Abstract
Free full text

Chlamydia trachomatis Plasmid-Encoded Pgp4 Is a Transcriptional Regulator of Virulence-Associated Genes
Abstract
Chlamydia trachomatis causes chronic inflammatory diseases of the eye and genital tract and has global medical importance. The chlamydial plasmid plays an important role in the pathophysiology of these diseases, as plasmid-deficient organisms are highly attenuated. The cryptic plasmid carries noncoding RNAs and eight conserved open reading frames (ORFs). To understand plasmid gene function, we generated plasmid shuttle vectors with deletions in each of the eight ORFs. The individual deletion mutants were used to transform chlamydiae and the transformants were characterized phenotypically and at the transcriptional level. We show that pgp1, -2, -6, and -8 are essential for plasmid maintenance, while the other ORFs can be deleted and the plasmid stably maintained. We further show that a pgp4 knockout mutant exhibits an in vitro phenotype similar to its isogenic plasmidless strain, in terms of abnormal inclusion morphology and lack of glycogen accumulation. Microarray and qRT-PCR analysis revealed that Pgp4 is a transcriptional regulator of plasmid-encoded pgp3 and multiple chromosomal genes, including the glycogen synthase gene glgA, that are likely important in chlamydial virulence. Our findings have major implications for understanding the plasmid's role in chlamydial pathogenesis at the molecular level.
INTRODUCTION
Chlamydia trachomatis is an obligate intracellular human pathogen with a unique biphasic developmental growth cycle (1). It is the etiological agent of trachoma, the world's leading cause of preventable blindness and the most common cause of bacterial sexually transmitted disease (2–4). C. trachomatis isolates maintain a highly conserved plasmid of approximately 7.5 kb with copy numbers ranging from 4 to 10 copies per cell (5). Naturally occurring plasmidless clinical isolates are rare, indicating the plasmid's importance in chlamydial pathogenesis (6). Increased knowledge of the plasmid's role in biology and pathogenesis will enhance our understanding of chlamydial growth and development and will be important for guiding the design of live attenuated vaccine strains for the prevention of chlamydial disease.
Plasmid-deficient C. trachomatis strains and the murine pathogen Chlamydia muridarum have been studied in both nonhuman primate and murine infection models. A common theme emerging from these investigations is that in vivo infections with plasmid-deficient organisms either are asymptomatic (7, 8) or exhibit significantly reduced pathology (9), providing evidence that the plasmid plays an essential role in chlamydial pathogenesis. The molecular basis of plasmid-mediated virulence is poorly understood but is linked to enhanced proinflammatory cytokine stimulation by engagement of Toll-like receptors (TLR) in murine models (10–12). Notably, in a macaque model of trachoma, ocular infection with plasmid-deficient organisms generated no clinical pathology but induced strong protective immunity against challenge with fully virulent plasmid-bearing organisms, findings that support the use of plasmid-deficient organisms as novel live-attenuated chlamydial vaccines (7).
The chlamydial plasmid contains both noncoding RNAs (13–15) and eight open reading frames (ORFs). All eight ORFs, designated pgp1 to -8 (16), have been shown to be expressed in infected cells (11). Putative functions for several ORFs have been assigned based on homology to known proteins in the public databases (17–19). These are Pgp1, which is a DnaB like helicase; Pgp7 and -8, which are integrase/recombinase homologues; and Pgp5, which is a homologue of partitioning protein ParA. Pgp3, the most intensely studied plasmid protein, has been studied serologically (20), is secreted into the host cell cytosol (11, 21), and has been implicated as a potential TLR4 agonist (11) and a vaccine target (22, 23). Pgp2, -4, and -6 are chlamydia-specific proteins, showing little or no homology to proteins in the public databases. Importantly, the plasmid functions as a transcriptional regulator of various chromosomal genes which may be virulence factors important to chlamydial pathogenicity (10, 24, 25). The mechanism underlying this transcriptional regulation of chromosomal genes by the plasmid remains unknown.
The understanding of chlamydial pathogenesis at the molecular level has been hindered in the past by the lack of genetic tools. Very recently, chlamydial forward and reverse genetics systems, and chlamydial transformation, were described (26–29). However, there have been no reports of studies using these tools to characterize chlamydial genes of unknown function. Here, we describe the use of deletion mutagenesis and chlamydial transformation to genetically characterize the function of plasmid ORFs. We found that the ORFs can be grouped into two sets: ORFs that are essential (pgp1, -2, -6, and -8) and ORFs that are nonessential (pgp3 to -5 and -7) for stable plasmid maintenance in tissue culture. Transcriptional analyses using individual plasmid ORF deletion mutants showed that pgp4 is required for transcriptional regulation of both plasmid and chromosomal genes.
MATERIALS AND METHODS
Cell culture and chlamydial strains.
McCoy cells were grown at 37°C with 5% CO2 in high-glucose-containing Dulbecco's modified Eagle medium (DMEM) (Mediatech, Inc.) supplemented with 10% fetal bovine serum. C. trachomatis L2 (L2/LGV-434/Bu) and L2R [L2(25667R)] (30) elementary bodies (EBs) were purified from infected HeLa 229 cells by Renografin density gradient centrifugation as previously described (31). Strain L2R was originally isolated by Schachter and Osoba (32) and was kindly provided by Luis de la Maza (University of California, Irvine, CA).
Construction of pBRCT.
Plasmid pL2 was purified from L2 EBs using a QIAquick plasmid miniprep kit (Qiagen). pL2 was partially digested with SspI and amplified with primers JHC389 and JHC390 (see Table S1 in the supplemental material) using an Expand long-range PCR amplification kit (Roche). pBR322 and the PCR product were doubly digested with EagI and SalI, and the desired products were gel purified using a QIAEX II kit (Qiagen). The restriction-digested PCR product was ligated into the pBR322 backbone and transformed into Escherichia coli TOP10 (Invitrogen). Clone pBRCT#5 was sequenced, and six mutations were identified in the pL2 portion. All six mutations were replaced using three separately cloned EagI/BglII, BglII/MfeI, and MfeI/SalI fragments of the chlamydial plasmid. Sequence analysis of the final pBRCT clone identified a single nucleotide polymorphism (SNP) relative to the predicted sequence. It is a C-to-T transition located 190 bp downstream of the EagI site in the 3′ truncated portion of the pBR322 tet ORF.
Generation of pBRCT deletion derivatives.
Deletion derivatives of pBRCT were constructed using a modified PCR-based approach (33). Primer pairs for knocking out each individual ORF of pL2 were as shown in Table S1 in the supplemental material. Briefly, PCR amplifications were performed in 100-μl reaction volumes containing 10 ng pBRCT, a 200 nM concentration of each deoxynucleoside triphosphate (dNTP), a 0.1 nM concentration of each primer, 2 U Phusion high-fidelity (HF) DNA polymerase (New England BioLabs [NEB]), and 20 μl Phusion HF buffer. PCR conditions were as follows: initial denaturation at 98°C for 1 min, followed by 25 cycles of amplification at 98°C for 10 s, 60°C for 15 s, and 72°C for 3.5 min, followed by final extension at 72°C for 10 min. PCR products were purified by a QIAquick PCR purification kit (Qiagen) and doubly digested with DpnI (to remove template DNA) and NcoI-HF (NEB). Digested products were repurified using the QIAquick PCR purification kit. The resulting digested PCR products were ligated using Quick T4 DNA ligase (NEB) and transformed into E. coli TOP10. Clones were screened for plasmids containing all the pL2 ORFs but lacking the deleted region by PCR. The resulting plasmids were purified using a plasmid maxikit (Qiagen) and subjected to sequencing analysis and transformation.
Transformation of C. trachomatis L2R.
The initial transformation experiments with pBRCT and its eight derivatives followed the procedures described by Wang et al. (28). Subsequent transformations using pBRCT and the pgp1, -2, -6, and -8 knockout plasmids were performed following a simplified transformation procedure. Briefly, 10 μl density gradient-purified EBs (1 × 108 infection-forming units [IFU]) and 6 μl plasmid DNA (6 μg) were added to 200 μl CaCl2 buffer. After gentle pipetting 10 times, the mixture was incubated for 30 min at room temperature. The plasmid-EB mix was added to 12 ml ice-cold sucrose phosphate glutamate buffer and gently mixed. Two ml of the EB-plasmid mixture was then added to each well of McCoy cells (1.5 × 106 cells per well) grown on a 6-well plate. After centrifugation at 545 × g for 1 h at 20°C, the inoculum was removed and 3 ml of medium without cycloheximide or penicillin was added to each well. The infected cells were incubated at 37°C in 5% CO2 for 40 h. The cultures were then passaged with penicillin selection as described by Wang et al. (28).
Chlamydial culture and purification.
All stable C. trachomatis transformants were plaque purified as described previously by Matsumoto et al. (34). Recombinant plasmids, isolated from plaque-purified chlamydial transformants, were resequenced to confirm the expected sequence of each vector. Chlamydial transformants were then propagated in McCoy cells to make EB stocks. Large-scale cultures of Chlamydia were harvested and EBs purified by 30% density gradient centrifugation and stored at −80°C as previously described (31).
Microscopy and glycogen staining.
Monolayers of McCoy cells grown in 24-well plates were infected with C. trachomatis strains at a multiplicity of infection (MOI) of 0.3. Infected cultures were incubated at 37°C for 40 h. Phase-contrast images were captured at a magnification of ×200 using a Nikon eclipse TS100 inverted microscope to record the inclusion morphology of C. trachomatis strains. The growth media were removed and the monolayers directly stained with 1 ml 1% Lugol's iodine solution for photomicroscopy.
Transcription analysis.
Six 6-well plates containing McCoy cells (1.5 × 106 cells per well) were infected with L2, L2R, L2Rp+, L2RpΔpgp4, or L2RpΔpgp5 (MOI of 1.5) or mock infected and grown for 24 h at 37°C with 5% CO2. Each infection group consisted of six replicates. The medium was removed by aspiration. RNA and genomic DNA were extracted in random order using TRIzol LS (Life Technologies) according to the manufacturer's recommendations. DNA microarray targets were prepared as described by Omsland et al. (35). Chip hybridizations with Affymetrix GeneChip arrays (24) were as done described previously (36, 37).
Microarray data were analyzed using Expression Console (Affymetrix), Partek Genomics Suite (Partek), and Excel (Microsoft) as previously described (24, 38). Quantile normalized values were subjected to a principal-components analysis. A two-way analysis of variance (ANOVA) was performed within Partek to obtain multiple-test corrected P values using the false-discovery-rate method at the 0.05 significance level and was combined with a 2-fold-change cutoff, signal above background, and call consistency as calculated using custom Excel templates for each comparison. qRT-PCR validation was performed using the primers listed in Table S1 in the supplemental material as previously described (24). A two-tailed Student t test was used for qRT-PCR analysis.
Microarray data accession number.
The microarray data were deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE40979.
RESULTS
Chlamydial shuttle vector and open reading frame mutagenesis.
The shuttle vector pBRCT (Fig. 1A), similar to the previously described pBR325::L2 (28), was constructed. The difference between these two chimeric plasmids is that pBRCT retains all plasmid ORFs intact while pBR325::L2 has a disrupted pgp7. A PCR-based deletion mutagenesis approach was adopted to generate specific deletions in each of the eight chlamydial ORFs (Fig. 1B). In an attempt to minimize the chance of polar effects on downstream gene expression, the transcriptional start site (TSS) and an ~100-bp region upstream (5′) of the TSS was left intact for each ORF targeted for knockout. Discrepancies exist in the reported TSSs identified by two different groups (Table 1) (13, 39). For ORFs where conflicting TSSs have been reported, we chose the TSSs located the farthest upstream from the start codon for primer design. Another strategy employed was to introduce frameshift mutations in partially deleted ORFs with otherwise conserved 5′ ends (i.e., pgp1, -2, -6, and -8). The noncoding RNA (ncRNA) antisense to the 3′ end of pgp8 (15) was left intact in the pgp8 knockout. Conversely, another noncoding RNA (pL2-sRNA1), antisense to the pgp5 gene (13), was codeleted in the pgp5 knockout.
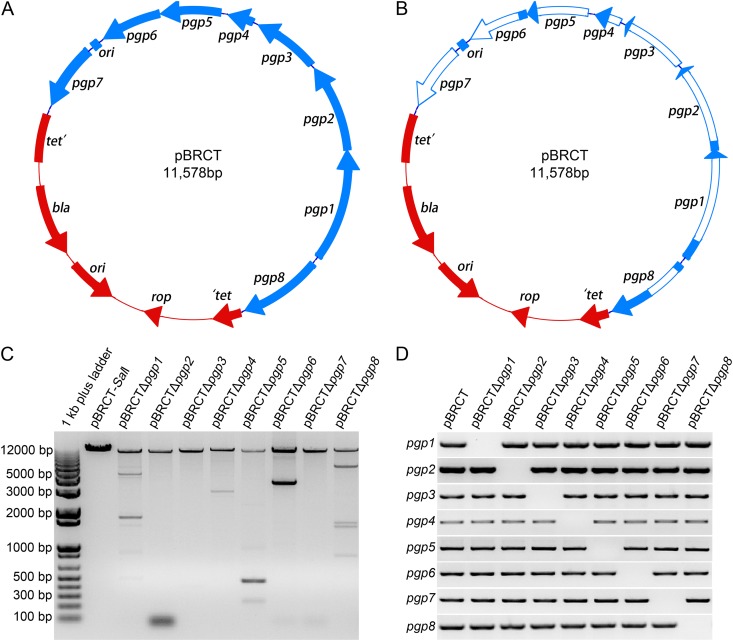
Construction of chlamydial shuttle vector and eight individual ORF knockout vectors. (A and B) The circles represent the plasmid from C. trachomatis L2 (blue) and the vector pBR322 (red). The coding sequences and the direction of transcription are represented by the arrows on the circle. Functional elements of pBRCT, including β-lactamase gene (bla), gene coding for the ROP protein (rop), origin of replication (ori), inactivated tetracycline resistance gene (tet′), and eight pL2 ORFs (pgp1 to -8) are shown. (A) Vector pBRCT harbors eight intact pL2 plasmid ORFs. (B) Schematic representation of deleted regions (white) of pBRCT in eight individual plasmid ORF knockout vectors. (C) Agarose electrophoresis of SalI digestion products of pBRCT and eight cleaned PCR products. The top band in each lane represents the expected-size amplicon for the construction of each individual plasmid ORF knockout vector. (D) Whole-length PCR products of eight pL2 ORFs show that each individual plasmid ORF knockout vector lacks one correct-size amplicon at its corresponding position in agarose electrophoresis, in comparison to pBRCT, which has all expected-size amplicons.
Table 1
Comparison of different transcriptional start sites of plasmid ORFs and deletion positions of eight individual plasmid-ORF knockouts
| Plasmid ORF (18) | TSS(s) identified by: | ORF positiona | Deleted region | |
|---|---|---|---|---|
| Albrecht et al. (13) | Ricci et al. (39) | |||
| pgp1 | 4140b | 4107, 4108 | 4068–2713 | 3827–2804 |
| pgp2 | 2703b | 4107, 4108 | 2719–1655 | 2603–1712 |
| pgp3 | 1611b | 1593–799 | 1593–857 | |
| pgp4 | 756 | 758b | 729–421 | 729–577 |
| pgp5 | 476 | 478b | 330–7098 | 330–7218 |
| pgp6 | 7117b | 7101–6358 | 6997–6383 | |
| pgp7 | 6157b | 6157 | 6126–5344 | 6126–5344 |
| pgp8 | 3926,b 4108 | 4079 | 4167–5159 | 4241–4668 |
We then identified the desired deletion endpoints for each ORF (Table 1), designed primers with a unique NcoI site at their 5′ ends (see Table S1 in the supplemental material) flanking the desired deletion region, and performed PCRs with pBRCT using each primer pair as described in Materials and Methods (Fig. 1C). After digestion with DpnI and NcoI, purified PCR products were ligated and transformed into E. coli TOP10. Positive transformants were screened by PCR with primer pairs specific for each ORF (Fig. 1D), and the desired plasmid construct was confirmed by sequencing. The plasmid ORF knockout vectors are referred to as pBRCTΔpgp1, -Δpgp2, -Δpgp3, -Δpgp4, -Δpgp5, -Δpgp6, -Δpgp7, and -Δpgp8. pgp3 to -5 and -7 are completely deleted, while the other four ORFs contain internal deletions.
pgp1, -2, -6, and -8 are essential for plasmid maintenance.
Chlamydiae are obligate intracellular parasites with a biphasic developmental cycle, and as a result, it is difficult to accurately calculate two parameters, transformation frequency and stability of transformants, typically used to assess the success of transformation with free-living bacteria. Here we define the stability of the transformed chlamydiae as the ability to pass the infection three times in cell culture in the presence of antibiotic selection. The stability of the transformants is assessed by the visual appearance of chlamydial inclusions. Stable transformants have normal-appearing inclusion morphology and growth kinetics, with abundant EB movement typical of L2 infection, on each successive passage in the presence of penicillin selection. In contrast, the transient transformants lose their penicillin-resistant phenotype altogether within two passages, and the unstable transformants always give a mixture of normal-appearing inclusions and aberrant inclusions, typical of wild-type penicillin-sensitive chlamydiae, on successive passes in the presence of penicillin selection.
Transformations of a naturally occurring plasmidless C. trachomatis strain L2R with pBRCT and its derivatives were performed. Transformation results are summarized in Table 2. Stable transformants carrying pBRCT and pBRCTΔpgp3, -Δpgp4, -Δpgp5, and -Δpgp7, referred to as L2Rp+, L2RpΔpgp3, L2RpΔpgp4, L2RpΔpgp5, and L2RpΔpgp7, respectively, were isolated and plaque cloned. These results indicate that Pgp3, -4, -5, and -7 are not required for stable plasmid maintenance in chlamydial growth in vitro.
Table 2
Viability and stability of C. trachomatis L2R transformed with pBRCT and eight pBRCT deletion derivatives
| Vector | Presence of penicillin-resistant transformants | Phenotypea |
|---|---|---|
| pBRCT | + | S |
| pBRCTΔpgp1 | − | NA |
| pBRCTΔpgp2 | − | NA |
| pBRCTΔpgp3 | + | S |
| pBRCTΔpgp4 | + | S |
| pBRCTΔpgp5 | + | S |
| pBRCTΔpgp6 | + | T |
| pBRCTΔpgp7 | + | S |
| pBRCTΔpgp8 | + | U |
Transformation with the other four deletion vectors yielded either transient/unstable transformants (pBRCTΔpgp6 and -Δpgp8) or no transformants (pBRCTΔpgp1 and -Δpgp2), suggesting that pgp1, -2, -6, and -8 are essential genes for the isolation of stable transformants. This result is in keeping with the assigned functions of Pgp1 and -8, based on homology to proteins known to be involved in plasmid maintenance in other bacterial systems, a helicase and integrase/recombinase, respectively. The inability to obtain stable transformants with pBRCTΔpgp2 suggests that Pgp2 is also involved in plasmid maintenance, although surprisingly, as Pgp2 is a chlamydia-specific protein, it has little or no homology to nonchlamydial proteins deposited in the protein databases.
pgp4 null mutants exhibit an L2R-like in vitro phenotype.
When grown in cell culture, L2 and L2R display two distinguishing phenotypes: inclusion morphology and glycogen accumulation (24). Of the four stable mutants, (pBRCTΔpgp3, -Δpgp4, -Δpgp5, and -Δpgp7), only the pgp4 deletion mutant exhibits an L2R-like phenotype, including late infection inclusion morphology, with a central dense amorphous structure surrounded by empty space, and dramatically decreased glycogen accumulation, as demonstrated by a lack of iodine staining (Fig. 2).
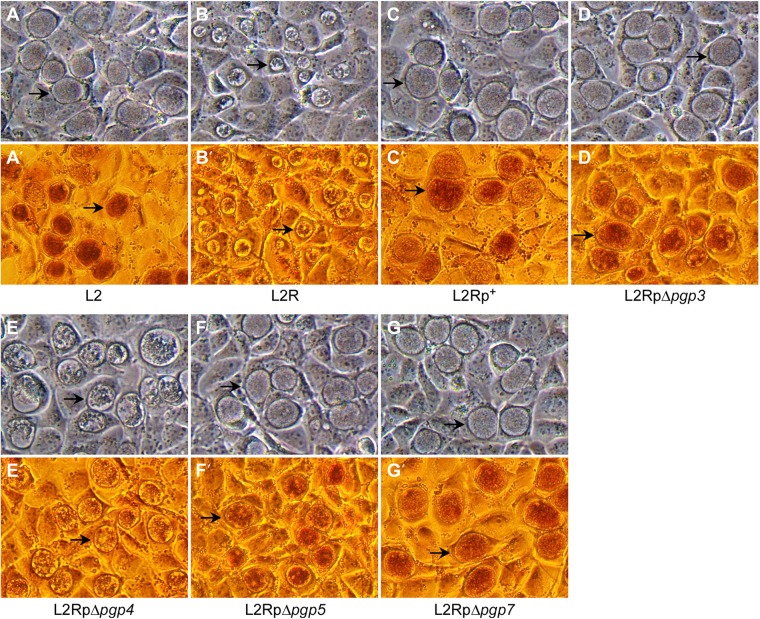
Phase microscopy of McCoy cell monolayers infected with L2, L2R, and stable transformants of L2R at an MOI of 0.3 after 40 h p.i. (magnification, ×200). Arrows indicate mature inclusions. (A to G) The inclusions of L2R and L2RpΔpgp4 mutants are morphologically distinct from those of L2, L2Rp+, L2RpΔpgp3, L2RpΔpgp5, and L2RpΔpgp7. (A′ to G′) The inclusions of L2R and L2RpΔpgp4 mutants stain negative for glycogen.
Pgp4 is a transcriptional regulator of plasmid and chromosomal genes.
We next compared the transcriptional profiles of L2-, L2R-, L2Rp+-, L2RpΔpgp4-, and L2RpΔpgp5-infected McCoy cells by microarray analyses. L2RpΔpgp5 was included as we surmised that pgp5 and its antisense ncRNA, one of the most abundant RNAs in L2 (13), could also play a transcriptional regulatory role. Microarray data were collected from six independent culture replicates representing each of the plasmid constructs. Principal component analysis demonstrated tight clustering of replicates within each plasmid construct group and good distance between replicate groups generating three distinct clusters composed of (i) L2, L2Rp+, and L2RpΔpgp5, (ii) L2R, and (iii) L2RpΔpgp4 (Fig. 3A).
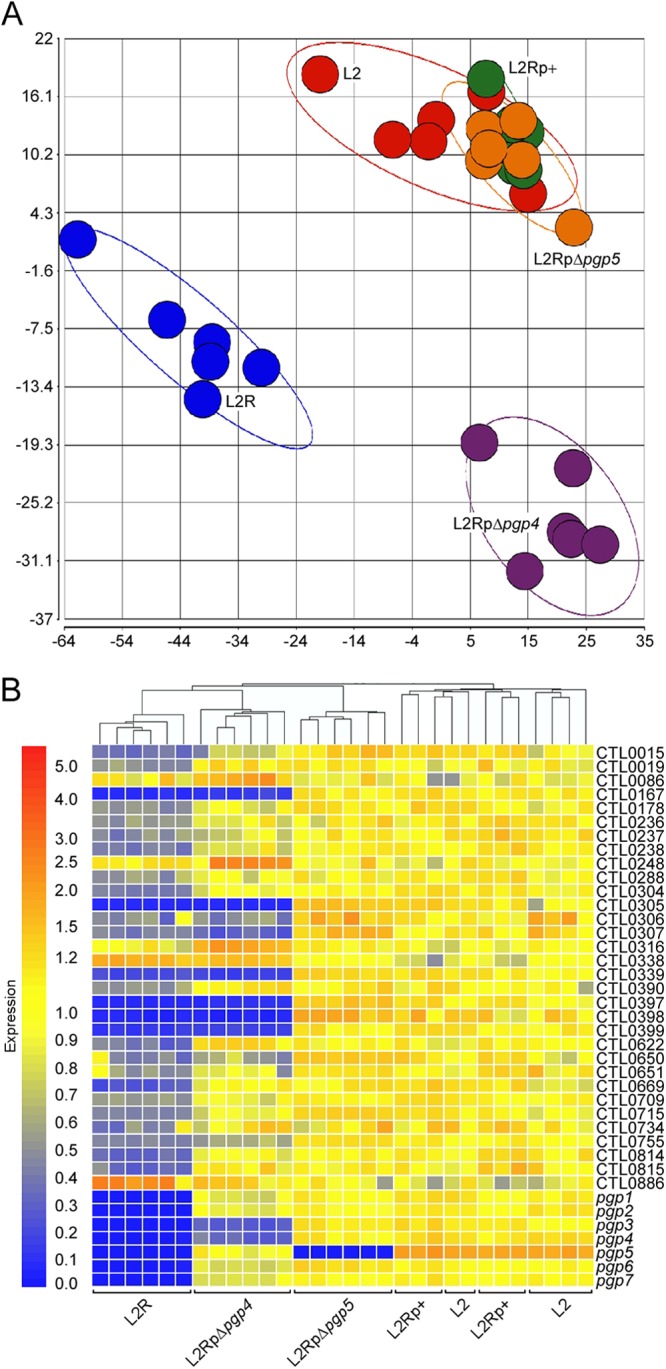
Microarray analyses identify Pgp4 as a transcriptional regulator of plasmid and chromosomal genes. (A) Principal components analysis of quantile-normalized microarray data of strains L2, L2R, L2p+, L2RpΔpgp4, and L2RpΔpgp5 at 24 h p.i. Each colored circle represents all C. trachomatis data produced from a single chip, and grouping and separation of replicates and strains, respectively, are demonstrated by each colored ellipse (red, L2; blue, L2R; green, L2Rp+; purple, L2RpΔpgp4; orange, L2RpΔpgp5). (B) Hierarchical cluster analysis of microarray results showed a close association in gene expression between L2RpΔpgp4 and L2R. Thirty-nine genes that passed the test criteria in at least one microarray comparison analysis were selected. Four pairwise comparisons were performed: L2 versus L2R, L2 versus L2Rp+, L2Rp+ versus L2pΔpgp4, and L2Rp+ versus L2RpΔpgp5. Yellow indicates signal intensity equal to the reference intensity of L2; red indicates a greater intensity, and blue indicates a lower intensity.
Hierarchical clustering analysis based on differentially expressed genes was then performed to compare the gene expression patterns between the five groups (Fig. 3B). No significant transcriptional differences were observed in the L2-versus-L2Rp+ and L2Rp+-versus-L2RpΔpgp5 comparisons, indicating that pBRCT complemented the wild-type endogenous plasmid at the transcript level. This also demonstrated that neither Pgp5 nor its antisense ncRNA has a role in regulating chromosomal gene expression. Differentially expressed genes were observed in the L2-versus-L2R and L2Rp+-versus-L2RpΔpgp4 pairwise comparisons. Overall, 39 transcripts exhibiting a ≥2-fold differential passed all test criteria (Table 3). In addition to the chromosomal genes, the pgp4 mutant also exhibited decreased expression levels of Pgp3, a potential virulence factor (11, 21). Clustering analysis of these 39 genes generated a hierarchical tree showing a strong correlation between L2RpΔpgp4 and L2R. Noticeably, the six genes showing the greatest differential in expression (>5-fold change) were consistently found in both L2-versus-L2R and L2Rp+-versus-L2RpΔpgp4 comparisons. One discrepancy between the present and our previous L2-versus-L2R comparison (24) is the expression of CTL0638, which was previously identified as a downregulated gene and was found here to be <2-fold decreased (−1.54-fold) in L2R. This discrepancy was addressed in the qRT-PCR experiments described below.
Table 3
Genes demonstrating a 2-fold or greater transcript differential by microarray analysis in pairwise comparisonsa
| L2 gene or CTL no. | Gene annotation | Fold change | |
|---|---|---|---|
| L2 vs L2R | L2Rp+ vs L2RpΔpgp4 | ||
| pgp1 | −1666.80 | −1.21 | |
| pgp2 | −340.04 | −1.11 | |
| pgp3 | −282.78 | −4.03 | |
| pgp4 | −65.26 | −3.65 | |
| pgp5 | −1890.48 | −1.83 | |
| pgp6 | −205.90 | −1.23 | |
| pgp7 | −607.15 | −1.18 | |
| 0015 | Hypothetical | −3.17 | −1.66 |
| 0019 | Hypothetical | −2.21 | 1.24 |
| 0086 | fliI | 1.18 | 2.00 |
| 0167 | glgA | −18.32 | −8.57 |
| 0178 | Hypothetical | −2.36 | −1.14 |
| 0236 | copB2 | −2.38 | −1.19 |
| 0237 | lcrH2 | −2.82 | −1.44 |
| 0238 | Hypothetical | −2.97 | −1.31 |
| 0248 | pmpE | 1.03 | 2.69 |
| 0288 | recD | −2.14 | −1.20 |
| 0304 | Hypothetical | −2.84 | −1.21 |
| 0305 | Hypothetical | −15.93 | −18.47 |
| 0306 | Hypothetical | −2.99 | −2.03 |
| 0307 | Hypothetical | −3.32 | −3.38 |
| 0316 | flhA | −1.16 | 2.13 |
| 0338 | Hypothetical | 1.58 | 2.19 |
| 0339 | pld | −5.92 | −7.13 |
| 0390 | Hypothetical | −2.12 | 1.20 |
| 0397 | Hypothetical | −17.55 | −13.25 |
| 0398 | Hypothetical | −25.41 | −22.71 |
| 0399 | Hypothetical | −10.57 | −6.61 |
| 0622 | aroC | −2.87 | 1.32 |
| 0650 | hrcA | −1.99 | −2.04 |
| 0651 | grpE | −2.07 | −1.32 |
| 0669 | pmpA | −4.32 | −1.11 |
| 0709 | Hypothetical | −2.21 | −1.17 |
| 0715 | murA | −2.53 | −1.12 |
| 0734 | Hypothetical | −2.42 | −1.04 |
| 0755 | sohB | −2.59 | −1.71 |
| 0814 | Hypothetical | −3.74 | −1.19 |
| 0815 | Hypothetical | −3.42 | −1.05 |
| 0886 | CHLPN | 2.27 | 1.93 |
To verify the microarray results, the same RNA samples used for microarray analyses were used to examine the expression of nine transcripts (pgp3, glgA, CTL0071, CTL0305, CTL0339, CTL0638, and CTL0397 to -0399) by qRT-PCR. Given that pgp3 expression is downregulated in the L2RpΔpgp4 mutant, it was necessary to establish whether the effects on chromosome-encoded gene expression were caused directly by pgp4 or indirectly by pgp3. To differentiate the regulatory role of pgp3 and pgp4, RNA samples of a pgp3 mutant were also included in the qRT-PCR analysis. The target transcripts were normalized against a constitutively expressed transcript (lpdA). We found significant changes in transcript levels of all nine genes in L2-versus-L2R and L2Rp+-versus-L2RpΔpgp4 comparisons (Fig. 4), which verified CTL0638 as a regulated gene in both L2R and pgp4 mutants and confirmed the microarray results for other genes. No significant changes in expression of the eight chromosomal genes were observed in the Δpgp3 mutant, indicating that the observed changes in expression were a direct result of pgp4 and not an indirect result of pgp3. Collectively, the microarray and qRT-PCR data show that pgp4 is the master plasmid gene involved in transcriptional regulation of chromosomal genes.
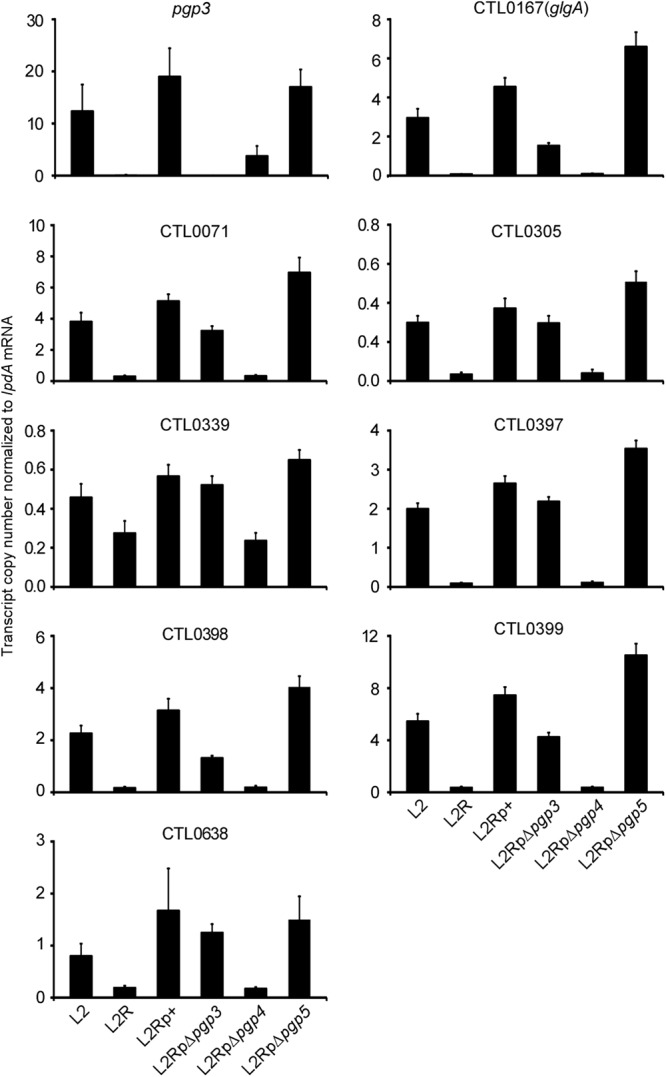
Validation of microarray data by quantitative RT-PCR of plasmid-borne and eight chromosomal genes, including glgA, CTL0071, CTL0305, CTL0339, CTL0397 to CTL0399, and CTL0638, significantly (P < 0.01) differentially expressed in L2-versus-L2R and L2Rp+-versus-L2pΔpgp4 comparisons. Transcript copy numbers derived from three replicates were normalized to those of lpdA mRNA.
DISCUSSION
Studies in both C. trachomatis (7, 8) and C. muridarum (9) have implicated the chlamydial plasmid as a key virulence factor in vivo. The chlamydial plasmid carries both noncoding RNAs and eight ORFs (17). Several studies (10, 24, 34) have implicated the plasmid in regulating the expression of a set of chromosomal genes which result in the unusual inclusion morphology and glycogen-negative phenotype, characteristic of plasmid-deficient isolates. In our nonhuman primate trachoma model, infections with plasmid-cured organisms, but not the parental plasmid-bearing organisms, are short-lived, resolve without inducing measurable pathology, and paradoxically induce superior levels of protective immunity. These findings clearly implicate plasmid-borne ORFs as important chlamydial virulence factors in vivo. In the present study, we took advantage of the recently developed plasmid-based transformation procedure for chlamydiae (28) to carry out deletion mutagenesis on all eight plasmid-borne genes. Our goal was to identify the genes needed for plasmid maintenance and, more importantly, those required for regulating the expression of the chromosomal genes responsible for the characteristic in vitro phenotype displayed by plasmid-deficient organisms.
Most plasmids are mosaic, containing genes necessary for plasmid maintenance (replication, copy number control, and partitioning) and various additional genes which, for pathogenic microbes, are typically associated with virulence and/or antibiotic resistance. Maintenance of plasmids having fewer than 10 copies/cell requires an active partitioning system (40). In the era of mass genomic sequencing, most maintenance-related genes are identified by sequence homology to well-characterized ORFs, required for plasmid replication and partitioning in E. coli. For chlamydiae, previous annotation of plasmid genes has suggested that Pgp1, -5, -7, and -8 could be involved in plasmid maintenance. Our data indicating that Pgp1 and -8, a putative helicase and integrase/recombinase, respectively, are essential for stable plasmid maintenance support the roles suggested for these ORFs based on homology searches.
Our results indicating that Pgp7, a predicted integrase/recombinase, is not essential for plasmid maintenance is in agreement with the following findings. First, pgp7 is interrupted in the plasmids of some naturally occurring chlamydiae, for example, the equine Chlamydia pneumoniae plasmid (18) and the Swedish new variant plasmid, which has a deletion which prevents detection using plasmid based PCR diagnostics that target pgp7 (41). Second, pgp7 is the site where Wang et al. (28) inserted antibiotic resistances markers in shuttle vectors designed for chlamydial transformation. Pgp7 and -8 share substantial sequence homology (32 to 35% amino acid identity) with their C-terminal domains, which is typical of the phage integrase/recombinase family of proteins (15). Taken together, the data suggest that of Pgp7 and -8, Pgp8 is more important for plasmid maintenance.
Given that Pgp5 has homology to ParA, a plasmid-encoded protein involved in plasmid partitioning, it was unexpected that it would not be required for stable plasmid maintenance in chlamydiae. This leaves open the possibility that there is a chromosomally encoded gene product capable of complementing this function or that another plasmid-borne ORF is responsible for partitioning. It is also surprising that the Pgp5 knockout transformants showed no phenotype, considering that pgp5 encodes an antisense ncRNA, one of the most abundant RNAs in L2 (13). Our microarray data exclude a regulatory role for pgp5 and its antisense ncRNA at the transcript level. However, we cannot determine whether pgp5 itself is regulated by its antisense ncRNA.
The chlamydial plasmid's copy number ranges from 4 to 10 copies per cell (5). The ori for the chlamydial plasmid resembles those of E. coli plasmids pSC101, mini-F, and RK2 (42). In other bacteria, the plasmid maintenance systems of such low-copy-number plasmids, comprised of replication, partitioning, and copy number control, have been extensively studied (40, 43, 44). These E. coli plasmids' replication and copy numbers are controlled by interactions of iterons and cognate Rep proteins, which are typically plasmid encoded (43). None of the predicted chlamydial plasmid ORFs, nor any chromosomal ORF, shows any homology to known Rep proteins. It is possible that chlamydiae use a unique method of initiating plasmid replication. Alternatively, one of the plasmid-encoded hypothetical proteins could perform this function. We found that Pgp2 and -6, both hypotheticals, were essential for stable plasmid maintenance. We are currently doing experiments to determine if Pgp2 and or -6 functions as a Rep protein controlling chlamydial plasmids' replication and copy number by binding to the ori. Intriguingly, of all the plasmid-borne ORFs, only the Pgp6 ORF has a homologue in the chromosome (CTL0846), and that homologue is present in all chlamydiae, including those that do not carry a plasmid.
It is also interesting that ColE2 plasmid replication requires a plasmid-encoded Rep, the expression of which is controlled by antisense RNA (45). As mentioned, pgp5 encodes a highly expressed ncRNA, and Pgp5 has homology to proteins (ParA) involved in partitioning. However, our data indicate that Pgp5 is not essential for plasmid maintenance. Clearly, further experimentation is needed to definitively determine which, if any, of these gene products function in plasmid replication, partitioning, and copy number control.
Our results provide new information on the function of the ORFs annotated as encoding hypothetical proteins likely involved in accessory functions related to virulence. Stable transformants were isolated for both pgp3 and -4 deletion mutants, indicating that neither gene is essential for in vitro growth, a trait common to many virulence factors. This suggests that these proteins may have a particular role related to chlamydial pathogenicity and, in the case of Pgp3, is in keeping with findings implicating it as an important virulence factor (11, 20, 21). Clearly, it is necessary to test the in vivo growth characteristics of these two mutants to positively identify them as new virulence factors. In contrast to the pgp3 mutant, which displayed no in vitro phenotype, the pgp4 mutant displayed a phenotype of atypical late inclusion morphology and lack of glycogen accumulation, characteristic of plasmidless L2R and all other plasmid-deficient chlamydial strains studied to date (24, 34, 46). Of particular importance, our microarray and qRT-PCR results indicate that pgp4 is the gene responsible for regulating the expression of pgp3 and chromosomal genes associated with this well-characterized phenotype. The association of both late infection inclusion morphology and lack of glycogen accumulation suggests that glycogen metabolism correlates with inclusion morphology, which is consistent with altered late infection morphology caused by mutations in the glycogen-branching gene glgB (26).
Besides the secreted effector Pgp3 gene on the plasmid, several Pgp4-regulated chromosomal genes could also provide insight into the key role of the plasmid in chlamydial pathogenesis. One is glgA, the gene associated with excessive glycogen accumulation within the inclusion lumen. We hypothesize that glycogen could serve as an essential carbon source within a nutritionally limiting inflammatory environment, which would be an advantage for chlamydial survival in mucosal tissues. Alternatively, chlamydial glycogen may be a TLR2 ligand or immune modulator, as has been shown for enzymatically synthesized glycogen (47) and Mycobacterium tuberculosis glucans (48), respectively. Consistent with this reasoning, plasmid-dependent TLR2 activation has been shown to be associated with both the early production of inflammatory mediators and the development of chronic oviduct pathology in the murine model of C. muridarum infection (9). Besides glgA, all the other chromosomal genes regulated by pgp4 are hypothetical ORFs. Our understanding of these genes is limited, but a few studies suggest that they are virulence genes. For example, alterations in CTL0399 (CT144) are highly associated with rectal tropism in serovar G isolates (49), CTL0305 (CT049) is secreted into the lumen of the C. trachomatis inclusion (50), and CTL0339 (CT084) has been proposed as a modulator of mitochondrial function (51). These studies suggest that Pgp4 is responsible for regulating the expression of genes important for chlamydial virulence. The precise role that these Pgp4-regulated genes play in chlamydial pathogenicity is an area of intense interest and the focus of our current studies.
How might Pgp4 regulate the expression of its target genes? Pgp4 is the smallest ORF on the plasmid, encoding 102 amino acids. Results from genomic transcriptional profiling by DNA microarray (52) show that pgp4 is expressed very early (3 h postinfection [p.i.]) in the developmental cycle. Secondary structural analysis and sequence comparison reveal that Pgp4 has a putative helix-loop-helix domain. Pgp4 may be a novel member of the helix-loop-helix family of transcriptional regulators; however, our initial attempts to detect binding of purified recombinant Pgp4 to target DNA (~150 bp upstream sequences of CTL0071, CTL0305, CTL0339, CTL0397, CTL0638, and glgA) were not successful. It is possible that Pgp4-DNA interactions require additional protein partners for successful DNA binding to occur. Of note, it has recently been reported that efficient transcription activation of several promoters by a chlamydial specific transcription factor, general regulator of genes A (GrgA), in vitro requires contact with both DNA and a portion of a nonconserved region of the primary σ factor, σ66, of C. trachomatis (53). Studies are under way to determine the molecular mechanism of Pgp4-mediated gene regulation.
In conclusion, we employed deletion mutagenesis and chlamydial transformation to define the functions of individual C. trachomatis plasmid genes. We propose two plasmid ORF clusters, pgp1 and -2 and pgp6 to -8, that function in plasmid maintenance. The pgp3-5 cluster therefore likely represents the primary virulence genes that function in chlamydial pathogenicity and may account for the profound in vivo infection attenuation characteristics displayed by plasmid-deficient isolates. We present convincing evidence that pgp4 is the gene that regulates the transcription of plasmid encoded pgp3 and multiple chromosomal genes, including glgA, CTL0071, CTL0638, CTL0305, CTL0339, and CTL0397 to -0399. Our pgp4 mutant will allow future genetic studies to define the mechanism of Pgp4-regulated gene expression. Furthermore, our findings have important practical applications for the design of novel live-attenuated vaccines that are capable of targeting immunization of mucosal surfaces. For example, a chlamydial plasmid shuttle vector with deleted major virulence genes (e.g., pgp4) could be engineered to express multiple C. trachomatis serovariable protective antigens, such as the major outer membrane protein, or to overexpress protective T cell antigens in a single transformed chlamydial strain.
ACKNOWLEDGMENTS
We thank Kelly Matteson and Anita Mora for editorial and graphic assistance.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print 14 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01305-12.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.01305-12
Read article for free, from open access legal sources, via Unpaywall:
https://iai.asm.org/content/iai/81/3/636.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Diverse animal models for Chlamydia infections: unraveling pathogenesis through the genital and gastrointestinal tracts.
Front Microbiol, 15:1386343, 28 Mar 2024
Cited by: 0 articles | PMID: 38605708 | PMCID: PMC11007077
Review Free full text in Europe PMC
Expression activation of over 70% of Chlamydia trachomatis genes during the first hour of infection.
Infect Immun, 92(3):e0053923, 01 Feb 2024
Cited by: 4 articles | PMID: 38299827 | PMCID: PMC10929459
Metabolic dormancy in Chlamydia trachomatis treated with different antibiotics.
Infect Immun, 92(2):e0033923, 12 Jan 2024
Cited by: 4 articles | PMID: 38214508 | PMCID: PMC10863404
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance.
mBio, 15(1):e0203623, 19 Dec 2023
Cited by: 1 article | PMID: 38112466 | PMCID: PMC10790707
Molecular pathogenesis of Chlamydia trachomatis.
Front Cell Infect Microbiol, 13:1281823, 18 Oct 2023
Cited by: 2 articles | PMID: 37920447 | PMCID: PMC10619736
Review Free full text in Europe PMC
Go to all (118) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Plasmid Negative Regulation of CPAF Expression Is Pgp4 Independent and Restricted to Invasive Chlamydia trachomatis Biovars.
mBio, 9(1):e02164-17, 30 Jan 2018
Cited by: 11 articles | PMID: 29382731 | PMCID: PMC5790913
The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor.
Infect Immun, 76(6):2273-2283, 17 Mar 2008
Cited by: 120 articles | PMID: 18347045 | PMCID: PMC2423098
Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum.
Infect Immun, 79(3):1044-1056, 03 Jan 2011
Cited by: 71 articles | PMID: 21199910 | PMCID: PMC3067522
Chlamydial Plasmid-Dependent Pathogenicity.
Trends Microbiol, 25(2):141-152, 03 Oct 2016
Cited by: 61 articles | PMID: 27712952 | PMCID: PMC5272858
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Intramural NIH HHS
Wellcome Trust (1)
Understanding the genetic basis of common human diseases: core funding for the Wellcome Trust Centre for Human Genetics.
Professor Peter Donnelly, University of Oxford
Grant ID: 090532
 a
a




