetz & Mollereau, 2014), therefore the fidelity of UPR activation must be tightly regulated. Accordingly, a role for NMD in regulating the UPR has been proposed (Karam et al, 2015; Goetz & Wilkinson, 2017). In particular, mRNAs encoding UPR components, including the UPR sensor IRE1α, as well as ATF‐4 and CHOP that are activated by PERK branch signaling, are regulated by NMD and thus control the threshold of cellular stress that is necessary to activate the UPR (Gardner, 2008; Karam et al, 2015; Sieber et al, 2016). A feed‐back loop mechanism operates, wherein NMD ensures a correct activation threshold for the UPR response and the activity of NMD is in turn down‐regulated by the UPR (Karam et al, 2015). It is likely that limiting the chronic activation of the UPR has a protective effect in neurodegenerative diseases. Along those lines, a protective role for UPF1 was shown in rodent primary neuronal models of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD; Barmada et al, 2015) and in a rat ALS paralysis model (Jackson et al, 2015).
It is also possible that modulation of ER‐NMD activity could be crucial to regulate ER stress (Longman et al, 2020). In such a scenario, NMD‐mediated degradation of RNA targets would decrease ER load by suppressing aggregation of truncated and/or misfolded proteins in the ER, limiting UPR activation. The newly identified UPF1LL that, upon ISR activation and impaired translation, regulates a new subset of NMD targets, can be a key to the appropriate cellular stress response (Fritz et al, 2022).
NMD in human disease
In vertebrates, NMD activity is essential for proper development, and loss of NMD leads to embryonic lethality (Chousal et al, 2022). There are several reports of mutations in core NMD factors that lead to human disease. Two consanguineous families with homozygous loss‐of‐function mutations in SMG9, a component of the SMG1c complex, display a multiple congenital anomaly syndrome, that includes heart and eye defects, and brain malformation (Shaheen et al, 2016). Four consanguineous families with four deleterious homozygous variants in SMG8 display a phenotype that resembles that found in patients with SMG9 mutations, including global developmental delay, microcephaly, facial dysmorphism, and variable congenital heart and eye malformations. These patients display increased phosphorylation of UPF1, which most likely reflect the loss of SMG8‐mediated inhibition of SMG1 kinase activity (Alzahrani et al, 2020). NMD has also an important role in neuronal development as demonstrated by the fact that mutations, as well as copy number variations in UPF2 and UPF3B, lead to intellectual disability and/or neurodevelopmental disorders in humans, including schizophrenia and autism spectrum disorder (Jolly et al, 2013; Nguyen et al, 2013; Jaffrey & Wilkinson, 2018). Altered levels of NMD activity were also implicated in the pathogenesis of C9orf72‐linked ALS/FTD (Xu et al, 2019; Ortega et al, 2020; Sun et al, 2020) and other neurodegenerative diseases (Kurosaki et al, 2021). For example, loss of the fragile X protein FMRP leads to intellectual disability and autism, and FMRP deficiency results in a hyperactivated NMD response in human cells (Kurosaki et al, 2021). UPF3B mutations cause intellectual disability with impairment of neural stem cell differentiation and reduction in neuronal branching, through a loss of UPF2 interaction leading to NMD abrogation (Bufton et al, 2022). Altogether, these data suggest that an imbalance in NMD activity could lead to neurodegeneration. Interestingly, NGD and NSD components Pelo and HBs1l are critical for cerebellum neurogenesis in mice but expendable for survival of these neurons after development (Terrey et al, 2021). Similar effects were observed upon deletion of the core NMD factor, Upf2. This suggests that several RNA quality control pathways may interact or have compensatory roles to drive early development.
The role of NMD in cancer is complex, since it can display both tumor suppressing and tumor enhancing roles (Wang et al, 2011; Tan et al, 2022). Cancer cells can exploit the NMD response through introduction of selective tumor suppressor mutations that initiate mRNA decay or via the introduction of NMD‐insensitive mutations in oncogenes to prevent their targeting (Lindeboom et al, 2016). For example, tumor cells are known to hijack the NMD system to suppress the expression of potent tumor suppressors such as WT1, BRCA1/2 and p53, inducing uncontrolled cell growth (Mort et al, 2008; Nogueira et al, 2021). A therapeutic potential for the treatment of cancer was suggested in a number of studies. Abrogating NMD in cancer may lead to the expression of tumor‐specific proteins that can increase natural immune responses directed against the tumor (Pastor et al, 2010). Furthermore, attenuation of NMD facilitates the response to cancer therapeutics, as shown in human breast cancer cells, subject to NMD inhibition. Combining this with the front‐line chemotherapeutic doxorubicin promotes a faster and more robust cancer‐cell killing by apoptosis (Popp & Maquat, 2015).
Conclusions
Translation‐coupled RNA quality control pathways play a central role in ensuring accurate gene expression. There is a great variety of such mechanisms that link defective mRNA translation with mRNA decay and degradation of truncated proteins. We have highlighted here three main pathways, NGD, NSD, and NMD. The first two share common features and trans‐acting factors and mainly differ in features present in the target mRNAs that trigger these pathways. NMD is the most studied mRNA surveillance system, both in terms of mechanism and trans‐acting factors, mostly due to its high impact as a potent buffering system for human disease. Given that the NMD pathway affects the phenotype of approximately one third of all genetic diseases (Holbrook et al, 2004; Mort et al, 2008), a better understanding of its regulation and factors that influence its function in vivo could be of importance for designing strategies to modulate the NMD response for therapeutic use. Challenges for the future involve the identification of all factors required for NMD regulation in a physiological setting, as well as a better understanding of what constitutes an NMD target.
Author contributions
Javier F Caceres: Conceptualization; writing – original draft; writing – review and editing. Laura Monaghan: Conceptualization; writing – original draft; writing – review and editing. Dasa Longman: Conceptualization; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
Javier F Caceres is a member of the Advisory Editorial Board of The EMBO Journal. This has no bearing on the editorial consideration of this article for publication.
Acknowledgements
We thank Craig Nicol (MRC IGC core facility) for help with preparation of figures. This work was supported by Core funding to the MRC Human Genetics Unit from the Medical Research Council (JFC).
Notes
The EMBO Journal (2023) 42: e114378 [Europe PMC free article] [Abstract] [Google Scholar]
References
- Alalam H, Zepeda‐Martínez JA, Sunnerhagen P (2022) Global SLAM‐seq for accurate mRNA decay determination and identification of NMD targets. RNA 28: 905–915 [Europe PMC free article] [Abstract] [Google Scholar]
- Alexandrov A, Colognori D, Shu M‐D, Steitz JA (2012) Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense‐mediated decay. Proc Natl Acad Sci USA 109: 21313–21318 [Europe PMC free article] [Abstract] [Google Scholar]
- Alexandrov A, Shu M‐D, Steitz JA (2017) Fluorescence amplification method for forward genetic discovery of factors in human mRNA degradation. Mol Cell 65: 191–201 [Europe PMC free article] [Abstract] [Google Scholar]
- Alzahrani F, Kuwahara H, Long Y, Al‐Owain M, Tohary M, AlSayed M, Mahnashi M, Fathi L, Alnemer M, Al‐Hamed MH et al (2020) Recessive, deleterious variants in SMG8 expand the role of nonsense‐mediated decay in developmental disorders in humans. Am J Hum Genet 107: 1178–1185 [Europe PMC free article] [Abstract] [Google Scholar]
- Anastasaki C, Longman D, Capper A, Patton EE, Cáceres JF (2011) Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res 39: 3686–3694 [Europe PMC free article] [Abstract] [Google Scholar]
- Annibaldis G, Domanski M, Dreos R, Contu L, Carl S, Klaÿ N, Mühlemann O (2020) Readthrough of stop codons under limiting ABCE1 concentration involves frameshifting and inhibits nonsense‐mediated mRNA decay. Nucleic Acids Res 48: 10259–10279 [Europe PMC free article] [Abstract] [Google Scholar]
- Aoki T, Ichimura S, Itoh A, Kuramoto M, Shinkawa T, Isobe T, Tagaya M (2009) Identification of the neuroblastoma‐amplified gene product as a component of the syntaxin 18 complex implicated in Golgi‐to‐endoplasmic reticulum retrograde transport. Mol Biol Cell 20: 2639–2649 [Europe PMC free article] [Abstract] [Google Scholar]
- Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439 [Abstract] [Google Scholar]
- Baird TD, Cheng KC‐C, Chen Y‐C, Buehler E, Martin SE, Inglese J, Hogg JR (2018) ICE1 promotes the link between splicing and nonsense‐mediated mRNA decay. eLife 7: e33178 [Europe PMC free article] [Abstract] [Google Scholar]
- Balasubramanian M, Hurst J, Brown S, Bishop NJ, Arundel P, DeVile C, Pollitt RC, Crooks L, Longman D, Caceres JF et al (2017) Compound heterozygous variants in NBAS as a cause of atypical osteogenesis imperfecta. Bone 94: 65–74 [Europe PMC free article] [Abstract] [Google Scholar]
- Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H (2012) Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol 19: 983–990 [Abstract] [Google Scholar]
- Barmada SJ, Ju S, Arjun A, Batarse A, Archbold HC, Peisach D, Li X, Zhang Y, Tank EMH, Qiu H et al (2015) Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci USA 112: 7821–7826 [Europe PMC free article] [Abstract] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense‐mediated mRNA decay. RNA 6: 1226–1235 [Europe PMC free article] [Abstract] [Google Scholar]
- Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE (2010) NMD: RNA biology meets human genetic medicine. Biochem J 430: 365–377 [Abstract] [Google Scholar]
- Boehm V, Kueckelmann S, Gerbracht JV, Kallabis S, Britto‐Borges T, Altmüller J, Krüger M, Dieterich C, Gehring NH (2021) SMG5‐SMG7 authorize nonsense‐mediated mRNA decay by enabling SMG6 endonucleolytic activity. Nat Commun 12: 3965 [Europe PMC free article] [Abstract] [Google Scholar]
- Brandman O, Hegde RS (2016) Ribosome‐associated protein quality control. Nat Struct Mol Biol 23: 7–15 [Europe PMC free article] [Abstract] [Google Scholar]
- Bufton JC, Powers KT, Szeto JA, Toelzer C, Berger I, Schaffitzel C (2022) Structures of nonsense‐mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder‐causing mutation. Nucleic Acids Res 50: 5934–5947 [Europe PMC free article] [Abstract] [Google Scholar]
- Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O (2006) EJC‐independent degradation of nonsense immunoglobulin‐mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 [Abstract] [Google Scholar]
- Cacciottolo M, Numitone G, Aurino S, Caserta IR, Fanin M, Politano L, Minetti C, Ricci E, Piluso G, Angelini C et al (2011) Muscular dystrophy with marked Dysferlin deficiency is consistently caused by primary dysferlin gene mutations. Eur J Hum Genet 19: 974–980 [Europe PMC free article] [Abstract] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [Europe PMC free article] [Abstract] [Google Scholar]
- Casadio A, Longman D, Hug N, Delavaine L, Vallejos Baier R, Alonso CR, Cáceres JF (2015) Identification and characterization of novel factors that act in the nonsense‐mediated mRNA decay pathway in nematodes, flies and mammals. EMBO Rep 16: 71–78 [Europe PMC free article] [Abstract] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E (2011) Molecular mechanisms for the RNA‐dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703 [Abstract] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93 [Abstract] [Google Scholar]
- Chan W‐K, Huang L, Gudikote JP, Chang Y‐F, Imam JS, MacLean JA, Wilkinson MF (2007) An alternative branch of the nonsense‐mediated decay pathway. EMBO J 26: 1820–1830 [Europe PMC free article] [Abstract] [Google Scholar]
- Chandrasekaran V, Juszkiewicz S, Choi J, Puglisi JD, Brown A, Shao S, Ramakrishnan V, Hegde RS (2019) Mechanism of ribosome stalling during translation of a poly(A) tail. Nat Struct Mol Biol 26: 1132–1140 [Europe PMC free article] [Abstract] [Google Scholar]
- Chapman JH, Craig JM, Wang CD, Gundlach JH, Neuman KC, Hogg JR (2022) UPF1 mutants with intact ATPase but deficient helicase activities promote efficient nonsense‐mediated mRNA decay. Nucleic Acids Res 50: 11876–11894 [Europe PMC free article] [Abstract] [Google Scholar]
- Chen C, Shen Y, Li L, Ren Y, Wang Z‐Q, Li T (2023) UPF3A is dispensable for nonsense‐mediated mRNA decay in mouse pluripotent and somatic cells. Life Sci Alliance 6: e202201589 [Europe PMC free article] [Abstract] [Google Scholar]
- Cho H, Abshire ET, Popp MW, Pröschel C, Schwartz JL, Yeo GW, Maquat LE (2022) AKT constitutes a signal‐promoted alternative exon‐junction complex that regulates nonsense‐mediated mRNA decay. Mol Cell 82: 2779–2796.e10 [Europe PMC free article] [Abstract] [Google Scholar]
- Chousal JN, Sohni A, Vitting‐Seerup K, Cho K, Kim M, Tan K, Porse B, Wilkinson MF, Cook‐Andersen H (2022) Progression of the pluripotent epiblast depends upon the NMD factor UPF2. Development 149: dev200764 [Europe PMC free article] [Abstract] [Google Scholar]
- Colak D, Ji S‐J, Porse BT, Jaffrey SR (2013) Regulation of axon guidance by compartmentalized nonsense‐mediated mRNA decay. Cell 153: 1252–1265 [Europe PMC free article] [Abstract] [Google Scholar]
- Colombo M, Karousis ED, Bourquin J, Bruggmann R, Mühlemann O (2017) Transcriptome‐wide identification of NMD‐targeted human mRNAs reveals extensive redundancy between SMG6‐ and SMG7‐mediated degradation pathways. RNA 23: 189–201 [Europe PMC free article] [Abstract] [Google Scholar]
- D’Orazio KN, Green R (2021) Ribosome states signal RNA quality control. Mol Cell 81: 1372–1383 [Europe PMC free article] [Abstract] [Google Scholar]
- D’Orazio KN, Wu CC‐C, Sinha N, Loll‐Krippleber R, Brown GW, Green R (2019) The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. Elife 8: e49117 [Europe PMC free article] [Abstract] [Google Scholar]
- Dave P, Roth G, Griesbach E, Mateju D, Hochstoeger T, Chao JA (2023) Single‐molecule imaging reveals translation‐dependent destabilization of mRNAs. Mol Cell 83: 589–606.e6 [Europe PMC free article] [Abstract] [Google Scholar]
- Doma MK, Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [Europe PMC free article] [Abstract] [Google Scholar]
- Eberle AB, Lykke‐Andersen S, Mühlemann O, Jensen TH (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [Abstract] [Google Scholar]
- Feng Q, Jagannathan S, Bradley RK (2017) The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol Cell 67: 239–251.e6 [Europe PMC free article] [Abstract] [Google Scholar]
- Fernández IS, Yamashita A, Arias‐Palomo E, Bamba Y, Bartolomé RA, Canales MA, Teixidó J, Ohno S, Llorca O (2011) Characterization of SMG‐9, an essential component of the nonsense‐mediated mRNA decay SMG1C complex. Nucleic Acids Res 39: 347–358 [Europe PMC free article] [Abstract] [Google Scholar]
- Fiorini F, Bagchi D, Le Hir H, Croquette V (2015) Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat Commun 6: 7581 [Europe PMC free article] [Abstract] [Google Scholar]
- Franks TM, Singh G, Lykke‐Andersen J (2010) Upf1 ATPase‐dependent mRNP disassembly is required for completion of nonsense‐mediated mRNA decay. Cell 143: 938–950 [Europe PMC free article] [Abstract] [Google Scholar]
- Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [Abstract] [Google Scholar]
- Fritz SE, Ranganathan S, Wang CD, Hogg JR (2022) An alternative UPF1 isoform drives conditional remodeling of nonsense‐mediated mRNA decay. EMBO J 41: e108898 [Europe PMC free article] [Abstract] [Google Scholar]
- Ganesan R, Mangkalaphiban K, Baker R, He F, Jacobson A (2022) Ribosome‐bound Upf1 forms distinct 80S complexes and conducts mRNA surveillance. RNA 28: 1621–1642 [Europe PMC free article] [Abstract] [Google Scholar]
- Gardner LB (2008) Hypoxic inhibition of nonsense‐mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol 28: 3729–3741 [Europe PMC free article] [Abstract] [Google Scholar]
- Gehring NH, Kunz JB, Neu‐Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE (2005) Exon‐junction complex components specify distinct routes of nonsense‐mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [Abstract] [Google Scholar]
- Glover ML, Burroughs AM, Monem PC, Egelhofer TA, Pule MN, Aravind L, Arribere JA (2020) NONU‐1 encodes a conserved endonuclease required for mRNA translation surveillance. Cell Rep 30: 4321–4331.e4 [Europe PMC free article] [Abstract] [Google Scholar]
- Goetz AE, Wilkinson M (2017) Stress and the nonsense‐mediated RNA decay pathway. Cell Mol Life Sci 74: 3509–3531 [Europe PMC free article] [Abstract] [Google Scholar]
- Gowravaram M, Bonneau F, Kanaan J, Maciej VD, Fiorini F, Raj S, Croquette V, Le Hir H, Chakrabarti S (2018) A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform‐specific manner. Nucleic Acids Res 46: 2648–2659 [Europe PMC free article] [Abstract] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M (2014) MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585 [Abstract] [Google Scholar]
- Haack TB, Staufner C, Köpke MG, Straub BK, Kölker S, Thiel C, Freisinger P, Baric I, McKiernan PJ, Dikow N et al (2015) Biallelic mutations in NBAS cause recurrent acute liver failure with onset in infancy. Am J Hum Genet 97: 163–169 [Europe PMC free article] [Abstract] [Google Scholar]
- Hall GW, Thein S (1994) Nonsense codon mutations in the terminal exon of the beta‐globin gene are not associated with a reduction in beta‐mRNA accumulation: a mechanism for the phenotype of dominant beta‐thalassemia. Blood 83: 2031–2037 [Abstract] [Google Scholar]
- Hanson G, Coller J (2018) Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19: 20–30 [Europe PMC free article] [Abstract] [Google Scholar]
- Harigaya Y, Parker R (2010) No‐go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA 1: 132–141 [Abstract] [Google Scholar]
- He F, Jacobson A (2015) Nonsense‐mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet 49: 339–366 [Europe PMC free article] [Abstract] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, Von Haeseler A, Zuber J, Ameres SL (2017) Thiol‐linked alkylation of RNA to assess expression dynamics. Nat Methods 14: 1198–1204 [Europe PMC free article] [Abstract] [Google Scholar]
- Hetz C, Mollereau B (2014) Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci 15: 233–249 [Abstract] [Google Scholar]
- Hickey KL, Dickson K, Cogan JZ, Replogle JM, Schoof M, D’Orazio KN, Sinha NK, Hussmann JA, Jost M, Frost A et al (2020) GIGYF2 and 4EHP inhibit translation initiation of defective messenger RNAs to assist ribosome‐associated quality control. Mol Cell 79: 950–962.e6 [Europe PMC free article] [Abstract] [Google Scholar]
- Hoek TA, Khuperkar D, Lindeboom RGH, Sonneveld S, Verhagen BMP, Boersma S, Vermeulen M, Tanenbaum ME (2019) Single‐molecule imaging uncovers rules governing nonsense‐mediated mRNA decay. Mol Cell 75: 324–339.e11 [Europe PMC free article] [Abstract] [Google Scholar]
- Hogg JR, Goff SP (2010) Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [Europe PMC free article] [Abstract] [Google Scholar]
- Holbrook JA, Neu‐Yilik G, Hentze MW, Kulozik AE (2004) Nonsense‐mediated decay approaches the clinic. Nat Genet 36: 801–808 [Abstract] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R (2002) Exosome‐mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [Abstract] [Google Scholar]
- Huang L, Lou C‐H, Chan W, Shum EY, Shao A, Stone E, Karam R, Song H‐W, Wilkinson MF (2011) RNA homeostasis governed by cell type‐specific and branched feedback loops acting on NMD. Mol Cell 43: 950–961 [Europe PMC free article] [Abstract] [Google Scholar]
- Hug N, Cáceres JF (2014) The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay‐inducing complex. Cell Rep 8: 1845–1856 [Europe PMC free article] [Abstract] [Google Scholar]
- Hug N, Longman D, Cáceres JF (2016) Mechanism and regulation of the nonsense‐mediated decay pathway. Nucleic Acids Res 44: 1483–1495 [Europe PMC free article] [Abstract] [Google Scholar]
- Hug N, Aitken S, Longman D, Raab M, Armes H, Mann AR, Rio‐Machin A, Fitzgibbon J, Rouault‐Pierre K, Caceres JF (2022) A dual role for the RNA helicase DHX34 in NMD and pre‐mRNA splicing and its function in hematopoietic differentiation. RNA 28: 1224–1238 [Europe PMC free article] [Abstract] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [Europe PMC free article] [Abstract] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB (2013) Global analyses of UPF1 binding and function reveal expanded scope of nonsense‐mediated mRNA decay. Genome Res 23: 1636–1650 [Europe PMC free article] [Abstract] [Google Scholar]
- Huth M, Santini L, Galimberti E, Ramesmayer J, Titz‐Teixeira F, Sehlke R, Oberhuemer M, Stummer S, Herzog V, Garmhausen M et al (2022) NMD is required for timely cell fate transitions by fine‐tuning gene expression and regulating translation. Genes Dev 36: 348–367 [Europe PMC free article] [Abstract] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, Inada T (2019) Collided ribosomes form a unique structural interface to induce Hel2‐driven quality control pathways. EMBO J 38: e100276 [Europe PMC free article] [Abstract] [Google Scholar]
- Inglis AJ, Guna A, Gálvez‐Merchán Á, Pal A, Esantsi TK, Keys HR, Frenkel EM, Oania R, Weissman JS, Voorhees RM (2023) Coupled protein quality control during nonsense‐mediated mRNA decay. J. Cell Sci 136: jcs261216 [Europe PMC free article] [Abstract] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JWB, Maquat LE (2008) Upf1 phosphorylation triggers translational repression during nonsense‐mediated mRNA decay. Cell 133: 314–327 [Europe PMC free article] [Abstract] [Google Scholar]
- Ito‐Harashima S, Kuroha K, Tatematsu T, Inada T (2007) Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev 21: 519–524 [Europe PMC free article] [Abstract] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [Europe PMC free article] [Abstract] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S (2010) AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense‐mediated mRNA decay. Sci Signal 3: ra27 [Abstract] [Google Scholar]
- Jackson KL, Dayton RD, Orchard EA, Ju S, Ringe D, Petsko GA, Maquat LE, Klein RL (2015) Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP‐43‐induced motor paralysis. Gene Ther 22: 20–28 [Europe PMC free article] [Abstract] [Google Scholar]
- Jaffrey SR, Wilkinson MF (2018) Nonsense‐mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat Rev Neurosci 19: 715–728 [Europe PMC free article] [Abstract] [Google Scholar]
- Jolly LA, Homan CC, Jacob R, Barry S, Gecz J (2013) The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet 22: 4673–4687 [Abstract] [Google Scholar]
- Juszkiewicz S, Hegde RS (2017) Initiation of quality control during poly(A) translation requires site‐specific ribosome ubiquitination. Mol Cell 65: 743–750.e4 [Europe PMC free article] [Abstract] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS (2018) ZNF598 is a quality control sensor of collided ribosomes. Mol Cell 72: 469–481.e7 [Europe PMC free article] [Abstract] [Google Scholar]
- Juszkiewicz S, Speldewinde SH, Wan L, Svejstrup JQ, Hegde RS (2020) The ASC‐1 complex disassembles collided ribosomes. Mol Cell 79: 603–614.e8 [Europe PMC free article] [Abstract] [Google Scholar]
- Karam R, Lou C‐HC‐H, Kroeger H, Huang L, Lin JH, Wilkinson MF (2015) The unfolded protein response is shaped by the NMD pathway. EMBO Rep 16: 599–609 [Europe PMC free article] [Abstract] [Google Scholar]
- Karousis ED, Mühlemann O (2022) The broader sense of nonsense. Trends Biochem Sci 47: 921–935 [Abstract] [Google Scholar]
- Karousis ED, Gurzeler L‐A, Annibaldis G, Dreos R, Mühlemann O (2020) Human NMD ensues independently of stable ribosome stalling. Nat Commun 11: 4134 [Europe PMC free article] [Abstract] [Google Scholar]
- Karousis ED, Gypas F, Zavolan M, Mühlemann O (2021) Nanopore sequencing reveals endogenous NMD‐targeted isoforms in human cells. Genome Biol 22: 223 [Europe PMC free article] [Abstract] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG‐1‐Upf1‐eRF1‐eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense‐mediated mRNA decay. Genes Dev 20: 355–367 [Europe PMC free article] [Abstract] [Google Scholar]
- Katsioudi G, Dreos R, Arpa ES, Gaspari S, Liechti A, Sato M, Gabriel CH, Kramer A, Brown SA, Gatfield D (2023) A conditional Smg6 mutant mouse model reveals circadian clock regulation through the nonsense‐mediated mRNA decay pathway. Sci. Adv 9: eade2828 [Europe PMC free article] [Abstract] [Google Scholar]
- Kaygun H, Marzluff WF (2005) Regulated degradation of replication‐dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [Abstract] [Google Scholar]
- Kervestin S, Jacobson A (2012) NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712 [Europe PMC free article] [Abstract] [Google Scholar]
- Kim YK, Maquat LE (2019) UPFront and center in RNA decay: UPF1 in nonsense‐mediated mRNA decay and beyond. RNA 25: 407–422 [Europe PMC free article] [Abstract] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [Abstract] [Google Scholar]
- Kim YJ, Nomakuchi T, Papaleonidopoulou F, Yang L, Zhang Q, Krainer AR (2022) Gene‐specific nonsense‐mediated mRNA decay targeting for cystic fibrosis therapy. Nat Commun 13: 2978 [Europe PMC free article] [Abstract] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R (2015) Ribosomes slide on lysine‐encoding homopolymeric A stretches. Elife 4: e05534 [Europe PMC free article] [Abstract] [Google Scholar]
- Kuroha K, Ando K, Nakagawa R, Inada T (2013) The Upf factor complex interacts with aberrant products derived from mRNAs containing a premature termination codon and facilitates their proteasomal degradation. J Biol Chem 288: 28630–28640 [Europe PMC free article] [Abstract] [Google Scholar]
- Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE (2014) A post‐translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 28: 1900–1916 [Europe PMC free article] [Abstract] [Google Scholar]
- Kurosaki T, Miyoshi K, Myers JR, Maquat LE (2018) NMD‐degradome sequencing reveals ribosome‐bound intermediates with 3′‐end non‐templated nucleotides. Nat Struct Mol Biol 25: 940–950 [Europe PMC free article] [Abstract] [Google Scholar]
- Kurosaki T, Popp MW, Maquat LE (2019) Quality and quantity control of gene expression by nonsense‐mediated mRNA decay. Nat Rev Mol Cell Biol 20: 406–420 [Europe PMC free article] [Abstract] [Google Scholar]
- Kurosaki T, Imamachi N, Pröschel C, Mitsutomi S, Nagao R, Akimitsu N, Maquat LE (2021) Loss of the fragile X syndrome protein FMRP results in misregulation of nonsense‐mediated mRNA decay. Nat Cell Biol 23: 40–48 [Europe PMC free article] [Abstract] [Google Scholar]
- Langer LM, Bonneau F, Gat Y, Conti E (2021) Cryo‐EM reconstructions of inhibitor‐bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8. Elife 10: e72353 [Europe PMC free article] [Abstract] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000a) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon‐exon junctions. EMBO J 19: 6860–6869 [Europe PMC free article] [Abstract] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE (2000b) Pre‐mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon‐exon junctions. Genes Dev 14: 1098–1108 [Europe PMC free article] [Abstract] [Google Scholar]
- Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke‐Andersen J (2015) Target discrimination in nonsense‐mediated mRNA decay requires Upf1 ATPase activity. Mol. Cell 59: 413–425 [Europe PMC free article] [Abstract] [Google Scholar]
- Lindeboom RGH, Supek F, Lehner B (2016) The rules and impact of nonsense‐mediated mRNA decay in human cancers. Nat Genet 48: 1112–1118 [Europe PMC free article] [Abstract] [Google Scholar]
- Loh B, Jonas S, Izaurralde E (2013) The SMG5‐SMG7 heterodimer directly recruits the CCR4‐NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27: 2125–2138 [Europe PMC free article] [Abstract] [Google Scholar]
- Longman D, Plasterk RHA, Johnstone IL, Cáceres JF (2007) Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085 [Europe PMC free article] [Abstract] [Google Scholar]
- Longman D, Hug N, Keith M, Anastasaki C, Patton EE, Grimes G, Cáceres JF (2013) DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans . Nucleic Acids Res 41: 8319–8331 [Europe PMC free article] [Abstract] [Google Scholar]
- Longman D, Jackson‐Jones KA, Maslon MM, Murphy LC, Young RS, Stoddart JJ, Hug N, Taylor MS, Papadopoulos DK, Cáceres JF (2020) Identification of a localized nonsense‐mediated decay pathway at the endoplasmic reticulum. Genes Dev 34: 1075–1088 [Europe PMC free article] [Abstract] [Google Scholar]
- López‐Perrote A, Castaño R, Melero R, Zamarro T, Kurosawa H, Ohnishi T, Uchiyama A, Aoyagi K, Buchwald G, Kataoka N et al (2016) Human nonsense‐mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res 44: 1909–1923 [Europe PMC free article] [Abstract] [Google Scholar]
- López‐Perrote A, Hug N, González‐Corpas A, Rodríguez CF, Serna M, García‐Martín C, Boskovic J, Fernandez‐Leiro R, Caceres JF, Llorca O (2020) Regulation of RUVBL1‐RUVBL2 AAA‐ATPases by the nonsense‐mediated mRNA decay factor DHX34, as evidenced by Cryo‐EM. Elife 9: e63042 [Europe PMC free article] [Abstract] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K et al (2017) Ubiquitination of stalled ribosome triggers ribosome‐associated quality control. Nat. Commun 8: 159 [Europe PMC free article] [Abstract] [Google Scholar]
- Matsuo Y, Tesina P, Nakajima S, Mizuno M, Endo A, Buschauer R, Cheng J, Shounai O, Ikeuchi K, Saeki Y et al (2020) RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1. Nat Struct Mol Biol 27: 323–332 [Abstract] [Google Scholar]
- Melero R, Hug N, Lopez‐Perrote A, Yamashita A, Caceres JF, Llorca O (2016) The RNA helicase DHX34 functions as a scaffold for SMG1‐mediated UPF1 phosphorylation. Nat. Commun 7: 10585 [Europe PMC free article] [Abstract] [Google Scholar]
- Metze S, Herzog VA, Ruepp M‐D, Mühlemann O (2013) Comparison of EJC‐enhanced and EJC‐independent NMD in human cells reveals two partially redundant degradation pathways. RNA 19: 1432–1448 [Europe PMC free article] [Abstract] [Google Scholar]
- Monem PC, Vidyasagar N, Piatt AL, Sehgal E, Arribere JA (2023) Ubiquitination of stalled ribosomes enables mRNA decay via HBS‐1 and NONU‐1 in vivo. PLoS Genet 19: e1010577 [Europe PMC free article] [Abstract] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA (2008) A meta‐analysis of nonsense mutations causing human genetic disease. Hum Mutat 29: 1037–1047 [Abstract] [Google Scholar]
- Nagy E, Maquat LE (1998) A rule for termination‐codon position within intron‐containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [Abstract] [Google Scholar]
- Neu‐Yilik G, Raimondeau E, Eliseev B, Yeramala L, Amthor B, Deniaud A, Huard K, Kerschgens K, Hentze MW, Schaffitzel C et al (2017) Dual function of UPF3B in early and late translation termination. EMBO J 36: 2968–2986 [Europe PMC free article] [Abstract] [Google Scholar]
- Nguyen LS, Kim H‐G, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gécz J (2013) Contribution of copy number variants involving nonsense‐mediated mRNA decay pathway genes to neuro‐developmental disorders. Hum Mol Genet 22: 1816–1825 [Abstract] [Google Scholar]
- Nguyen LS, Wilkinson MF, Gecz J (2014) Nonsense‐mediated mRNA decay: inter‐individual variability and human disease. Neurosci Biobehav Rev 46: 175–186 [Europe PMC free article] [Abstract] [Google Scholar]
- Nogueira G, Fernandes R, García‐Moreno JF, Romão L (2021) Nonsense‐mediated RNA decay and its bipolar function in cancer. Mol Cancer 20: 72 [Europe PMC free article] [Abstract] [Google Scholar]
- Ortega JA, Daley EL, Kour S, Samani M, Tellez L, Smith HS, Hall EA, Esengul YT, Tsai Y‐H, Gendron TF et al (2020) Nucleocytoplasmic proteomic analysis uncovers eRF1 and nonsense‐mediated decay as modifiers of ALS/FTD C9orf72 toxicity. Neuron 106: 90–107.e13 [Europe PMC free article] [Abstract] [Google Scholar]
- Palma M, Leroy C, Salomé‐Desnoulez S, Werkmeister E, Kong R, Mongy M, Le Hir H, Lejeune F (2021) A role for AKT1 in nonsense‐mediated mRNA decay. Nucleic Acids Res 49: 11022–11037 [Europe PMC free article] [Abstract] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE (2013) Staufen2 functions in Staufen1‐mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci USA 110: 405–412 [Europe PMC free article] [Abstract] [Google Scholar]
- Pastor F, Kolonias D, Giangrande PH, Gilboa E (2010) Induction of tumour immunity by targeted inhibition of nonsense‐mediated mRNA decay. Nature 465: 227–230 [Europe PMC free article] [Abstract] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV, Pisarev AV (2011) Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30: 1804–1817 [Europe PMC free article] [Abstract] [Google Scholar]
- Popp MW, Maquat LE (2015) Attenuation of nonsense‐mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun 6: 6632 [Europe PMC free article] [Abstract] [Google Scholar]
- Powers KT, Szeto J‐YA, Schaffitzel C (2020) New insights into no‐go, non‐stop and nonsense‐mediated mRNA decay complexes. Curr Opin Struct Biol 65: 110–118 [Abstract] [Google Scholar]
- Rio‐Machin A, Vulliamy T, Hug N, Walne A, Tawana K, Cardoso S, Ellison A, Pontikos N, Wang J, Tummala H et al (2020) The complex genetic landscape of familial MDS and AML reveals pathogenic germline variants. Nat Commun 11: 1044 [Europe PMC free article] [Abstract] [Google Scholar]
- Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M et al (2015) Effect of predicted protein‐truncating genetic variants on the human transcriptome. Science 348: 666–669 [Europe PMC free article] [Abstract] [Google Scholar]
- Saito S, Hosoda N, Hoshino SI (2013) The Hbs1‐Dom34 protein complex functions in non‐stop mRNA decay in mammalian cells. J Biol Chem 288: 17832–17843 [Europe PMC free article] [Abstract] [Google Scholar]
- Sato H, Singer RH (2021) Cellular variability of nonsense‐mediated mRNA decay. Nat Commun 12: 7203 [Europe PMC free article] [Abstract] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG‐4). Mol Cell Biol 21: 209–223 [Europe PMC free article] [Abstract] [Google Scholar]
- Shaheen R, Anazi S, Ben‐Omran T, Seidahmed MZ, Caddle LB, Palmer K, Ali R, Alshidi T, Hagos S, Goodwin L et al (2016) Mutations in SMG9, encoding an essential component of nonsense‐mediated decay machinery, cause a multiple congenital anomaly syndrome in humans and mice. Am J Hum Genet 98: 643–652 [Europe PMC free article] [Abstract] [Google Scholar]
- Shao S, von der Malsburg K, Hegde RS (2013) Listerin‐dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell 50: 637–648 [Europe PMC free article] [Abstract] [Google Scholar]
- Shoemaker CJ, Green R (2012) Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601 [Europe PMC free article] [Abstract] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R (2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl‐tRNA drop‐off to initiate no‐go decay. Science 330: 369–372 [Europe PMC free article] [Abstract] [Google Scholar]
- Sieber J, Hauer C, Bhuvanagiri M, Leicht S, Krijgsveld J, Neu‐Yilik G, Hentze MW, Kulozik AE (2016) Proteomic analysis reveals branch‐specific regulation of the unfolded protein response by nonsense‐mediated mRNA decay. Mol Cell Proteomics 15: 1584–1597 [Europe PMC free article] [Abstract] [Google Scholar]
- Simms CL, Thomas EN, Zaher HS (2016) Ribosome‐based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev RNA 8: 10.1002/wrna.1366 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sinha NK, Ordureau A, Best KM, Saba JA, Zinshteyn B, Sundaramoorthy E, Fulzele A, Garshott DM, Denk T, Thoms M et al (2020) EDF1 coordinates cellular responses to ribosome collisions. Elife 9: 1–84 [Europe PMC free article] [Abstract] [Google Scholar]
- Staufner C, Peters B, Wagner M, Alameer S, Barić I, Broué P, Bulut D, Church JA, Crushell E, Dalgıç B et al (2020) Defining clinical subgroups and genotype‐phenotype correlations in NBAS‐associated disease across 110 patients. Genet Med 22: 610–621 [Abstract] [Google Scholar]
- Steckelberg A‐L, Boehm V, Gromadzka AM, Gehring NH (2012) CWC22 connects pre‐mRNA splicing and exon junction complex assembly. Cell Rep 2: 454–461 [Abstract] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE (1998) A mutated human homologue to yeast Upf1 protein has a dominant‐negative effect on the decay of nonsense‐containing mRNAs in mammalian cells. Proc Natl Acad Sci USA 95: 10009–10014 [Europe PMC free article] [Abstract] [Google Scholar]
- Sun Y, Eshov A, Zhou J, Isiktas AU, Guo JU (2020) C9orf72 arginine‐rich dipeptide repeats inhibit UPF1‐mediated RNA decay via translational repression. Nat Commun 11: 3354 [Europe PMC free article] [Abstract] [Google Scholar]
- Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino S, Kontani K, Nishina H, Katada T (2008) Upf1 potentially serves as a RING‐related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14: 1950–1958 [Europe PMC free article] [Abstract] [Google Scholar]
- Tan K, Stupack DG, Wilkinson MF (2022) Nonsense‐mediated RNA decay: an emerging modulator of malignancy. Nat Rev Cancer 22: 437–451 [Europe PMC free article] [Abstract] [Google Scholar]
- Terrey M, Adamson SI, Chuang JH, Ackerman SL (2021) Defects in translation‐dependent quality control pathways lead to convergent molecular and neurodevelopmental pathology. Elife 10: e66904 [Europe PMC free article] [Abstract] [Google Scholar]
- Tesina P, Lessen LN, Buschauer R, Cheng J, Wu CC, Berninghausen O, Buskirk AR, Becker T, Beckmann R, Green R (2020) Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J 39: e103365 [Europe PMC free article] [Abstract] [Google Scholar]
- Thermann R, Neu‐Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [Europe PMC free article] [Abstract] [Google Scholar]
- Trcek T, Sato H, Singer RH, Maquat LE (2013) Temporal and spatial characterization of nonsense‐mediated mRNA decay. Genes Dev 27: 541–551 [Europe PMC free article] [Abstract] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T (2012) Dom34: Hbs1 plays a general role in quality‐control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell 46: 518–529 [Abstract] [Google Scholar]
- Tuck AC, Rankova A, Arpat AB, Liechti LA, Hess D, Iesmantavicius V, Castelo‐Szekely V, Gatfield D, Bühler M (2020) Mammalian RNA decay pathways are highly specialized and widely linked to translation. Mol Cell 77: 1222–1236.e13 [Europe PMC free article] [Abstract] [Google Scholar]
- Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [Abstract] [Google Scholar]
- Veltri AJ, D’Orazio KN, Lessen LN, Loll‐Krippleber R, Brown GW, Green R (2022) Distinct elongation stalls during translation are linked with distinct pathways for mRNA degradation. Elife 11: e76038 [Europe PMC free article] [Abstract] [Google Scholar]
- Wallefeld W, Krause S, Nowak KJ, Dye D, Horváth R, Molnár Z, Szabó M, Hashimoto K, Reina C, De Carlos J et al (2006) Severe nemaline myopathy caused by mutations of the stop codon of the skeletal muscle alpha actin gene (ACTA1). Neuromuscul Disord 16: 541–547 [Abstract] [Google Scholar]
- Wallmeroth D, Lackmann J, Kueckelmann S, Altmüller J, Dieterich C, Boehm V, Gehring NH (2022) Human UPF3A and UPF3B enable fault‐tolerant activation of nonsense‐mediated mRNA decay. EMBO J 41: e109191 [Europe PMC free article] [Abstract] [Google Scholar]
- Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086 [Abstract] [Google Scholar]
- Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB (2011) Inhibition of nonsense‐mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol 31: 3670–3680 [Europe PMC free article] [Abstract] [Google Scholar]
- Wu Q, Bazzini AA (2023) Translation and mRNA stability control. Annu Rev Biochem 92: 227–245 [Abstract] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH (2016) Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435 [Europe PMC free article] [Abstract] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, Devore ML, Castellano LA, Hand JM, Wright M, Bazzini AA (2019) Translation affects mRNA stability in a codon‐dependent manner in human cells. Elife 8: e45396 [Europe PMC free article] [Abstract] [Google Scholar]
- Xu W, Bao P, Jiang X, Wang H, Qin M, Wang R, Wang T, Yang Y, Lorenzini I, Liao L et al (2019) Reactivation of nonsense‐mediated mRNA decay protects against C9orf72 dipeptide‐repeat neurotoxicity. Brain 142: 1349–1364 [Europe PMC free article] [Abstract] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S (2001) Human SMG‐1, a novel phosphatidylinositol 3‐kinase‐related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense‐mediated mRNA decay. Genes Dev 15: 2215–2228 [Europe PMC free article] [Abstract] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T et al (2009) SMG‐8 and SMG‐9, two novel subunits of the SMG‐1 complex, regulate remodeling of the mRNA surveillance complex during nonsense‐mediated mRNA decay. Genes Dev 23: 1091–1105 [Europe PMC free article] [Abstract] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O (2011) Autoregulation of the nonsense‐mediated mRNA decay pathway in human cells. RNA 17: 2108–2118 [Europe PMC free article] [Abstract] [Google Scholar]
- Yi Z, Arvola RM, Myers S, Dilsavor CN, Abu Alhasan R, Carter BN, Patton RD, Bundschuh R, Singh G (2022) Mammalian UPF3A and UPF3B can activate nonsense‐mediated mRNA decay independently of their exon junction complex binding. EMBO J 41: e109202 [Europe PMC free article] [Abstract] [Google Scholar]
- Zetoune AB, Fontanière S, Magnin D, Anczuków O, Buisson M, Zhang CX, Mazoyer S (2008) Comparison of nonsense‐mediated mRNA decay efficiency in various murine tissues. BMC Genet 9: 83 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhu X, Zhang H, Mendell JT (2020) Ribosome recycling by ABCE1 links lysosomal function and iron homeostasis to 3′ UTR‐directed regulation and nonsense‐mediated decay. Cell Rep 32: 107895 [Europe PMC free article] [Abstract] [Google Scholar]
- Zinshteyn B, Sinha NK, Enam SU, Koleske B, Green R (2021) Translational repression of NMD targets by GIGYF2 and EIF4E2. PLoS Genet 17: e1009813 [Europe PMC free article] [Abstract] [Google Scholar]
- Zünd D, Gruber AR, Zavolan M, Mühlemann O (2013) Translation‐dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943 [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/153237989
Article citations
Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635(8037):237-242, 09 Oct 2024
Cited by: 0 articles | PMID: 39385025 | PMCID: PMC11540850
Calibrated ribosome profiling assesses the dynamics of ribosomal flux on transcripts.
Nat Commun, 15(1):7061, 26 Aug 2024
Cited by: 1 article | PMID: 39187487 | PMCID: PMC11347596
Selectively advantageous instability in biotic and pre-biotic systems and implications for evolution and aging.
Front Aging, 5:1376060, 16 May 2024
Cited by: 0 articles | PMID: 38818026 | PMCID: PMC11137231
Review Free full text in Europe PMC
UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2.
Nucleic Acids Res, 52(10):6036-6048, 01 Jun 2024
Cited by: 1 article | PMID: 38709891
Mechanisms and Delivery of tRNA Therapeutics.
Chem Rev, 124(12):7976-8008, 27 May 2024
Cited by: 1 article | PMID: 38801719
Review
Go to all (14) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
New insights into the interplay between the translation machinery and nonsense-mediated mRNA decay factors.
Biochem Soc Trans, 46(3):503-512, 06 Apr 2018
Cited by: 24 articles | PMID: 29626148 | PMCID: PMC6008592
Review Free full text in Europe PMC
Divergent effects of translation termination factor eRF3A and nonsense-mediated mRNA decay factor UPF1 on the expression of uORF carrying mRNAs and ribosome protein genes.
RNA Biol, 17(2):227-239, 17 Oct 2019
Cited by: 9 articles | PMID: 31619139 | PMCID: PMC6973328
From Yeast to Mammals, the Nonsense-Mediated mRNA Decay as a Master Regulator of Long Non-Coding RNAs Functional Trajectory.
Noncoding RNA, 7(3):44, 27 Jul 2021
Cited by: 13 articles | PMID: 34449682 | PMCID: PMC8395947
Review Free full text in Europe PMC
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.
Mol Biol Rep, 45(1):39-55, 27 Dec 2017
Cited by: 19 articles | PMID: 29282598
Review
Funding
Funders who supported this work.

 1
1