Abstract
Free full text

MicroRNA control of bone formation and homeostasis
Abstract
MicroRNAs (miRNAs) repress cellular protein levels to provide a sophisticated parameter of gene regulation that coordinates a broad spectrum of biological processes. Bone organogenesis is a complex process involving the differentiation and crosstalk of multiple cell types for formation and remodeling of the skeleton. Inhibition of mRNA translation by miRNAs has emerged as an important regulator of developmental osteogenic signaling pathways, osteoblast growth and differentiation, osteoclast-mediated bone resorption activity and bone homeostasis in the adult skeleton. miRNAs control multiple layers of gene regulation for bone development and postnatal functions, from the initial response of stem/progenitor cells to the structural and metabolic activity of the mature tissue. This Review brings into focus an emerging concept of bone-regulating miRNAs, the evidence for which has been gathered largely from in vivo mouse models and in vitro studies in human and mouse skeletal cell populations. Characterization of miRNAs that operate through tissue-specific transcription factors in osteoblast and osteoclast lineage cells, as well as intricate feedforward and reverse loops, has provided novel insights into the supervision of signaling pathways and regulatory networks controlling normal bone formation and turnover. The current knowledge of miRNAs characteristic of human pathologic disorders of the skeleton is presented with a future goal towards translational studies.
Introduction
Bone, a mineralized mesenchymal tissue, serves at least two essential biological roles. As an endocrine organ that regulates mineral homeostasis and energy metabolism, bone responds and signals to other vital tissue systems (for example, the parathyroid gland, kidney, vasculature, adipose tissue, hypothalamus). As the principal structural connective tissue, bone supports locomotion and protects key organs. In response to mechanical forces and levels of calcium and phosphate in the blood, bone tissue is continuously turned over and remodeled through the activities of two main cell populations, the bone-resorbing osteoclasts that arise from the hematopoietic lineage and the bone-forming osteoblasts of mesenchymal origin (Figure 1a).

The bone remodeling cycle and regulation of bone tissue homeostasis. a | Cellular activities supporting bone remodeling. A remodeling cycle, regulated by parathyroid hormone and 1,25-dihydroxyvitamin D3, is initiated with a resorption phase by activated osteoclasts that solubilize bone mineral and degrade the matrix. Osteoclasts originate from hematopoietic stem cells which differentiate first (dotted arrow) to a committed mononuclear preosteoclast cell that fuses to form the multinucleated cells. Attachment of osteoclasts to the bone surface creates a local acidic environment forming a resorption pit. The activities of monocytes or macrophages remove debris (reversal phase), followed by a bone formation phase by osteoblasts, producing osteoid matrix which will mineralize. Growth factors are released from the bone matrix during resorption, which increases the preosteoblast population to replace eroded bone surfaces. b | Regulation of bone programs. The skeletal landscape is illustrated as an integration of genetic (expressed genes) and epigenetic (microRNAs [miRNA]) factors that leads to the effector programs to support the three major activities of bone (lower boxes). The integration of these regulators is facilitated by post-transcriptional control that appears to be highly regulated by miRNAs.
The activity of these cells is under hormonal regulation and is tightly coupled by several key signaling pathways,1–5 including the RANKL–RANK pathway, which is essential for osteoclast differentiation (as detailed below), and the EPHB4–EFNB2 pathway, which mediates a switch from resorption to bone formation. These ligand– receptor interactions support crosstalk between osteoblast and osteoclast lineage cells to regulate the balance between resorption and formation. In addition, in each population, molecular controls oversee the recruitment of the precursors into their respective osteoclast or osteoblast lineages (see below). MicroRNAs (miRNAs; Box 1) have a role in the regulation of bone remodeling.6
The formation of bone by osteoblast lineage cells (Box 2) and its principal functional activities involve multiple layers of regulation that utilize morphogens and developmental signals (including BMPs, Wnt ligands, hormones, growth factors and cytokines) for induction of osteogenesis (Figure 1b). For specification of distinct cell populations, a cohort of tissue-restricted transcription factors and co-regulators mediate expression of genes for biosynthesis and mineralization of the bone matrix, as well as for the remodeling and repair of bone throughout life. These bone functions are also controlled at several genetic and epigenetic levels, which include transcriptional control of genes by DNA methylation, modifications of nucleosomal and chromatin architecture, and retention of proteins on chromosomes during mitosis to bookmark genes for postproliferative expression.7
Equally important, gene expression is regulated post-transcriptionally. Cellular protein levels and activities can be regulated by modifications, including phosphorylation and ubiquitin-mediated degradation. Modulation of protein levels is also contributed by miRNAs, which are considered a key epigenetic mechanism for control of expressed genes.8,9 Figure 1b illustrates the convergence of these levels of genetic and epigenetic control for bone homeostasis. miRNAs provide a novel dimension to post-transcriptional control8 by directly inhibiting mRNA translation. Our understanding of how miRNAs control the regulatory interplay between the different cell types in the bone remodeling unit is minimal. In addition, the roles of miRNAs in the differentiation and recruitment of mature cells derived from mesenchymal stem cells (MSCs) or hematopoietic stem cells remains to be established.
miRNAs represent a sophisticated level of gene regulation that coordinates a broad spectrum of biological processes. Mechanisms that control miRNA biogenesis and binding to mRNAs to repress cellular protein levels have been elucidated in some detail.9 The temporal levels of many miRNAs change only moderately in different biological contexts and the attenuation of protein levels by miRNAs can be modest. However, because miRNAs control existing mRNAs, regulation of mRNA and/or protein levels can proceed faster than transcriptional mechanisms. Consequently, miRNAs mediate rapid fine-tuning of gene expression.
Hundreds of miRNAs have been functionally linked to development of specific tissues, for example, skin, muscle, nerve, fat, cartilage and bone.10–15 Many miRNAs have been characterized that mediate cell cycle control, apoptosis and survival, whereas distinct miRNAs also mark various diseases,16–19 cancer cells and pathological progression of solid tumors and leukemias.20 However, miRNA control of biological processes beyond simple models invoking single gene targets remains a compelling area of investigation. More complex models must incorporate the two principal facts that one miRNA can target hundreds of mRNAs and can act selectively in different tissues, whereas one mRNA can have multiple binding sites for different miRNAs. These two fundamental properties predict that miRNAs function as powerful molecular managers that operate by controlling several gene regulatory networks simultaneously.
miRNAs contribute to every step of osteogenesis from embryonic bone development to maintenance of adult bone tissue, by regulating the growth, differentiation and functional activity of cells that constitute bone tissue. This Review first describes evolving biological concepts for the roles of miRNAs in bone formation, bone resorption and chondrogenesis (the process by which cartilage is developed), that clarify how miRNAs control tissue homeostasis. The following sections focus on miRNA control over the well-defined multistep processes of osteoblast and osteoclast differentiation, and the concept of miRNA allocation of MSCs to the osteogenic cell fate. The Review integrates current information about how miRNAs support tissue-specific activities through their regulation of phenotypic transcription factors and supervision of signaling pathways and regulatory networks in the skeleton. The broad functions of miRNAs emphasized in this Review provide the cornerstone for advancing bone biology and pathology. Finally, the few pathological disorders of the skeleton that have been examined for miRNA dysregulation are presented as a starting point for future studies with patient material.
miRNAs control the skeletal landscape
Bone forms by two fundamentally distinct processes. Intramembranous bone formation produces flat bones and endochondral ossification generates long-bone growth (Box 2). Understanding the contribution of miRNAs to bone growth and repair requires cell-type-specific disruption of miRNA biogenesis. This disruption can be achieved by deletion of the endoribonuclease Dicer, which is required to produce functionally mature miRNAs (Box 1).21 Because Dicer null mouse embryos do not survive beyond embryonic day 7.5 (E7.5),22,23 conditional Dicer null alleles have been used to construct more viable mouse models. Mice which harbor loxP sites within the Dicer gene express Dicer normally; however, when these ‘floxed’ mice are crossed with mice expressing the Cre recombinase under the control of a skeletal gene, the Dicer gene is excised in their offspring, which then lack cell-type-specific protein expression. Conditional deletion of Dicer in the cell types of the mouse skeleton demonstrates that miRNAs contribute to prenatal bone development, and convincing new data reveal that miRNAs control postnatal bone formation and turnover in the adult skeleton.
Skeletal morphogenesis and chondrogenesis
Dicer deletion in neural crest cells, from which the craniofacial skeleton arises, results in impaired morphogenesis and deformities in tissue development of the skull.24 Conditional deletion of Dicer alleles by Prx-Cre during early stages of limb morphogenesis at E9.5 results in severely impaired development of mesodermal tissues and fetal lethality. From E10.5, limb formation is stunted due to apoptosis of cells, yet, normal limb patterning occurs. By contrast, disruption of a specific miRNA that targets a Hox gene, miR-196,25,26 was found to compromise skeletal patterning. These findings directly establish that miRNAs contribute to early stages of fetal bone formation.
Long bones and vertebrae are formed from a cartilage template that develops into bone through the endochondral process of ossification. In one study, cartilage-specific Dicer excision in mice affected all sub-populations of chondrocytes in organized zones at the growth plate (Box 2), from the proliferating chondroprogenitor to the mature hypertrophic cell.14 The loss of miRNAs results in two abnormalities at the growth plate. The number of columnar proliferating cells is reduced, and differentiation of the mature hypertrophic chondrocytes accelerates. This finding suggests that the proliferating and hypertrophic zone chondrocytes could potentially be regulated by different sets of miRNAs or by the same miRNA targeting different proteins in each population. Although this study revealed critical roles for mature miRNAs in supporting the cellular activities in the growth plate,14 specific miRNAs involved in the observed abnormalities of bone growth and post-natal lethality in this mouse model remain unknown. However, in other studies, miRNAs have been identified in the regulation of chondrogenesis.
The miRNA miR-140 is a positive effector of chondrogenesis that contributes to palatogenesis in zebrafish through PDGF signaling.27 In mouse models, miR-14028 contributes to craniofacial development and endochondral bone formation through a mechanism related to miR-140 repression of both histone deactylase 4 (HDAC4; a known inhibitor of chondrocyte hypertrophy)29–31 and a splicing factor which reduces BMP signaling. Both factors can impair osteogenesis,32 and miR-140 prevents this negative effect on bone formation. miR-140 is also involved in a double negative feedback loop that directly suppresses Smad3 and TGF-β signaling genes in chondrocytes; however, TFG-β can limit accumulation of miR-140.33 miR-675 increases expression of cartilage-specific collagen type IIa through a pathway initiated by the cartilage-specific Sox9 transcription factor, which increases expression of the primary miR-H19 from which the mature miR-675 is derived.34 Thus, in mice, both direct and indirect effects of miRNAs can promote chondrogenesis.
In vitro studies have to date identified two negative regulators of chondrogenesis—miR-199a, which inhibits Smad1,35 and miR-145, which targets Sox9, the essential transcription factor for chondrogenesis (Supplementary Table 1 online).36 Studies using microRNA arrays are revealing sets of miRNAs expressed at different stages of chondrocyte differentiation.37,38 Given the complexity of regulatory events at the growth plate by transcription factors and secreted signaling factors,39 exploring the targets of these miRNAs will in future studies reveal higher order regulatory networks.
miRNAs regulate osteogenesis and bone mass
The subpopulations of bone-forming cells and the progressive principal stages of osteoblast differentiation with indicated markers are shown in Figure 2. Mouse studies in which mature miRNA production was ablated by conditional deletion of Dicer alleles at different stages of bone formation have established that miRNAs control both early and late steps of osteogenesis.15 Ablation of Dicer in osteoprogenitor cells by collagen type 1a40-Cre (Col1a-Cre) inhibits their maturation to cells that produce a mineralized tissue observed at E14.5.15 This mid-gestation defect indicates an early requirement for miRNAs in bone formation. However, subsequent embryonic lethality at E15.5 precludes analysis of bone maturation defects at later stages of skeletal development.
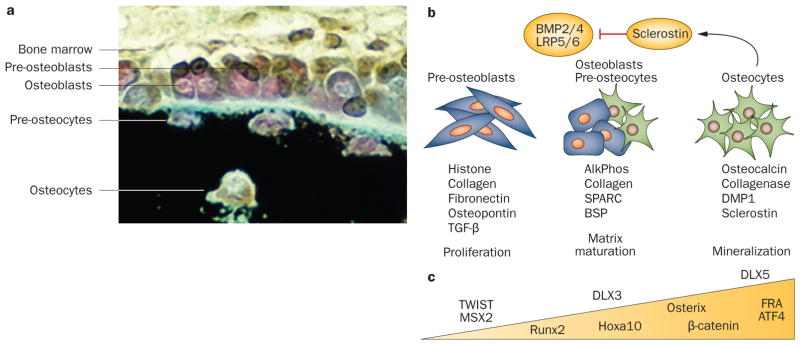
The osteoblast differentiation program. a | In vivo: bone surface shows organization of indicated osteoblast lineage cells (black, mineralized tissue). Mesenchymal stem cells and osteoprogenitor cells cannot be seen. b | In vitro: stages of differentiation of committed preosteoblast cells isolated from newborn rodent calvarium or bone marrow stromal cells. Peak expression of genes that are markers for the three major stages are shown. At mineralization, a feedback signal from sclerostin secreted by osteocytes inhibits BMP and Wnt osteogenic-mediated bone formation by regulating the number of cells entering the osteoblast lineage. c | Examples of transcription factors regulating osteoblast differentiation and in vivo bone formation are shown. Within the triangle are those that increase during differentiation, whereas those above the triangle are functional on gene promoters at the indicated stages of maturation. Permission obtained from American Society for Bone and Mineral Research © Favus, M. J. (Ed.) Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism, 6th edn (2006).
Deletion of Dicer in the mature osteoblast identifies the importance of Dicer processing of mature miRNAs for regulation of postnatal bone growth. In one of these studies,41 Cre was expressed under control of a different Col1a-Cre with restricted expression in committed osteoblasts.42 However, the resulting mice were only examined at birth and did not exhibit an overt bone defect.41 More definitive studies using the osteocalcin-Cre expressed in mature osteoblasts and osteocytes43 established that miRNAs in these cells have major roles in regulating the size of the bone through control of bone matrix protein levels in the adult skeleton.15 Specifically, a striking increase in the volume of trabecular and solid bone became evident between 4 weeks and 8 weeks after birth, and this increase continued for at least 8 months. The activity of the osteoblasts was stimulated and bone tissue exhibited elevated expression of bone-related collagens and noncollagenous proteins in the extracellular matrix that are directly linked to miRNA loss. The observed high bone mass phenotype (accompanied by normal BMD) was found to be strictly related to loss-of-function of miRNAs in mature osteoblasts and not to inhibition of bone resorption. Osteoclast activity was stimulated, indicating normal bone remodeling in the presence of accelerated bone formation.
Regulated osteoclastogenesis by miRNAs
Bone remodeling is a continuous process requiring the physiological coupling of osteoclast with osteoblast activities (Figure 1). Thus, the effects of Dicer excision in osteoclasts must also be considered, to achieve a full understanding of how miRNAs influence bone homeostasis. Each stage of osteoclast differentiation requires regulatory factors that also serve as markers of osteoclast function and bone resorbing activity (Figure 3a).1–5 The principal signaling pathway regulating osteoclast differentiation involves RANKL in osteoblast lineage cells that is coupled to the RANK receptor present on the osteoclast mononuclear precursors that permits their fusion to multi-nucleated osteoclasts. The RANK–RANKL interaction is regulated by osteoprotegerin, a soluble form of RANKL that acts as a decoy (receptor) to inhibit osteoclastogenesis (Figure 3a). The ratio of RANKL to osteoprotegerin is a marker of bone-resorbing activity.
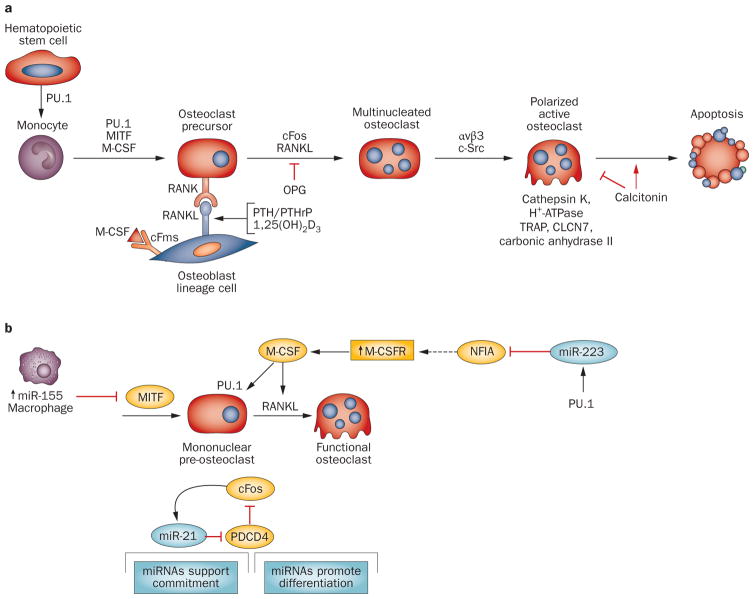
Osteoclast differentiation sequence and effect of microRNAs. a | Stages in the differentiation of the multinucleated osteoclast from its hematopoietic precursor are illustrated with key transcription factors and regulatory proteins established as critical for progression to the activated osteoclast. These include the RANK–RANKL interaction regulated by the indicated hormones and the inhibitor of RANK signaling osteoprotegerin. Integrin (αvβ3) mediates attachment of osteoclasts to bone surface and c-Src signaling induces polarization of the osteoclast and formation of the characteristic ruffled border for active bone resorption. b | MicroRNAs regulating commitment to osteoclastogenesis have been identified. Indicated are three different mechanisms. miR-155 functions as an inhibitor of osteoclastogenesis, being highly expressed in macrophages to support robust expression of this phenotype by inhibiting MITF, essential for preosteoclast differentiation. PU.1 initiates a feedforward mechanism increasing miR-223 which downregulates an inhibitor of osteoclast differentiation, NFIA, resulting in an increase in M-CSFR and thereby M-CSF functional activity. Also shown is a regulatory loop between miR-21 and cFos (AP-1), which activates many osteoclast genes essential for multinucleated cell formation and promotes resorptive activity.133 Abbreviations: αvβ3, integrin αvβ3; CLNC7, chloride channel; OPG, osteoprotegerin; TRAP, tartrate-resistant acid phosphatase.
The absence of mature miRNAs in mononuclear pre-osteoclasts (in CD11b-Cre mutant mice) and in mature multinucleated osteoclasts (in cathepsin-K-Cre mutant mice) results in a similar phenotype of increased bone mass due to decreased number and activity of osteoclasts.41,44,45 Furthermore, osteoclasts could not be formed from bone marrow hematopoietic lineage cells from either mouse model.41,45 Nor could they be formed from bone marrow precursor cells in which short interfering RNA (siRNA) was used to silence components of the Drosha enzyme complex (which initiates miRNA biosynthesis) or the RISC complex (which is involved in binding of the miRNA to its target messenger; Box 1).46 These studies confirm that loss of miRNA activity in osteoclast precursor cells causes a block in osteoclastogenesis.
To date, only a few miRNAs are known to contribute to osteoclast differentiation and the in vivo phenotype from Dicer loss of function in osteoclast lineage cells. The few studies available already indicate that microRNAs are operative at the commitment stage of osteoclasto genesis (Figure 3b). A regulatory pathway is indicated by the action of miR-223, first identified as a myeloid factor potentially regulated by the transcription factor PU.147 and also found to be expressed in a mononuclear osteoclast precursor cell line of hematopoietic origin.46 PU.1, which is induced by M-CSF in the osteoclast lineage, increases miR-223 and RANK levels in bone marrow osteoclast precursors (Figure 3b). The mechanism for miR-223 stimulating osteoclastogenesis was later established,44 in that miR-223 represses NFIA, a suppressor of osteoclastogenesis. The net effect is increased expression of the M-CSF receptor, necessary for osteoclastogenesis.44
A second molecular event for regulation of osteoclastogenesis was revealed by studies of miR-155, which supports commitment of hematopoietic cells to become macrophages (Figure 3b). miR-155 downregulates MITF (an essential transcription factor in osteoclast differentiation).48 These findings reflect positive and negative regulation by miRNAs at the commitment stage of two different cell types (Figure 3b); thus miR-155 functions as a molecular switch between the macrophage and osteoclast lineage. Beyond this early stage, miR-21 was identified in a miRNA profiling study as being upregulated during RANKL-induced differentiation of mononuclear to multinucleated cells.45 This miRNA inhibited PDCD4 protein, which derepressed c-Fos (Figure 3a,b). Although broadly expressed, miR-21 is regulated by a promoter with functional AP1 elements,45 and c-Fos, another critical regulator of osteoclastogenesis,49 upregulates miR-21 in a feedforward mechanism in order to drive osteoclast differentiation.
In summary, in vivo studies in mice show that compromised biogenesis of functional miRNAs by loss-of-function of Dicer in bone have several ramifications. The phenotypes clearly indicate that miRNAs have both early developmental and postnatal effects on the murine skeleton. It is likely that distinct sets of mature miRNAs exist that guide normal development of bone tissue, whereas other sets of miRNAs are important for homeostasis of adult bone. miRNAs have major biological functions in each of the three principal cell types, chondrocytes (Supplementary Table 1 online), osteoblasts (Table 1) and osteoclasts that support bone functions.
Table 1
Functional microRNAs in the skeleton
| miRNA | Protein encoded by target gene(s) | Description | Reference |
|---|---|---|---|
| BMP-induced osteogenesis
| |||
| Downregulated miRNAs: | |||
| miR-125 | Osterix | Osterix is required for bone mineralization | 140 |
| miR-125b | ERBβ2 | Target inhibits proliferation | 141 |
| miR-133 | Runx2 | miRNA inhibits osteoblast differentiation | 89 |
| miR-135 | Smad5, BMP-R1a | miRNA inhibits BMP2-induced osteoblast commitment, Bsp and osterix | 89,142 |
| miR-138 | PTK2, osterix | Highly expressed in human MSC; inhibits osteoblast focal adhesion kinase pathway and osterix, which is required for osteoblast differentiation | 88 |
| miR-141, miR-200a | Dlx5 | Several osteoblast genes require Dlx5 for activation | 143 |
| miR-181 | Hoxa11 | Establishes muscle phenotype by releasing MyoD from Hoxa11 repression | 144 |
| miR-206 | Connexin 43 | miR expression results in bone loss in vivo; miR-206 is also muscle-related | 93,95,109 |
| miR-208 | ETS-1 | ETS-1 contributes to transactivation of several osteoblast-related genes | 145 |
| Upregulated miRNAs:miR-9 | MyoD, Myf5 | Downregulating muscle transcription factors promotes osteogenesis | 89 |
|
| |||
| Osteogenesis-promoting
| |||
| miR-20a | BAMB1, CRIM PPARγ | Increases BMP signaling by targeting antagonists of the pathway Represses adipogenesis | 115 |
|
| |||
| miR-27 | APC | miR-27 expression increases during differentiation of human osteoblasts and enhances Wnt signaling by repressing APC | 63 |
|
| |||
| miR-29b | HDAC4, DUSP2, TGFβ3, AcvR2b, CTNNIBP1 | Increases osteoblast differentiation in pre-osteoblasts by repressing inhibitors of osteogenesis | 54 |
|
| |||
| miR-29c | Dkk1, Kremen, SFRP2, Osteonectin | Promotes Wnt signaling in osteoblasts by targeting inhibitors Regulates matrix proteins | 56,64 |
|
| |||
| miR-196a | Hoxc8 | miRNA overexpression in adipose derived MSCs promotes osteoblast differentiation | 146 |
|
| |||
| miR-210 | AcvR1b | BMP4 induces preosteoblasts to differentiate and increases miR-210 | 147 |
|
| |||
| miR-335-5p | DKK1, a Wnt inhibitor | Wnt signaling is stimulated in mature osteoblasts, which is anabolic for bone; miR regulates Runx2 in MSCs | 65,66 |
|
| |||
| miR-378 | Nephronectin GaINT-7 | Nephronectin is an extracellular matrix protein that competes for miR binding, which promotes osteoblast differentiation | 148 |
|
| |||
| miR-2861/miR-3960 | HDAC5, Hoxa2 | Both targets of these miRNAs repress Runx2 activity In a feed-forward loop, Runx2 upregulates the clustered miRNAs to inhibit target expression | 70,122 |
|
| |||
| Osteoblast-inhibitory
| |||
| miR-23a-27a-24-2 | SatB2, Runx2 Hoxa10 | Each miRNA in cluster inhibits osteoblast differentiation Cluster is downregulated by Runx2 to promote osteogenesis | 62 |
|
| |||
| miR-26a | Smad1 | Expressed late during hADSC differentiation to osteoblasts; mechanisms not clear | 149 |
|
| |||
| miR-29a,b | Collagens, Col1A, Col3A, Col5 | miR-29 attenuates collagen synthesis in mineralized bone and is linked to sclerosis during tissue repair | 54,57,58 |
|
| |||
| miR-204/miR-211 | Runx2 | Present in undifferentiated C3H10T1/2 and human MSCs; promotes adipogenesis | 91 |
|
| |||
| miR-335 | Runx2 | Highly expressed in human MSCs and downregulated during osteogenesis induction | 65 |
|
| |||
| 11 miRs (see Figure 7) | Runx2 | Overexpression inhibits/delays osteoblast differentiation | 90 |
|
| |||
| miR-637 | Osterix | Promotes adipocyte and inhibits osteoblast differentiation via osterix | 150 |
Abbreviations: hADSC, human adipose tissue-derived stem cell; miRNAs, microRNAs; MSCs, mesenchymal stem cells.
Although signaling pathways that are critical molecular mediators of cell communication between the mesenchymal osteoblasts and hematopoietic-derived osteoclasts are known, future studies need to address miRNA control of the regulatory crosstalk between these two metabolically opposing cell types—osteoblasts and osteoclasts. In a study by Yu et al., a role of miRNAs in crosstalk regulation was suggested by the significant decrease in levels of 10 miRNAs when conditioned medium from osteoclastic cells was added to induce osteogenic differentiation of spinal ligament fibroblasts.50 In addition to secreted miRNAs with systemic effects, a potential mechanism for more direct miRNA-mediated crosstalk involves the capacity of microvesicles to ‘shuttle’ microRNAs to another cell.51 Thus, two cell types that are coupled through receptor and ligand interactions could be regulated through an exchange of miRNAs. From a translational perspective, the studies with conditional Dicer null mutants indicate that a subset of miRNAs expressed in osteoblasts in the postnatal skeleton have the remarkable function of limiting bone mass. In addition, miRNAs can inhibit essential factors for differentiation of bone resorbing osteoclasts. Potentially, therapeutic cocktails of miRNAs or inhibitors of miRNAs could be identified for the treatment of severe disorders of bone that would target osteoblasts, osteoclasts or both cell types together.
miRNAs orchestrate bone programs
Timing expression of genes for osteogenesis
A committed preosteoblast progresses through principal stages of differentiation (Figure 2), each characterized by subsets of expressed genes and factors regulating crosstalk between the osteoprogenitors and mature osteocytes, which secrete sclerostin (encoded by the SOST gene).52,53 Studies have, therefore, addressed which miRNAs may be selectively expressed at particular stages to potentially support progression of the differentiation program. Profiling studies identified a cohort of miRNAs expressed early but highly upregulated at the late mineralization stage.54 These miRNAs might contribute to the high bone mass phenotype observed in mice with conditional deletion of Dicer in osteoblasts or osteocytes. A survey of the most probable targets of these miRNAs from profiling studies revealed that they include bone matrix proteins, regulators of metabolic activity and components of osteogenic signaling pathways.54 These bone-regulating miRNAs (designated ‘osteomiRs’) are most likely responsive to a broad spectrum of physiologic cues and represent a novel level of control for the rate of bone formation and/or for regulating the completion of bone formation at the terminal mineralization stage.
The biological activity of this group of bone-regulating miRNAs is illustrated by miR-29, which has multiple distinct activities at different stages of osteoblast differentiation (Figure 4). Consistent with the need to activate osteogenic pathways, miR-29b targets several well-characterized inhibitors of in vivo bone formation at the onset of osteoblastogenesis. These include HDAC4, which reduces transcriptional activity of the and a TGF-β receptor bone essential Runx2, TGF-β3 AcvR2a, as well as a dual-specific phosphatase (DUSP2) that dephosphorylates and inactivates ERK pathway signaling that is pro-osteogenic and antiadipogenic.55 In addition, miR-29 targets collagen type I, which forms ~90% of the bone organic matrix. At least five other families of miRNAs upregulated in the mineralization stage also target collagens (for example, let-7, miR-15/16 and miR-196).15,54 miR-29 also directly targets SPARC (also known as osteonectin), an extracellular matrix protein with properties that contribute to supporting osteoblast differentiation and bone mass.56
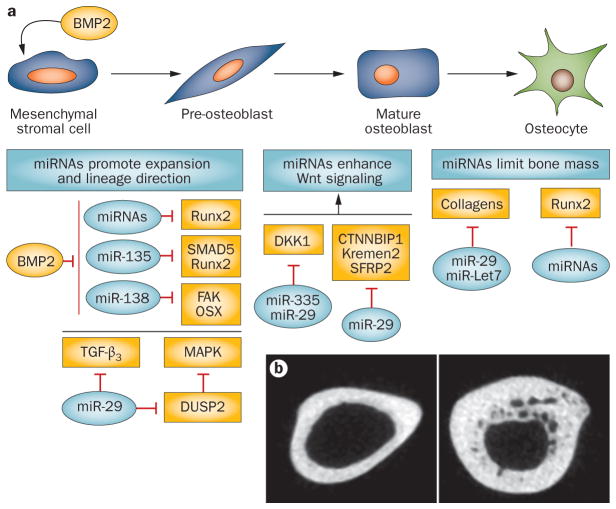
Effect of microRNAs (miRNAs) on osteoblast differentiation. a | BMP2 induces mesenchymal stromal cell commitment to osteogenesis. Below the subpopulations of osteoblasts (top row) are boxes indicating the major activities of miRNAs influencing the continuous maturation of the cells from the mesenchymal stromal cells to the osteocyte. Selected miRNAs (blue) in relation to their targets (yellow) influencing each stage to regulate progression of differentiation are indicated. Note that the miR-29 family targets several inhibitors of Wnt, MAPK signaling and collagens at different stages of differentiation.54,56 Other miRNAs, such as miR-335, also target Wnt inhibitors.66 b | The consequence of Dicer deletion in osteoblasts and osteocytes (left panel; Dicer-C/C) driven by osteocalcin (OC-Cre) (right panel; Dicer ΔOC/ΔOC). Loss-of-function of mature miRNAs in this population results in increased bone mass potentially by relieving repression of Runx2 miRNAs (n=11)90 and collagen protein levels. Courtesy of J. B. Lian, University of Massachusetts Medical School, USA.
Thus, attenuation of critical extracellular matrix components as miR-29 increases during late stages of differentiation may serve several functions in regulating bone formation. The reduced rate of collagen synthesis would facilitate formation of crosslinked fibrils for stability of bone structure. Given the association of miR-29 depletion and resulting fibrosis of damaged muscle and systemic sclerosis of lung and other tissues in both human diseases and mouse models,57,58 increased levels of miR-29 in osteoblasts would prevent formation of sclerotic bone. These studies elegantly illustrate that a single miRNA, such as miR-29, can have a broad range of activities in regulating progression of cell differentiation from the proliferating progenitor to its terminal stage by simultaneously targeting multiple signaling pathways.
The canonical Wnt pathway, mediated through the LRP5/6 co-receptor complex, is involved in the anabolic regulation of bone formation.59 Low β-catenin cellular levels facilitate chondrogenesis, whereas high β-catenin levels induce osteogenesis. Different Wnt ligands (for example, Wnt-7a and Wnt-10a) also contribute to cell phenotypes and support mineralization and accrual of bone mass.39,60,61 Thus, miRNAs that target Wnt inhibitors would be considered anabolic for bone formation. miR-27, which is part of a cluster for which expression increases during osteoblast differentiation,62 positively regulates osteogenesis by inhibiting the APC gene, which negatively controls Wnt signaling.63 The miR-29 family (described above) also targets several inhibitors of the Wnt signaling pathway to drive osteoblast differentiation. In early-stage osteoblasts, miR-29b represses protein LZIC (also known as CTNNBIP1), an inhibitor of β-catenin, whereas miRNAs 29a and 29c target DKK1, SFRP2 and Kremen2 inhibitors of the Wnt receptor complex.64 Moreover, miR-29 is upregulated in both MSCs and osteoblasts when Wnt signaling is activated, thus generating a feedforward mechanism during osteoblast differentiation.56 miR-335 was also identified as being upregulated by Wnt signaling65 and directly targets the DKK1 inhibitor to stimulate further differentiation.66
These few examples demonstrate a significant role for miRNAs in activating Wnt signaling for osteogenesis and involve multiple miRNAs targeting the same inhibitors to generate maximal effect on osteoblast cell fate and differentiation (Figure 4). The large number of miRNAs that are highly upregulated as mineral deposition begins appears to be needed to limit bone mass in order to support normal bone structure, probably in response to a broad spectrum of physiological cues.
Regulatory circuits in osteoblasts
Many miRNAs are embedded in introns and are expressed along with the genes in which they reside, whereas other miRNAs are clustered and/or are regulated by their own promoter. In principle, clustering permits coexpression of miRNAs with related functions. During osteoblast differentiation several clusters are expressed, such as the tumor suppressor miR15a/16 cluster.54 The characterization of one cluster, miR-23a~27a~24-2, revealed key regulatory circuits for induction of osteogenesis and regulation of the progression of differentiation. This cluster has also been associated with normal hematopoietic differentiation, leukemia67,68 and human diseases. The cluster’s promoter has a regulatory element bound by the Runx family of tissue-specific transcription factors. Expression of the cluster is downregulated by Runx1 in hematopoietic cells67,68 and Runx2 in osteoblasts.62 Importantly, separate overexpression of each miR in the cluster severely impairs osteoblast differentiation. Runx2-mediated reduction of miR-23a~27a~24-2 expression increases protein levels of two targets of each miRNA, Runx2 and Satb2, which are both required for osteogenesis.69 This example illustrates how clustered miRNAs orchestrate the regulation of several genes in a feedforward pathway for osteoblastogenesis (Figure 5a). In addition, the miRNA cluster miR-23a~27a~24-2 becomes robustly upregulated in terminal osteocytes as a consequence of miR-23a association with the Runx2 3' untranslated region to attenuate its protein levels.62 Furthermore, miR-27a in the cluster downregulates Hoxa10, an activator of Runx2. Hence, through suppression of these transcription factors, the miRNA cluster becomes de-repressed.
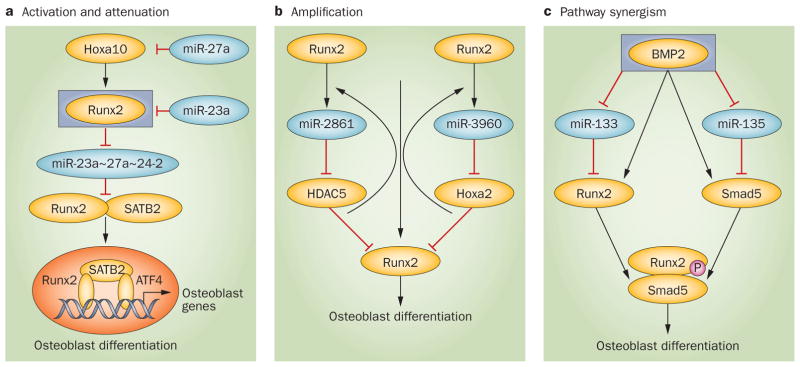
Regulatory circuits operating in osteoblast lineage cells. a | Activation and attenuation pathways. From top, BMP2 induces Hoxa10 which activates Runx2.134,135 Runx2 downregulates expression of the cluster miR23a~27a~24-2 directly through a Runx regulatory element in the promoter for this cluster, which results in increased Runx2 and its coregulator SATB2 protein, that are targets of the cluster miRNAs. Runx2, SATB2 and ATF4 form an activating complex that is essential for differentiation.69 The cluster becomes robustly elevated at mineralization through a feedback mechanism by which miR-27a suppresses Hoxa10 and miR-23a attenuates Runx2 protein. This feedforward and feed reverse circuit exemplifies the central role of this microRNA cluster in promoting and attenuating the progression of differentiation to regulate bone formation. b | Amplification mechanism. Two microRNAs form the cluster miR-2861~miR-3960, each of which targets inhibitors of Runx2 activity, HDAC5 and Hoxa2, respectively. This downregulation increases Runx2 transcriptional activity. The effect is amplified because Runx2 transactivates the cluster through a Runx2 binding site in the cluster’s promoter.70 Thus, a regulatory circuit exists where Runx2 and a microRNA cluster operate in a coordinate manner. c | Pathway synergism. BMP2 downregulates miRNAs that suppress two essential factors for osteogenesis that form a transcriptional complex. The integration of Runx2 and miRNAs occurs at two levels—Runx2 regulation of miRNA expression and miRNA blockade of Runx2 inhibitors.
This miR-23a feedback regulation of Runx2 during the mineralization stage of bone formation, together with the Runx2 feedforward attenuation of the miR cluster, provides a clear example of a transcription-factor–miRNA regulatory circuit for activation and attenuation of the osteoblast differentiation program.62 Such regulatory configurations involving clustered miRNAs and their cognate gene regulators may function to maintain physiological expression levels of genes that regulate the progression of osteoblast differentiation.
A second interesting miRNA regulatory loop that is linked to bone homeostasis involves the miR-2861~miR-3960 cluster (Figure 5b) which is induced by the BMP2–Runx2 pathway.70 Runx2 binding to the cluster’s promoter increases expression of both miRNAs. miR-2861 targets Hdac5, which is a transcriptional corepressor that antagonizes Runx2-mediated bone formation71 and, therefore, miR-2861 epigenetically promotes osteoblast differentiation by Hdac5 repression. miRNA-3960 targets Hoxa2, which also opposes the biological activity of Runx2. Consequently, the two Runx2 inhibitors, Hdac5 and Hoxa2, become repressed, thereby maximizing Runx2 activity to drive differentiation.70
Thus, the miR-23a~27a~24-2 and miR-2861~ miR-3960 clusters are each central components of two intersecting regulatory loops that are respectively repressed and activated by Runx2. Taken together, these studies demonstrate that miRNAs are the core of intricate circuits involving components of multiple pathways for promoting osteogenesis. Consequently, the biological significance of miRNAs extends far beyond their direct targeting of specific proteins. These regulatory circuits fulfill a requirement to manage the timing and level of operation of the many signaling networks that are integrated for ordered bone formation. In silico biological analysis (analysis of proteins using programs that define functional categories of miR targets combined with pathways analyses) indicates that many of the miRNAs expressed in both bone and cartilage are linked to intricate regulatory networks.72 How these multiple miRNAs and their corresponding networks support skeletal development and homeostasis remains to be experimentally addressed.
miRNAs manage cell fate pathways
Mesenchymal stem cells are allocated into differentiated lineages that include fibroblasts, adipocytes, muscle and nerve cells, chondrocytes and osteoblasts.73 The specification of these phenotypes is supported by multiple levels of control, which originate from the initial developmental signals that are transduced to intracellular mediators for activation of phenotypic gene expression. The distinct phenotypes are determined in part by tissue-specific transcription factors as well as miRNAs (Figure 6). Both have the ability to activate and repress gene expression as the cell marches through its differentiation program. Recognition now exists that both transcription factors and miRNAs together support cell commitment and differentiation through gene transcription and protein translation, respectively, and that these two classes of transregulators can functionally interact to coordinate gene regulatory networks.74
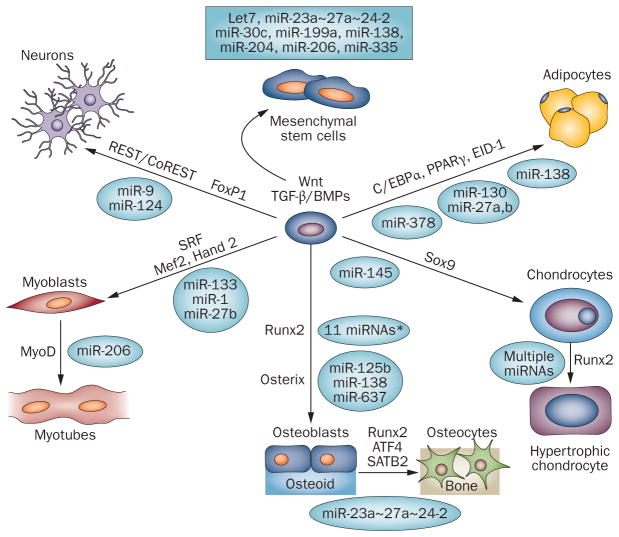
Allocation of mesenchymal stem cells (MSCs) to lineage-specific phenotypes by transcription factors and microRNAs. Schematic illustration of MSC lineages directed by cell-type specific transcription factors (arrows). Selected miRNAs highly expressed in MSCs are shown because they are downregulated during differentiation into phenotype-committed cells. Cell-type-related miRNAs targeting the transcription factors or their coregulatory proteins are indicated. Although the transcription factors are attenuated by miRNAs at different stages of maturation, they are critical for regulating a normal program of differentiation and for cell specification within a tissue. Relevant references to support this concept for the indicated tissues are as follows: muscle,10,17,136 nerve,78–80,137 fat,16,75,76,138,139 bone and cartilage (Table 1). *Many miRNAs repress Runx2 directly (shown in Figure 7), and to date three miRNAs have been shown to repress osterix: miR-125b140 in vascular smooth muscle cells prevents calcification and miR-138 in MSCs retains stemness,88 and miR-637 is expressed in adipocytes.150
Illustrated in Figure 6 are miRNAs that target or regulate indirectly the essential transcription factors for lineage commitment from an MSC or pre-phenotype progenitor cell. In adipocytes, miR-27b targets PPARγ,75,76 and miR-27a reduces levels of C/EBPα.75,76 Essential to neuronal differentiation are miR-9/miR-9*77 and miR-124, which together can establish the neurogenic phenotype; these miRNAs are implicated in regulation of REST and CoREST78 and components of the SWI/SNF remodeling complex.79 miR-9 also regulates the FoxP1 transcription factor and, therefore, contributes to the differentiation of motor neuron subtypes.80 The muscle-specific miRNAs play key roles in positive and negative regulation of transcription factors required for proliferation and differentiation of myoblasts and their maturation to myotubes (Figure 6).10 Overall, Figure 6 illustrates miRNAs that are negative regulators of the phenotypic essential transcription factors; these miRNAs are generally expressed in undifferentiated cells and downregulated by an inducer of the differentiation program.
Profiling studies of human MSC differentiation to osteoblasts81–83 and other phenotypes (for example, neuronal cells, adipocytes, skeletal and vascular smooth muscle cells),84,85 show a remarkable consistency in the many highly expressed miRNAs in MSCs (Figure 6), given that the studies examined MSCs derived from different patient and/or tissue sources. MSCs express miRNAs that target proteins to retain their stem cell property, such as Lif,81,85,86 and protect their survival. These cells have altered expression of miRNAs when exposed to conditions that induce apoptosis.87 Also apparent is that miRNAs highly expressed in MSCs repress differentiation to phenotypes (Figure 6 and Table 1). For example, the miR-27 isoforms prevent differentiation into both adipocytes75,76 and osteoblasts.62,75 However, in one study, miR-27 was reported to increase Wnt signaling in MSCs and stimulate osteoblast differentiation.63 This inconsistency most likely reflects the maturation stage of the cells being used in each study, because the effect of an miRNA is in part dependent on the mRNA level of the target in a cell type. Noteworthy is that miR-138 inhibits both osteogenesis88 and adipogenesis89 and is downregulated as human MSCs are differentiated to either lineage.
Several miRNAs also support ‘crossregulation’ in different cell types. miRNA suppression of tissue-specific transcription factors of alternate lineage has been indicated as a mechanism to facilitate commitment to one cell fate by repressing alternate cell fates and to maintain stability of the differentiated phenotype.17 For example, the muscle-specific miRNA-133 is highly expressed in premyocytes in a negative feedback loop to regulate muscle cell fate and survival.10 Perhaps equally important, miR-133 also potently downregulates the osteogenic phenotype by decreasing Runx2 levels.89 Furthermore, miR-204, which suppresses the osteoblast phenotype through Runx2 repression, promotes adipogenesis.90,91 During adipogenesis, miR-30 is a key regulator in promoting adipocyte differentiation partly by targeting Runx2.92 The muscle-specific miR-206 supports myotube differentiation, and its levels increase during skeletal muscle development,93,94 but it also inhibits osteo genesis by targeting connexin 43.95 miR-335 targets Runx265 and is upregulated during adipocyte differentiation.96,97 This mechanism may be operative to support both pluripotency in MSCs and the stability of adipocytes. These examples highlight both complementary and opposing functions of miRNAs dependent on the cell phenotype.
The armamentarium of Runx2 miRNAs
Runx2 (also known as AML3 or Cbfa1) is one of three mammalian runt-related transcription factors, each of which is essential for specification of distinct cell fates.98–100 Runx2 protein, which is required for phenotype commitment and differentiation of osteoblasts, is regulated by 12 validated and conserved miRNAs (Figure 7).62,65,90,92 The majority of miRNAs targeting Runx2 are strikingly increased in expression during the late mineralization stage of osteoblast differentiation as part of a requirement to tightly regulate Runx2 protein levels. Transgenic mouse models have demonstrated the importance of maintaining physiologic levels of Runx2. Overexpression of Runx2 in osteoblast lineage cells, rather than increasing bone formation, results in osteopenia.101 The bone loss is related to enhanced secretion of RANKL, the inducer of osteoclastogenesis.102 Thus, by attenuating Runx2 through miRNA inhibition, mechanisms are in place to prevent uncontrolled bone resorption.
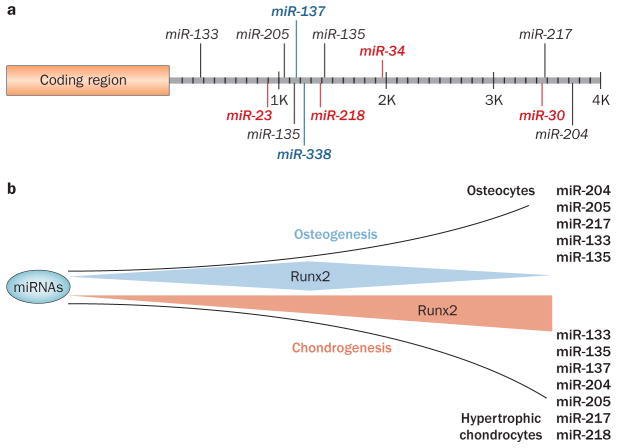
MicroRNAs (miRNAs) regulating Runx2. a | The mouse Runx2 3′ untranslated region. Binding sites are shown for validated miRNAs.90,91 MiRNAs are expressed at high (red), moderate (black) or low (blue) levels in murine MC3T3 preosteoblasts (at confluency in growth media).90 In addition, miR-335 targets the human Runx2 3′ untranslated region.65 b | Modification of Runx2 miRNAs during differentiation. The osteochondral progenitor is a bipotent cell and commitment to either the osteoblastic or chondrocytic lineage is regulated in part by miRNAs. A subset of Runx2-targeting miRNAs is downregulated as cells are induced into osteogenesis, while a subset is upregulated in differentiated osteocytes to support physiological regulation of Runx2. A group of miRNAs targeting Runx2 is highly expressed at the onset of chondrogenesis to inhibit the bone phenotype. However, with further differentiation to the hypertrophic chondrocyte, miRNAs targeting Runx2 are downregulated.90
The multiplicity of miRNAs that target Runx2 is biologically significant in that selective expression of each targeting miRNA occurs in distinct nonosseous cell types.90 This finding is consistent with a common theme by which tissue-specific transcription factors and miRNAs together regulate cell-fate decisions. As an example, the bipotential osteochondroprogenitor has several molecular switches in place for commitment of this cell to either the osteoblast or chondrocyte phenotype in developing bone and during fracture repair. Transcriptional mechanisms to downregulate Runx2 for chondrogenesis to proceed are well-documented;39,103–106 in addition, miRNAs are an important component of lineage direction as described above. Our studies,90 which used in vitro models, show that several miRNAs targeting Runx2 are highly expressed in the prechondrocyte in order to suppress Runx2 and favor chondrogenesis. Subsequently, cartilage cells differentiate into hypertrophic chondrocytes within the growth plate and miRNAs targeting Runx2 become downregulated. As a result, Runx2, which is required for hypertrophic chondrocyte differentiation, is elevated to drive progression of endochondral bone formation. Figure 7 shows that the subsets of miRNAs targeting Runx2 are differentially modified between osteoblasts and chondrocytes during their respective differentiation program.
These examples suggest a requirement for multiple miRNAs to regulate precise protein levels of Runx2 during osteoblast differentiation and in relation to differentiation of MSCs to nonosseous cell phenotypes. miRNAs have probably evolved to regulate Runx2 expression to limit its potent biological activity in pluripotent, multipotent or the bipotential osteochondroprogenitor. A remaining challenge is to unravel the mechanisms that contribute to the expression levels of the miRNAs that are at the center of regulating the networks controlling the differentiation program.
BMP/TGF-β regulates miRNAs for cell fate
The intricate molecular mechanisms that commit MSCs to many specialized cell phenotypes involve common signaling pathways, such as those involving BMP/TGF-β superfamily members and Wnt signaling. The intra-cellular machinery for mediating these external signals converges on a limited number of regulatory transducers (for example, Smads and β-catenin) that interact with cognate nuclear effectors (such as Runx2 and other transcriptional regulators). These mechanisms are enforced in osteoblasts by a requirement for formation of a complex of Smad and Runx2 for completion of osteoblast differentiation that provides a level of tissue specificity in response to BMP/TGF-β signaling.107 BMP and TGF-β are known to regulate miRNA biogenesis through ribonuclease 3 (also known as the Drosha enzyme).108 The regulation of specific miRNAs by the BMP/TGF-β family has been described. BMP2 blocks processing of pri-miR-206.109 TGF-β is a known inhibitor of muscle cell differentiation that downregulates key miRNAs for transcriptional control of myogenesis, miR-206 and miR-29.110 TGF-β also regulates miR-21 and miR-24 expression in order to stimulate vascular smooth muscle cell fate,108,111 or promotes skeletal smooth muscle cell differentiation by inhibiting miR-24 alone.112 Evidence from a number of studies that have characterized changes in miRNA levels in response to BMP induction of specific phenotypes supports the concept that BMP signaling functions through miRNAs to regulate myocardial,113 adipogenic114 and osteoblastic differentiation.89,91,115
As miRNAs can directly repress cell phenotypes by targeting transcriptional regulators or signaling pathway inducers, mechanisms are required to release this repression for commitment to one cell type. BMP2, a potent osteogenic factor, was shown to downregulate a group of miRNAs that target inhibitors of multiple osteogenic pathways in premyogenic cells, including BMP and Wnt receptors and their ligands, as well as transcriptional regulators and MAPK signaling components,89 all essential for osteoblast differentiation.116,117 Although attenuation of each target protein by the miRNAs may be modest, together they have a robust effect in preventing osteoblast differentiation in the absence of BMP2.89 Figure 5c illustrates this mechanism of pathway synergism to promote expression of osteoblast-related genes that is based on the functional characterization of two microRNAs, miR-133, which represses Runx2,89 and miR-135, which inhibits Smad589 and Runx2.90 These targets are established factors essential for osteoblast differentiation.118,119 The convergence of Runx2 and miRNA signaling in feedforward loops reflects the intricate circuits that are needed for osteoblast commitment and progression of differentiation.
Interestingly, the few miRNAs that are strongly upregulated by BMP2 in premyogenic cells target essential factors for muscle cell differentiation, thereby repressing differentiation to myocytes (Table 1). This finding further confirms a miRNA-based mechanism for lineage switching from premyoblasts to osteoblasts by BMP2 and on a broader scale reveals linkage of signaling cascades operating through miRNAs to support commitment to a phenotype from a stem cell or to change a phenotype.
miRNAs and skeletal disorders
The studies of miRNAs in skeletal disorders are in their infancy. Dysregulation of miRNAs is associated with bone degeneration in osteoporosis and osteoarthritis, disorders affecting millions of individuals worldwide. These diseases not only have a genetic component but can be secondary to trauma or other diseases (such as diabetes mellitus or Crohn’s disease) or drugs used for their treatments. Osteoporosis is bone loss that most commonly occurs after menopause in women, but also in aging men. Osteoarthritis is the result of destruction of the articular cartilage and degeneration of the underlying subchondral bone over many years.
Several studies have examined genome-wide associations for susceptibility to osteoarthritis and for correlations between BMD and fracture risk in osteoporosis.120 A study has identified an association of three polymorphisms predicted in miRNA targeting sites residing in the 3' untranslated region of FGF2 mRNA with low BMD in patients with osteoporosis,121 although direct regulation was not tested. Future studies addressing polymorphisms in miRNAs or their interaction sites may provide insight into disease progression.
Another study identified mutations in a miRNA in two related adolescents that contribute to juvenile osteoporosis.122 A homozygous mutation in pre-miR-2861 blocked expression of the mature miRNA. As a consequence, levels of HDAC5, a miR-2861 target, were increased, concomitant with low levels of RUNX2 in bone samples from the patients. The mutant miR-2861 failed to support in vitro osteoblast differentiation. This study establishes a miRNA-related mechanism for this particular clinical situation of osteoporosis. However, microRNA-mediated contributions to progression of osteoporosis in the general population have yet to be addressed.
Studies of normal and diseased cartilage have revealed that miRNA expression is altered in osteoarthritis, and the changes can both contribute to the disease and protect the tissue. Many of the identified miRNAs that are associated with osteoarthritis either indirectly affect expression or directly target inflammatory cytokines, cartilage matrix components and enzymes that degrade cartilage.18,123–128 miR-27a decreases MMP13, which may represent a protective mechanism against damaged cartilage, possibly to counter the indirect effects of miR-27 isoforms, whereas miR-27b is stimulated by the pro-inflammatory cytokine IL-1.125 Thus, in normal tissue, a subset of miRNAs suppresses inflammation,19,129,130 and their decreased expression in chronic inflammatory disorders exacerbates the disease process. Once the direct targets of these miRNAs are established, therapeutic approaches with miRNAs and anti-miRNAs can be designed.
One of the most well-studied miRNAs downregulated in human osteoarthritic tissue is miRNA-140, which is associated with the normal development of embryonic cartilage (see above).27,28 Osteoarthritic cartilage contains reduced miR-140 levels compared with normal tissue, likely suppressed by IL-1β.131 A loss of function of this miRNA in mice results in age-related, osteoarthritic-like features, including fibrillation of the articular cartilage that is associated with loss of proteoglycans and elevation of a cartilage matrix-degrading enzyme.28 Importantly, a transgenic mouse overexpressing miR-140 was protected from induced osteoarthritis.28 Targets of miR-140 that affect osteoarthritic cartilage include ADAMTS5,27,28 IGF-binding protein 5 and MMP13123 (Supplementary Table 1 online). Other miRNAs associated with osteoarthritis are also described in Supplementary Table 1 online. These few studies show promise that miRNAs and antagonistic miRNA mimics (antagomiRs) can potentially be developed as therapeutic strategies to inhibit inflammation and to stimulate phenotypic progression and physiologic functions.
Current challenges in developing miRNAs as therapeutics include the lack of understanding of the precise control of miRNA regulation of biological processes that can be modified in a very selective manner by therapeutic expression or antagonism of miRNAs. Expression of a single miRNA in a broad spectrum of cells and with a vast expanse of targets in interconnecting regulatory networks will demand delivery systems with a high degree of specificity. Nonetheless, further elaborating miRNA regulation and control at molecular levels for normal and deregulated biological processes will increase our understanding of pathological disorders.
Conclusions
Identification of miRNAs in bone has contributed to an emerging dimension of control for skeletal gene expression. The elimination of mature miRNAs in different cells that form bone and cartilage tissue and remodel the skeleton has demonstrated their powerful involvement in regulation of cell growth and differentiation for normal embryonic development, postnatal bone formation processes and regulation of bone mass in the adult skeleton. Although the skeletal phenotypes related to disrupted miRNA functions provide evidence for their biological relevance, the exact mechanisms are still poorly understood. There is a compelling requirement to identify the specific miRNA defects in Dicer-deleted cell populations and their causal relationship to the skeletal phenotypes in mice. It is estimated that miRNAs regulate ~60% of the human genome. Only a small fraction of the many miRNAs that have been identified through in vitro expression profiling of skeletal cells undergoing differentiation programs have been characterized for their functional activity and known targets.
During bone formation, subsets of miRNAs function at both early and late stages of osteoblast and osteoclast differentiation to regulate the progressive differentiation by repressing inhibitors of pathways operating in these cells. Clearly, miRNAs have both direct and important indirect effects on phenotype development through inhibition of positive and negative transcriptional and cytokine regulators that work in complex miRNA dependent networks. The convergence of multiple miRNAs on critical transcription factors for differentiation, extra-cellular matrix components and signaling factors is consistent with miRNA-based mechanisms for orchestrating the activities of key regulators that sustain a viable tissue. How the numerous miRNAs involved in the control of bone formation and remodeling coordinate the activities of these pathways is an area for investigation. A current hypothesis postulates that all types of RNA transcripts have a communication system to form large-scale regulatory networks.132
Studies to date have provided an important glimpse of the potential parameters of control to be addressed. Compelling future steps will include improving our understanding of the regulatory circuits controlled by individual miRNAs and the roles of the many miRNAs that are responsive to and regulate osteogenic signals for induction of bone; also important will be an assessment of how the networks operate temporally and how they integrate with other networks. Regardless of the molecular intricacies, these networks ultimately result in relatively simple anabolic or catabolic outputs. The challenge now is to identify miRNAs that are responsible for specific cell phenotypes by controlling the principal regulatory nodes that govern these outputs. Equally important, miRNAs associated with skeletal diseases need to be determined, as the capacity to modulate allocation of MSCs into different lineages using miRNAs and miRNA antagonists may translate into applications for tissue engineering and modification of genetic disorders.
Acknowledgments
The authors’ work was supported by NIH grants DE012528, AR039588, AR049069 and AR048818. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
All authors researched the data for the article and provided a substantial contribution to discussions of the content. J. B. Lian, G. S. Stein, A. J. van Wijnen and J. L. Stein wrote the article and reviewed/edited the manuscript before submission.Supplementary information
Supplementary information is linked to the online version of the paper at www.nature.com/nrendo
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrendo.2011.234
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3589914?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nrendo.2011.234
Article citations
From Genomics to Metabolomics: Molecular Insights into Osteoporosis for Enhanced Diagnostic and Therapeutic Approaches.
Biomedicines, 12(10):2389, 18 Oct 2024
Cited by: 0 articles | PMID: 39457701 | PMCID: PMC11505085
Review Free full text in Europe PMC
Low frequency sinusoidal electromagnetic fields promote the osteogenic differentiation of rat bone marrow mesenchymal stem cells by modulating miR-34b-5p/STAC2.
Commun Biol, 7(1):1156, 16 Sep 2024
Cited by: 0 articles | PMID: 39284881 | PMCID: PMC11405519
Genetic Background of Medication-Related Osteonecrosis of the Jaw: Current Evidence and Future Perspectives.
Int J Mol Sci, 25(19):10488, 29 Sep 2024
Cited by: 0 articles | PMID: 39408816 | PMCID: PMC11477157
Review Free full text in Europe PMC
Morinda Officinalis Polysaccharides Inhibit Osteoclast Differentiation by Regulating miR-214-3p/NEDD4L in Postmenopausal Osteoporosis Mice.
Calcif Tissue Int, 115(5):673-685, 28 Aug 2024
Cited by: 0 articles | PMID: 39198270 | PMCID: PMC11531433
Role of MicroRNA-204 in Regulating the Hallmarks of Breast Cancer: An Update.
Cancers (Basel), 16(16):2814, 10 Aug 2024
Cited by: 0 articles | PMID: 39199587 | PMCID: PMC11352763
Review Free full text in Europe PMC
Go to all (347) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MicroRNAs and Periodontal Homeostasis.
J Dent Res, 96(5):491-500, 09 Jan 2017
Cited by: 42 articles | PMID: 28068481 | PMCID: PMC5453493
Review Free full text in Europe PMC
Osteoblast recruitment to sites of bone formation in skeletal development, homeostasis, and regeneration.
Birth Defects Res C Embryo Today, 99(3):170-191, 01 Sep 2013
Cited by: 84 articles | PMID: 24078495
Review
MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review).
Mol Med Rep, 14(1):623-629, 24 May 2016
Cited by: 57 articles | PMID: 27222009 | PMCID: PMC4918597
Review Free full text in Europe PMC
The expression and function of microRNAs in bone homeostasis.
Front Biosci (Landmark Ed), 20(1):119-138, 01 Jan 2015
Cited by: 22 articles | PMID: 25553443
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P01 CA082834
Grant ID: R01 CA139322
NIAMS NIH HHS (6)
Grant ID: R01 AR039588
Grant ID: P01 AR048818
Grant ID: AR049069
Grant ID: AR048818
Grant ID: R01 AR049069
Grant ID: AR039588
NIDCR NIH HHS (4)
Grant ID: DE012528
Grant ID: R37 DE012528
Grant ID: R56 DE012528
Grant ID: R01 DE012528





