Abstract
Free full text

A Runx2/miR-3960/miR-2861 Regulatory Feedback Loop during Mouse Osteoblast Differentiation*
Abstract
Our recent study showed that miR-2861 promotes osteoblast differentiation by targeting histone deacetylase 5, resulting in increased runt-related transcription factor 2 (Runx2) protein production. Here we identified another new microRNA (miRNA) (miR-3960) that played a regulatory role in osteoblast differentiation through a regulatory feedback loop with miR-2861. miR-3960 and miR-2861 were found clustered at the same loci. miR-3960 was transcribed during bone morphogenic protein 2 (BMP2)-induced osteogenesis of ST2 stromal cells. Overexpression of miR-3960 promoted BMP2-induced osteoblastogenesis. However, the inhibition of miR-3960 expression attenuated the osteoblastogenesis. Homeobox A2 (Hoxa2), a repressor of Runx2 expression, was confirmed to be a target of miR-3960. Electrophoretic mobility shift assay and chromatin immunoprecipitation experiments confirmed that Runx2 bound to the promoter of the miR-3960/miR-2861 cluster. Furthermore, overexpression of Runx2 induced miR-3960/miR-2861 transcription, and block of Runx2 expression attenuated BMP2-induced miR-3960/miR-2861 transcription. Here we report that miR-3960 and miR-2861, transcribed together from the same miRNA polycistron, both function in osteoblast differentiation through a novel Runx2/miR-3960/miR-2861 regulatory feedback loop. Our findings provide new insights into the roles of miRNAs in osteoblast differentiation.
Introduction
MicroRNAs (miRNAs)3 are an abundant class of endogenous, small (~22-nucleotide), single-stranded, noncoding RNA molecules (1). MicroRNAs mediate translational repression or degradation of target transcript by binding to sites of complementarity in the 3′-UTRs of target mRNAs. During the past decade, they have emerged as powerful post-transcriptional regulators of gene expression (2). To date, more than 3% of the genes in the human genome have been found to encode for miRNAs, and over 30% of genes in the human genome are estimated to be regulated by miRNAs (3). It has been documented that miRNAs participate in various biological processes, including developmental timing, cellular differentiation, proliferation, metabolism, tissue development, protein expression, and tumor development (4–6). However, their role in bone metabolism has just begun to be understood.
The maintenance of bone mass depends on the balance between bone formation and bone resorption; bone morphogenic protein 2 (BMP2)-induced osteoblast differentiation is a crucial component of bone formation (7). The runt-related transcription factor 2 (Runx2), the main transcription factor required for osteoblast differentiation, is important for osteoblast lineage commitment of multipotent mesenchymal cells (8). A few studies have revealed the regulatory role of miRNAs in osteoblastogenesis. Recent studies have discovered some miRNAs as important regulators of bone forming genes, including transcription factors and signaling pathway molecules that are required for osteoblastogenesis (9, 10). Some miRNAs have been reported to act as negative regulators of osteoblast differentiation, such as miR-26a and miR-125b (11, 12). However, not all miRNAs are functional inhibitors of osteoblastogenesis. miR-29b promotes osteogenesis by directly down-regulating multiple inhibitors of osteoblast differentiation (13), and miR-210 positively regulates osteogenesis by inhibiting the TGF-β/activin signaling pathway (14).
Recently, we reported a novel miRNA (miR-2861) that plays a positive regulatory role in osteoblast differentiation by targeting histone deacetylase 5, an enhancer of Runx2 degradation (15). In vivo silencing of miR-2861 using a specific antagomir inhibited bone formation in mice. Our study also discovered a mutation in pre-miR-2861 that resulted in primary osteoporosis in patients. However, the regulatory mechanism of miRNAs in osteoblast differentiation still needs further exploration.
In the present study, we cloned and identified a new miRNA (termed miR-3960) from osteoblasts by a small RNA cloning method and investigated its regulatory mechanism in BMP2-induced osteoblast differentiation. miR-3960 and miR-2861 were found to be clustered at the same loci. We then investigated the transcriptional mechanism of this cluster and have proposed a unique autoregulatory feedback loop between Runx2 and the miR-3960/miR-2861 cluster.
EXPERIMENTAL PROCEDURES
Cell Cultures
Primary mouse calvarial osteoblasts were isolated from calvaria of newborn C57BL/6 mice as reported previously (16). The cells were incubated in α-minimum essential medium (Invitrogen) supplemented with 5% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin and maintained in a humidified 5% CO2 atmosphere at 37 °C. For small RNA isolation and cloning, osteoblasts were plated at 1 × 106 cells/25-cm2 flasks and cultured in 10% FBS and 50 μg/ml ascorbic acid for 9 days. Osteoblast differentiation was induced by changing to medium containing 10% FBS supplemented with 300 ng/ml BMP2 (Peprotech Inc., Rocky Hill, NJ,). Osteoclasts were obtained and cultured as described previously (15). The mouse stromal cell line ST2 was obtained from RIKEN BioResource Center (Ibaraki, Japan) and cultured in α-minimum Eagle's medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2.
Small RNA Isolation and Cloning
Small RNAs were isolated from mouse osteoblasts using a mirVanaTM miRNA isolation kit (Ambion, Austin, TX,) according to the manufacturer's instructions. Small RNA isolation and cloning were performed as described (17). Briefly, small RNAs were polyadenylated, and a 5′-adapter was ligated to poly(A)-tailed RNA using T4 RNA ligase (Invitrogen). The ligation products were reverse transcribed to produce small RNA cDNAs, which were then amplified using PCR. The PCR products were directly subcloned into pcDNA3.1 TOPO vector (Invitrogen) for sequencing analysis. The sequences used in miRNA cloning are the same as those in our previous study (15).
Bioinformatics Analysis
DNA sequences were analyzed to locate small RNA sequences in the cloning vector. RNA sequences were subjected to BLAST analyses against the mouse genome. To identify miRNAs, all small RNAs were initially searched in the miRBase (6–8). Secondary structures of RNA precursors were predicted by importing a fragment of ~200 bp of genomic sequence flanking the small RNA at both the 5′- and 3′ ends into the Mfold software (9). If a small RNA was 19–24 bases long, its precursor sequence could form a stem-loop structure, and it had not been registered in the miRBase, we then classified it as a novel miRNA. Genes encoding these miRNAs were then located on chromosomes. The sequences of the new miRNA candidates and their precursors were subjected to a BLASTN search against NCBI genomes to estimate the species conservation.
Northern Blot
Total RNA was extracted from cells and C57BL/6 mouse (8-week-old) tissues with TRIzol reagent (Invitrogen). Northern blotting was performed as described previously (18). 20 μg of RNA was separated on a 15% urea-polyacrylamide gel with 0.5× Tris borate-EDTA and transferred to a Hybond-N+ nylon membrane (Amersham Biosciences) using a semidry transfer cell (Bio-Rad). Hybridization was performed according to a standard protocol. 32P-Labeled oligonucleotide probes complementary to the mature miR-3960 were used in the hybridization. The probes for miR-3960 and U6 were 5′-GCCCCCGCCTCCGCCGCCGCC-3′ and 5′-ATATGGAACGCTTCACGAATT-3′, respectively.
Western Blot
Protein concentrations were determined using a Bradford protein assay. Protein (100 μg) from each sample was loaded onto a 7.5% polyacrylamide gel. After electrophoresis, the SDS-PAGE-separated proteins were transferred to a PVDF membrane (Millipore, Billerica, MA). The membrane was blocked with 5% nonfat milk in PBS and then incubated with antibodies against Runx2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), homeobox A2 (Hoxa2) (Santa Cruz Biotechnology, Inc.), Myc (Santa Cruz Biotechnology, Inc.), or β-actin (Abcam, Cambridge, MA) in PBS for 3 h. The membrane was then reprobed with appropriate secondary antibodies conjugated with horseradish peroxidase for 1 h. Blots were processed using an ECL kit (Santa Cruz Biotechnology, Inc.) and exposed to film.
qRT-PCR Analysis
qRT-PCR was performed as previously described using a Roche LightCycler (Roche Applied Science) (19). Total RNA from cultured cells or tissues was isolated using TRIzol reagent (Invitrogen), and reverse transcription was performed using 1.0 μg of total RNA and SuperScript II (Invitrogen). PCRs (25-μl total volume each) were set up using reverse transcribed cDNA as template, amplification primers, and SYBR Green PCR Master Mix (Applied Biosystems, Shanghai, China). The following primer sequences were used: type II Runx2: forward, 5′-AGCCTCTTCAGCGCAGTGAC-3′; reverse, 5′-CTGGTGCTCGGATCCCAA-3′; and Hoxa2: forward, 5′-GTCACTCTTTGAGCAAGCCC-3′; reverse, 5′-TAGGCCAGCTCCACAGTTCT-3′.
Amplification data were analyzed using the Sequence Detector System Software (Applied Biosystems). Relative quantification was calculated by normalizing the crossing threshold (Ct) values of the test samples with that of the amplified β-actin control.
Plasmid Constructs and Transfections
Plasmid constructs were performed as described previously (15). A 73-bp genomic sequence of the miR-3960 precursor (pre-miR-3960) was inserted into the pSilencer vector (Ambion). The primers were 5′-GATCCGGCCACGGCTTCCTGCGCCCCCGATCGGGGCCGCCAACAGCGCTGGCGGCGGCGGCGGAGGCGGGGGCAGTGGCGGAAA-3′ (forward) and 5′-AGCTTTTCCGCCACTGCCCCCGCCTCCGCCGCCGCCGCCAGCGCTGTTGGCGGCCCCGATCGGGGGCGCAGGAAGCCGTGGCCG-3′ (reverse). The oligos were annealed and subsequently ligated with T4 DNA ligase (Invitrogen) into the BamHI-HindIII site in the pSilencer 4.1-CMV vector containing the puromycin cassette. For stable transfection of pre-miR-3960, ST2 cells were seeded in 6-well plates at a density of 3 × 104 cells/well and transfected with this plasmid (pSilencer pre-miR-3960) using Lipofectamine 2000 (Invitrogen). Stably transfected cells were clonally selected using puromycin (1 μg/ml). Empty vector (pSilencer control) was also stably transfected into ST2 cells as a control.
A segment (nucleotides 281–733) of the mouse Hoxa2 coding sequence (CDS) including the predicted miR-3960 binding site was PCR-amplified using mouse cDNA as template. The sense primer was 5′-GGCTCTAGAGCCTGAGTATCCCTGGATG-3′, and the antisense primer was 5′-GGCCGGCCACCCTTCCCTCTCCAGAAG-3′. The PCR product was purified and then inserted into the XbaI-FseI site immediately downstream of the stop codon in the pGL3 basic luciferase reporter vector (Promega Corp., Madison, WI), resulting in WT-pGL3-Hoxa2. The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce mutations into the seed region of WT-pGL3-Hoxa2. The primers for Hoxa2 CDS mutagenesis were 5′-CCAAGAAAACCGCGCTGCCGCCCGCAGCAGCATCCACGGGCC-3′ (forward) and 5′-GGCCCGTGGATGCTGCTGCGGGCGGCAGCGCGGTTTTCTTGG-3′ (reverse). The oligonucleotide primers, each complementary to opposite strands of the vector, were extended during temperature cycling by using PfuTurbo DNA polymerase. Incorporation of the primers generated a mutated plasmid containing staggered nicks. Following temperature cycling, the products were treated with DpnI, which was used to digest the parental DNA template and to select for the mutation-containing synthesized DNA. The nicked vector DNA incorporating the desired mutations was named MUT-pGL3-Hoxa2. WT-pGL3-Hoxa2 and MUT-pGL3-Hoxa2 were used for the luciferase reporter assay.
To construct a Runx2 expression vector, pcDNA3.1-Runx2, the Runx2 coding sequence was RT-PCR-amplified from mouse mRNA using the primer sequences 5′-CCGGTACCTTTACAACAGAGGGCACAA-3′ (forward) and 5′-CGCTCGAGCACAGCCAACTCAAACACTA-3′ (reverse). The PCR product was digested with KpnI-XhoI and subcloned into the similarly digested pcDNA3.1 vector (Invitrogen). The expression vector (pcDNA3.1-Hoxa2) for the full-length mouse Hoxa2 was constructed by RT-PCR amplification from mouse mRNA using the primer sequences 5′-CCGGTACCCAGCAGCGATCTTCTATGA-3′ (forward) and 5′-CGGCTCGAGTTAAACGGAAAGTCCTCAA-3′ (reverse). The PCR product was digested with KpnI-XhoI and subcloned into the pcDNA3.1 vector. ST2 cells were seeded in 6-well plates at a density of 3 × 104 cells/well and transfected with pcDNA3.1-Runx2, pcDNA3.1-Hoxa2, or control (empty vector pcDNA3.1) using Lipofectamine 2000 (Invitrogen). After overnight stabilization in the maintenance medium, stably transfected cell clones were selected with G418. After 2 weeks of culture under the selection medium, cell colonies were subcultured. All plasmids above were sequenced to ensure authenticity.
The miRNA inhibitor 2′-O-methyl antisense oligonucleotides targeted toward miR-3960 (anti-miR-3960) and the microRNA inhibitor negative control (anti-miR-C) were purchased from GenePharma Co., Ltd. For transient transfection, complexes of Lipofectamine 2000 and miRNA inhibitors were prepared and directly mixed with cells in 24-well cell culture plates at a density of 3 × 104 cells/well. Then these cells were harvested after transfection for 48 h.
Alkaline Phosphatase (ALP) Activity and Osteocalcin Secretion Assay
ALP activity was measured as described previously (15). Osteocalcin released into the culture media was measured using a specific radioimmunoassay kit (DiaSorin S.p.A., Vercelli, Italy). To normalize protein expression to total cellular protein, a fraction of the lysate solution was used in a Bradford protein assay.
Luciferase Reporter Assay
Luciferase reporter assays were performed as described previously (15). ST2 cells were transfected using Lipofectamine 2000 with either the wild-type or mutant pcDNA3.1-Hoxa2 constructs (200 ng) and pre-miR-3960 or miR-C for 48 h. As a positive control, the modified pcDNA3.1 control vector was used without a CDS insert. Cells treated solely with Lipofectamine served as negative controls. Firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega) with firefly luciferase activities calculated as the mean ± S.D. after being normalized by Renilla luciferase activities from the cotransfected phRL-null vector (Promega).
Preparation of Nuclear Extracts
Nuclear extracts were prepared from BMP2-induced ST2 cells using the NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Beijing, China). Cells were rinsed in ice-cold PBS, harvested by scraping and centrifugation, and resuspended in extraction buffer. After microcentrifugation, nuclear pellets were resuspended in hypertonic buffer and incubated on ice for 40 min. The nuclear extract supernatant was obtained by centrifugation at 14,000 rpm for 30 min and stored at −80 °C. Protein concentrations were measured using the Bio-Rad protein assay kit.
Electrophoretic Mobility Shift Assay (EMSA)
For the gel shift assay, double-stranded oligonucleotide probes were derived from the miR-3960/miR-2861 cluster promoter (wild-type oligo D1 sequence: 5′-GTAGGACCACAGACCAGGAG-3′ the italic nucleotides are the potential binding site for Runx2). The oligonucleotide probe and the complementary strand were annealed and end-labeled with digoxin using T4 polynucleotide kinase according to the manufacturer's instructions (Roche Applied Science). Digoxin-labeled probes were incubated with or without 5–10 μg of nuclear extracts for 30 min at room temperature and separated on a 6% non-denaturing polyacrylamide gel with 0.5× Tris borate-EDTA running buffer. After electrophoresis, DNA-protein complexes were electroblotted onto positively charged nylon membranes (Bio-Rad). For competition, the EMSA was performed by adding a 1:10 or 1:100 excess of unlabeled wild-type or mutant oligonucleotide (mutant oligo D1 sequence, 5′-GTAGGACCgtAGACCAGGAG-3′ the italic and lowercase nucleotides are the mutant nucleotides of wild-type oligo D1). For the supershift assay, the nuclear extracts were incubated with Runx-2-specific antibodies (Santa Cruz Biotechnology, Inc.) or control antibodies (Santa Cruz Biotechnology, Inc.) for 30 min before addition of the labeled probe. EMSA was performed using a digoxin gel shift kit following the manufacturer's instructions (Roche Applied Science).
Chromatin Immunoprecipitation (ChIP)
Cells were grown to 70% confluence and cross-linked with 1% formaldehyde at 37 °C in the presence of 4% CO2 for 15 min. The cells were then lysed, DNA-protein complexes were immunoprecipitated, and the formaldehyde-cross-linked DNA was reverse cross-linked with a ChIP assay kit (Upstate, Charlottesville, VA) according to the manufacturer's protocol. DNA-chromatin complexes were immunoprecipitated with anti-Runx2 (Santa Cruz Biotechnology, Inc.), no antibody, or normal mouse IgG (Santa Cruz Biotechnology, Inc.) as an internal control. The precipitated DNA was analyzed by PCR. The primers used for this analysis were as follows: primer-A: forward, 5′-ACAGGAGGGATTGGGAAACT-3′; reverse, 5′-AGGACAGCCAGGAAAAACAA-3′; primer-B: forward, 5′-AACTCCTGTGCAGGGCTAAA-3′; reverse, 5′-GCCATATGACCCAGAGCCTA-3′; and primer-C: forward, 5′-CCTCGTCTGCTCTGCCTATC-3′; reverse, 5′-AGTGCCCTGCTCCTCCTC-3′.
Small Interfering RNA (siRNA) Duplex Preparation and Transfection
The Runx2 siRNA was designed using The BLOCK-iTTM RNAi Designer (Invitrogen). The sequences of siRNA specific to Runx2 are as follows: 5′-UAACAGCAGAGGCAUUUCGUAGCUC-3′ and 5′-GAGCUACGAAAUGCCUCUGCUGUUA-3′. A nonsense random sequence of siRNA was synthesized as a negative control. The siRNAs were transfected into 50% confluent ST2 cells with Oligofectamine (Invitrogen) according to the manufacturer's instructions. The siRNA experiment was carried out for 72 h. Total RNA and proteins from the specific siRNA oligonucleotide-treated and control siRNA oligonucleotide-treated cells were analyzed by Western blotting and qRT-PCR.
Statistical Analyses
Data are presented as mean ± S.D. Comparisons were made using a one-way analysis of variance. All experiments were repeated at least three times, and representative experiments are shown. Differences were considered significant at p < 0.05.
RESULTS
Identification of Novel miRNA from Primary Mouse Osteoblasts
Short RNA molecules were cloned from primary mouse osteoblasts cultured in six flasks by a small RNA cloning method described previously (17). We identified 163 clones by DNA sequencing and database searching. More than 50% of the cloned RNAs represented breakdown products of abundant non-coding RNAs such as tRNA, rRNA, snRNA, and small nucleolar RNA. Some of them are known miRNAs, including the newly discovered miR-2861 (15). The remaining candidate RNA sequences were compared in the BLAST database, and the Mfold software was used to predict the stem-loop secondary structure of possible miRNA precursors (pre-miRNA) (20).
By bioinformatics analysis described under “Experimental Procedures,” we focused on a putative novel 20-bp miRNA (Fig. 1A), which was located in non-coding regions of chromosome 2 and conserved in the human sequence, similar to miR-2861. Its predicted precursor sequences showed the classical pre-miRNA stem-loop secondary structure (Fig. 1B). The new miRNA was subjected to the microRNA database (miRBase, Cambridge, UK) and officially named miR-3960. When the predicted 73-bp genomic sequence of the miR-3960 precursor (pre-miR-3960) was inserted into vector and transfected into ST2 cells, the levels of mature miR-3960 were elevated (Fig. 2A). This result confirmed the predicted miR-3960 precursor. By bioinformatics analysis, miR-3960 and miR-2861 were found clustered on the same loci (Fig. 1C) where they were separated by only 71 bp.
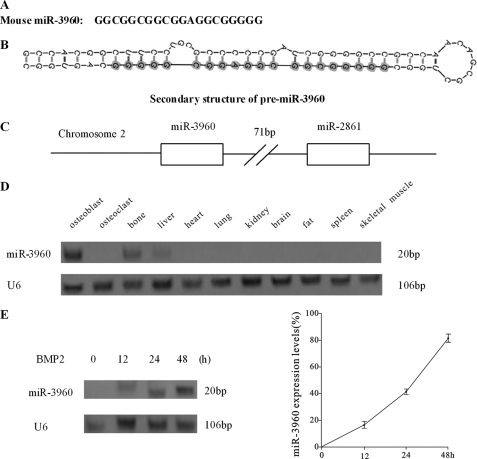
Expression profile of miR-3960. A, sequence of mouse miR-3960. B, schematic graph of the secondary structure of pre-miR-3960. The structure was predicted by Mfold, and the sequence of mature miR-3960 is shown by shadowed circles. C, graphic representation of the cluster of miR-3960 and miR-2861 on mouse chromosome 2. D, Northern blot analysis of the expression of miR-3960 in osteoblast, osteoclast, and different mouse tissues. E, Northern blot analysis showed the time-dependent expression of miR-3960 during BMP2-induced ST2 osteogenic differentiation after treatment with BMP2 (300 ng/ml) for the indicated times. U6 snRNA was used as a loading control. Bar, mean ± S.D. (*, p < 0.05 versus U6; n = 5).
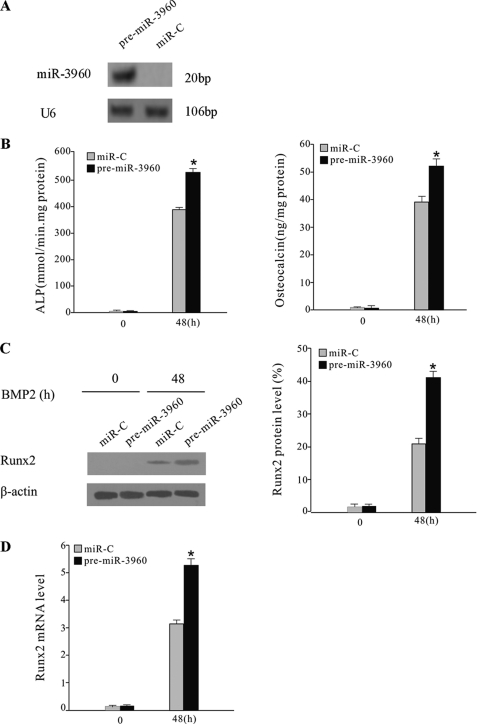
miR-3960 promoted BMP2-induced ST2 osteoblast differentiation. A, Northern blot analysis of miR-3960 expression in ST2 cells transfected with pre-miR-3960 or miR-C (control). B, overexpression of miR-3960 enhanced BMP2-induced ST2 osteoblast differentiation. ST2 cells were stably transfected with pre-miR-3960 or miR-C and then treated with BMP2 (300 ng/ml) for 48 h. The bar graphs show the increase of ALP activity and osteocalcin secretion after transfection of pre-miR-3960 (mean ± S.D.; *, p < 0.05 versus miR-C; n = 5). C, Runx2 protein expression was determined by Western blot. The bar graph represents the ratio of Runx2/β-actin by densitometry (mean ± S.D.; *, p < 0.05 versus miR-C; n = 5). D, levels of Runx2 mRNA were measured by qRT-PCR. The bar graph indicates the -fold induction of Runx2 mRNA expression in pre-miR-3960-transfected ST cells compared with the control (mean ± S.D.; *, p < 0.05 versus miR-C; n = 5).
Expression Profile of miR-3960
We assessed the expression profile of miR-3960 across different tissues by Northern blotting performed with total RNA extracted, respectively, from primary mouse osteoblast cells, primary mouse osteoclast cells, and mouse bone, liver, lung, heart, kidney, brain, fat, spleen, and skeletal muscle. The Northern blot analysis showed that there is tissue-specific expression of miR-3960. In cultured bone cells, miR-3960 was detected at a high level in primary mouse osteoblasts, but it was barely expressed in osteoclasts (Fig. 1D). In mouse tissues, miR-3960 was highly expressed in bone and detected at a lower level in liver but not found in other tissues (Fig. 1D).
To evaluate the expression profile of miR-3960 during osteoblast phenotype development, we treated mouse ST2 stromal cells with BMP2 and measured miR-3960 expression after 12–48 h. Northern blot analysis revealed that miR-3960 could be detected after treatment with BMP2 for 12 h and increased with time throughout osteoblast differentiation (Fig. 1E). These results showed that the expression profile of miR-3960 with or without BMP2 induction was similar to that of miR-2861. These results (Fig. 1, C and D) indicated that miR-3960 and miR-2861 might be transcribed together during osteoblast differentiation, and miR-3960 might play a similar regulatory role as miR-2861.
Regulation of miR-3960 in BMP2-induced ST2 Osteogenic Differentiation
We next determined the role of miR-3960 during osteoblast differentiation by changing the functional levels of miR-3960 in ST2 cells. After treatment with 300 ng/ml BMP2, ST2 cells were stably transfected with pre-miR-3960, and Northern blotting was performed to evaluate the levels of mature miRNAs (Fig. 2A). ST2 cells transfected with miRNA control (miR-C) were used as a control. ALP activity and osteocalcin secretion, as markers of osteoblast differentiation, were evaluated after 48 h. The levels of ALP and osteocalcin were both elevated in cells transfected with pre-miR-3960 compared with the control (Fig. 2B). The levels of Runx2 protein and mRNA were also elevated by miR-3960 transfection (Fig. 2, C and D). These results indicated that overexpression of miR-3960 promoted osteoblast differentiation of ST2 cells.
To further determine the role of miR-3960, 2′-O-methyl antisense miR-3960 oligoribonucleotides (anti-miR-3960), a specific blocker of miR-3960 function (21), were transfected into BMP2-induced ST2 cells. Anti-miRNA control (anti-miR-C)-transfected ST2 cells were used as a control. The increases of ALP activity and osteocalcin expression were attenuated after transfection with anti-miR-3960 (Fig. 3A). The levels of Runx2 protein and type II Runx2 mRNA were also reduced in anti-miR-3960-transfected cells (Fig. 3, B and C). Our data suggested that inhibition of endogenous miR-3960 inhibited BMP2-induced osteogenic differentiation.
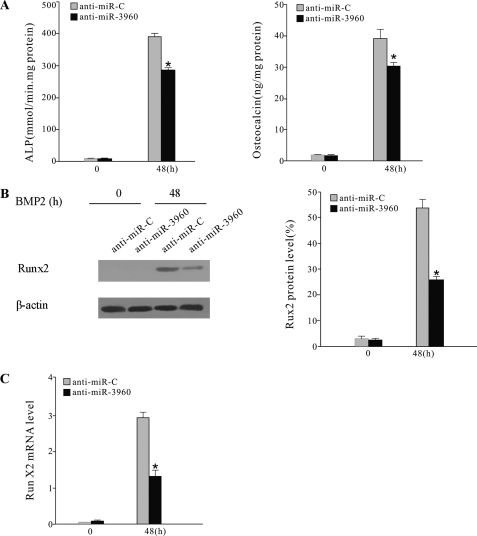
Inhibition of miR-3960 diminished BMP2-induced ST2 osteoblast differentiation. ST2 cells were treated with BMP2 and transiently transfected with anti-miR-3960 or anti-miR-C. A, ALP activity and osteocalcin secretion were reduced after transfection of anti-miR-3960 for 48 h. B, the levels of Runx2 protein were decreased by anti-miR-3960 transfection. C, the decreased expression of Runx2 mRNA after anti-miR-3960 transfection. The error bars represent the mean ± S.D. (*, p < 0.05 versus anti-miR-C; n = 5).
Hoxa2 Is the Target Gene of miR-3960
Some computational algorithms, such as Rna22, have been developed to predict the putative miRNA targets (22). Rna22 can identify putative miRNA binding sites along the length of the entire mRNA transcripts and does not rely upon cross-species conservation. We used Rna22 to predict the target of miR-3960. One putative target site of miR-3960 is predicted in the CDS of Hoxa2 (Fig. 4A). We presumed that miR-3960 promoted osteoblast differentiation by binding with the CDS of Hoxa2.
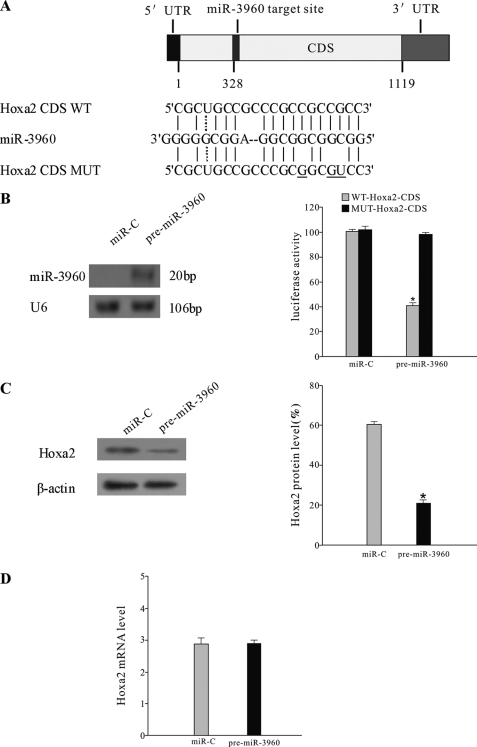
miR-3960 directly targeted Hoxa2. A, schematic representing the putative target site of miR-3960 in mouse Hoxa2 CDS and the base pairing of miR-3960 sequences with wild-type (WT) and mutant (MUT) CDS regions of Hoxa2. Three mutations are underlined. B, miR-3960 targeted the Hoxa2 CDS. ST2 cells were cotransfected with the luciferase reporters carrying wild-type Hoxa2 CDS (WT-Hoxa2-CDS) or mutated Hoxa2 CDS (MUT-Hoxa2-CDS) and pre-miR-3960 or miR-C for 48 h. On the left, Northern blot analysis showed that miR-3960 was overexpressed in ST2 cells transfected with pre-miR-3960. On the right, the bar chart shows the luciferase activities. Concurrent transfection of pre-miR-3960 decreased the reporter activity of WT-Hoxa2-CDS but not the reporter activity of MUT-Hoxa2-CDS. The error bars represent the mean ± S.D. (*, p < 0.05 versus MUT-Hoxa2-CDS; n = 3). C, miR-3960 repressed Hoxa2 expression post-transcriptionally. ST2 cells were transfected with 100 nm pre-miR-3960 or miR-C. Western blot showed the reduced Hoxa2 protein expression by miR-3960 overexpression. Results are indicated as the ratio of Hoxa2/β-actin by densitometry. D, quantitative RT-PCR was used to determine the levels of Hoxa2 mRNA 48 h after transfection. The error bars represent the mean ± S.D. (*, p < 0.05 versus miR-C; n = 5).
To experimentally validate the prediction, we produced a luciferase reporter plasmid in which the wild-type or mutated CDS of Hoxa2 was cloned into the 3′-UTR of the luciferase gene (Fig. 4A). ST2 cells were cotransfected with the Hoxa2 luciferase expression vector (WT-pGL3-Hoxa2) and pre-miR-3960 or miR-C. The luciferase expression vector containing mutant CDS of Hoxa2 (MUT-pGL3-Hoxa2) was designed as a control. We found that ectopic expression of miR-3960 significantly repressed luciferase activity of the wild-type Hoxa2 CDS construct (Fig. 4B). Mutation of three nucleotides within the putative target site in the Hoxa2 CDS (MUT-pGL3-Hoxa2) abolished the repression (Fig. 4B). Thus, these results demonstrated that miR-3960 directly targeted Hoxa2 by specifically binding with the target CDS of Hoxa2.
To directly identify the action of miR-3960 on Hoxa2, we introduced pre-miR-3960 into ST2 cells. Transfection of the pre-miR-3960, rather than the control, resulted in a decrease in the Hoxa2 protein levels after 48 h (Fig. 4C). However, the Hoxa2 mRNA levels were not affected (Fig. 4D). These results revealed that miR-3960 post-transcriptionally repressed Hoxa2 protein expression by inhibiting mRNA translation and not by mRNA degradation.
Some studies have revealed that Hoxa2 inhibits bone formation by repressing Runx2 expression (23). We confirmed the effects of Hoxa2 on Runx2 gene expression (5.4 kb) by experiment in which Hoxa2 was overexpressed. ST2 cells were stably transfected with Hoxa2 expression vector (pcDNA3.1-Hoxa2) or control empty vector (pcDNA3.1) followed by 48 h of culture in the presence of BMP2 (300 ng/ml). Western blotting confirmed the up-regulation of Hoxa2 protein levels (Fig. 5A). As Hoxa2 expression increased, the mRNA and protein levels of Runx2 were subsequently down-regulated in comparison with the control (Fig. 5, A and B). The ALP activity also decreased in cells transfected with the Hoxa2 expression vector (Fig. 5C). These results suggested that Hoxa2 suppressed Runx2 expression and thereby inhibited osteoblast differentiation. Therefore, we drew the conclusion that miR-3960 promoted osteogenesis by directly down-regulating the expression of Hoxa2, an inhibitor of osteoblast differentiation.
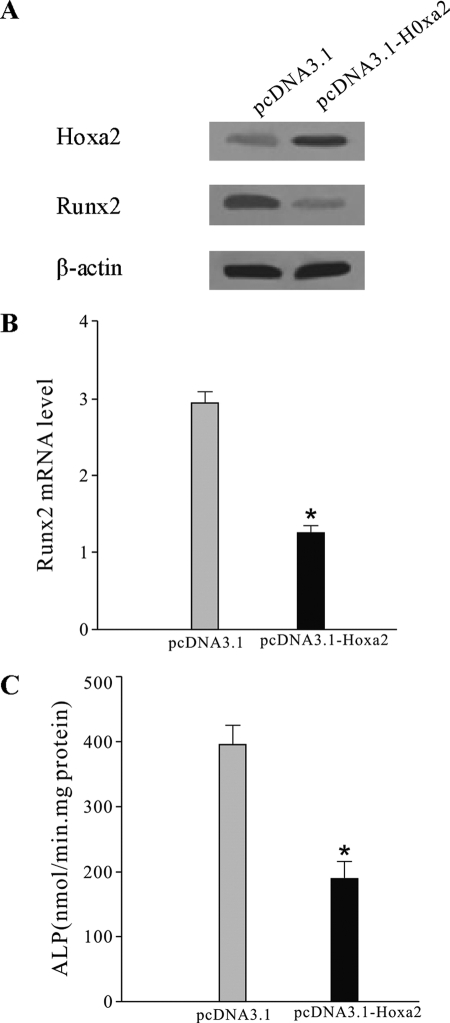
Effect of Hoxa2 overexpression on Runx2 production in ST2 cells. ST2 cells were stably transfected with Hoxa2 expression vector (pcDNA3.1-Hoxa2) or control empty vector (pcDNA3.1) and then treated with BMP-2 (300 ng/ml) for 48 h. A, effect of Hoxa2 overexpression on Hoxa2 and Runx2 expression. Hoxa2 and Runx2 expression was determined by Western blot analysis. B, the levels of Runx2 mRNA by qRT-PCR in cells transfected with pcDNA3.1-Hoxa2 or pcDNA3.1. C, ALP activity was reduced after transfection of Hoxa2 expression vector. The error bars represent the mean ± S.D. (*, p < 0.05 versus pcDNA3.1 control; n = 5).
Runx2 Binds to Promoter and Activates Transcription of miR-3960/miR-2861 Cluster
After showing the physical association between miR-3960 and miR-2861 loci, we wanted to explore the interaction of the two miRNAs in performing their regulatory functions. Based on the recently published data by Marson et al. (24), the genomic coordinates of the trimethylated Lys-4 of histone 3 (H3K4me3)-enriched loci and CpG islands found upstream of miR-3960 and miR-2861 allowed us to predict the transcription start sites (TSS) of miR-3960 and miR-2861 located on mouse chromosome 2: 10,171,258 (NCBI Reference Sequence accession number NT_039206.7).
We then examined 2 kb of the regions upstream of the miR-3960/miR-2861 cluster TSS. Using the TF-Search prediction program, we found a potential binding site for Runx2 located just 1103 nucleotides upstream from the TSS of the miR-3960/miR-2861 cluster. The sequence ACCACA was the core binding site for Runx2 (Fig. 6A).
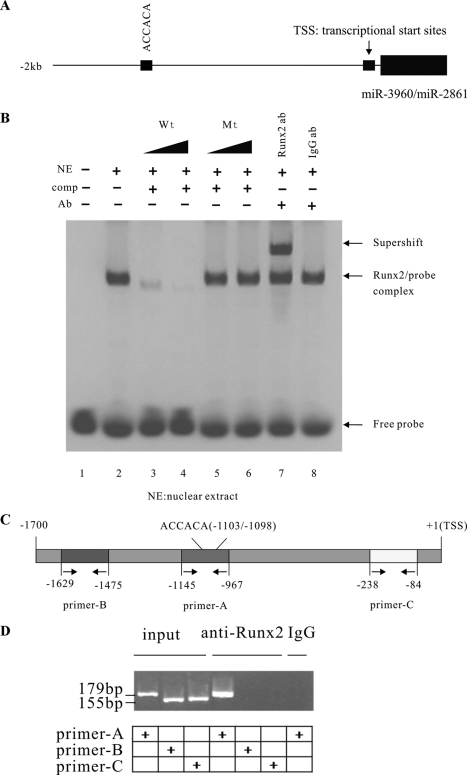
Analysis of Runx2 binding to promoter of miR-3960/miR-2861 cluster. A, a Runx2 binding site was located upstream of the miR-3960/miR-2861 cluster. The schematic represents the genomic region upstream of the miR-3960/miR-2861 cluster. One Runx2 putative binding site upstream of the miR-3960/miR-2861 cluster is indicated. B, EMSA for Runx2 binding to the promoter of the miR-3960/miR-2861 cluster. Electrophoretic mobility shift assays were performed using labeled oligonucleotide probes derived from the promoter of miR-3960/miR-2861 of BMP2-induced ST2 cells. The labeled oligonucleotide probes (WT Oligo 1) were incubated alone (lane 1), in combination with nuclear extracts (NE; lane 2), in the presence of 10- or 100-fold molar excess of specific unlabeled competitor probe (WT Oligo 1) (comp; lanes 3 and 4) or unlabeled mutant (Mt) competitor probe (comp; mutant Oligo 1) (lanes 5 and 6), and in the presence of the Runx2 antibody (anti-Runx2) (Ab; lane 7) or IgG control antibody (Ab; lane 8). C, schematic representation of the promoter region of the miR-3960/miR-2861 cluster. The positions of the Runx2 binding site and primer sites for ChIP assays are indicated. D, ChIP assay showed Runx2 binding to the miR-3960/miR-2861 cluster in BMP2-induced ST2 cells through the putative Runx2 binding site. ChIP assays were performed using no antibodies (input; lanes 1, 2, and 3), Runx2 antibodies (lanes 4, 5, and 6), and control IgG antibody (lane 7). Primer-B (−1629/−1475; lanes 2 and 5) and primer-C (−238/−84, lanes 3 and 6) were used as negative controls for PCR.
To validate whether Runx2 could physically bind with the putative binding sites, EMSA was performed using nuclear protein extracts from BMP2-induced ST2 cells. Double-stranded oligonucleotides (wild-type oligo D1) containing consensus binding sites for Runx2 in the miR-3960/miR-2861 promotor were labeled and used as probes in the EMSA (Fig. 6B). After incubation with the nuclear extracts of ST2 cells, the EMSA studies showed that the wild-type probes clearly formed a binding complex with the Runx2 protein (Fig. 6B, lane 2). The specificity of binding to the Runx2 sites was detected by the competition and supershift assays. An unlabeled wild-type probe competed with the binding complex in a dose-dependent manner (Fig. 6B, lanes 3 and 4). In contrast, a mutant oligo D1 showed no significant competition effect (Fig. 6B, lanes 5 and 6). The presence of Runx2 in the binding complex was shown by the formation of a supershift with specific anti-Runx2 antibody (Fig. 6B, lane 7), whereas control IgG antibodies had no such effect (Fig. 6B, lane 8).
ChIP assays were also performed to verify the possible association of the Runx2 protein with the promoter of the miR-3960/miR-2861 cluster in BMP2-induced ST2 cells. Cross-linked and fragmented DNA-protein complexes were immunoprecipitated with Runx2 antibody, no antibody, or control IgG antibody. After immunoprecipitation, PCR analysis on purified DNA was performed with primer-A that spans Runx2 potential binding sites (−1103/−1098) in the promoter of miR-3960/miR-2861 cluster. Primer-B and primer-C that span the 5′- and 3′-distal regions of putative Runx2 binding sites were used as negative controls in the PCR assays (Fig. 6, C and D). The antibodies against Runx2 could specifically immunoprecipitate the DNA fragment containing the potential Runx2 binding sites (Fig. 6D, lane 4). These findings further confirmed that Runx2 bound to the putative binding sites of the miR-3960/miR-2861 cluster promoter.
To further identify the impact of Runx2 levels on miR-3960/miR-2861 gene expression, we changed the functional levels of Runx2 in ST2 cells. First, we used a pcDNA-driven expression vector to overexpress Runx2 protein levels. The results showed that Runx2 overexpression could induce miR-3960 and miR-2861 expression in ST2 cells (Fig. 7A). Furthermore, we transfected siRNA designed against Runx2 into BMP2-induced ST2 cells to specifically knock down Runx2 protein levels, and siRNA control was used as a control. Western blot analysis revealed that si-Runx2 blocked Runx2 expression compared with the control. The expression of miR-3960 and miR-2861 was inhibited, partially due to the knockdown of Runx2 48 h after transfection (Fig. 7B). Taken together, all these results suggested that Runx2 could directly induce the expression of the miR-3960/miR-2861 cluster by binding to the putative binding site of its promoter.
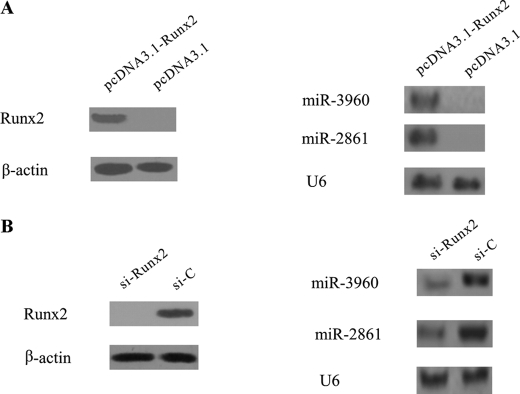
Effects of Runx2 overexpression or knockdown on miR-3960 and miR-2861 expression. A, ST2 cells were transfected with Runx2 pcDNA3.1 vector (pcDNA3.1-Runx2) or pcDNA3.1 vector control. Western blot analysis showed that Runx2 protein levels were elevated after transfection for 48 h. Northern blot represents miR-3960 and miR-2861 expression levels in control or Runx2-overexpressing cells at 48 h after transfection. B, knockdown of Runx2 attenuated BMP2-induced miR-3960 and miR-2861 expression. ST2 cells were transfected with si-Runx2 or siRNA control (si-C) and then cultured with BMP2 for 48 h. Total cellular protein was subjected to Western blot analysis using anti-Runx2 antibody. Northern blot revealed miR-3960 and miR-2861 expression using total RNA isolated from ST2 cells at 48 h after transfection.
DISCUSSION
Recently, several studies have discovered multiple miRNAs that are involved in osteoblast differentiation. In our study, we cloned and identified a new miRNA (miR-3960) clustered with miR-2861 and provided evidence that miR-3960 promotes osteoblast differentiation by targeting Hoxa2. Furthermore, our data showed that miR-3960 and miR-2861, which we reported previously, represents an autoregulatory loop in osteoblast differentiation.
Most miRNAs are expressed in a highly tissue-specific and developmental stage-specific manner (1, 25). In this study, we investigated the expression profile of miR-3960 in various tissues. In mouse tissues, miR-3960 was preferentially expressed in bone and was detected at a lower level in liver but not in other tissues and osteoclasts. We then showed that the expression of miR-3960 gradually was up-regulated during BMP2-induced osteoblast differentiation. Its expression profile was similar to that of miR-2861, another microRNA we reported previously that promotes osteoblast differentiation (15). Subsequent bioinformatics analysis revealed the close proximity of the miR-3960 and miR-2861 loci, suggesting that the two miRNAs are transcribed as a polycistronic unit by the same transcription factor and that miR-3960 may play a similar active role in osteoblast differentiation.
The functional role of miR-3960 was further confirmed by overexpression and knockdown experiments. In ST2 cells, overexpression of miR-3960 promoted BMP2-induced osteoblast differentiation, whereas knockdown of miR-3960 led to the opposite effect, that is an apparent attenuation of BMP2-induced osteoblastogenesis. We then explored the effects of miR-3960 on Runx2, the pivotal transcription factor of osteoblast differentiation. Unlike with miR-2861, miR-3960 overexpression increased the expression of Runx2 mRNA and protein. This result suggested that miR-3960 and miR-2861 regulate osteoblast differentiation via different mechanisms.
miRNAs mediate post-transcriptional gene silencing by base pairing to the complementary sites in the 3′-UTR of the target mRNA (1). However, recent studies showed that miRNAs could also exercise their control on mRNA by binding to the CDS region of transcription factors (26, 27). The bioinformatic algorithm Rna22 predicted the CDS of Hoxa2 as a potential target of miR-3960. The luciferase reporter assay confirmed the repressive effect of miR-3960 on the targeted site of Hoxa2 CDS, and the reduction of this repression by mutations of the target site in the Hoxa2 CDS further confirmed the function of miR-3960. In addition, overexpression of miR-3960 decreased the levels of Hoxa2 protein without changing Hoxa2 mRNA levels, demonstrating that miR-3960 directly targeted Hoxa2 by a post-transcriptional regulatory mechanism.
Hoxa2, a member of the Hox homeodomain family of transcription factors that regulate skeletal patterning, is found to control Runx2 expression and is required in skeletal morphogenesis (23). Hoxa2−/− mice show an up-regulation of the Runx2 level, and Hoxa2 inhibits bone formation by repressing Runx2 expression (23). SATB2, a nuclear matrix protein, represses Hoxa2 expression and thereby activates Runx2-dependent osteoblast differentiation (28). In our study, ST2 cells transfected with a Hoxa2 expression vector showed reduced Runx2 expression and ALP activity, confirming that Hoxa2 can suppress Runx2 expression and thereby inhibits osteoblast differentiation. Therefore, our study demonstrated that miR-3960 promotes osteoblast differentiation via down-regulation of Hoxa2 expression.
We previously identified that miR-2861 induces osteoblast differentiation by repressing histone deacetylase 5, an enhancer of Runx2 degradation (15). Therefore, we hypothesized that miR-3960 and miR-2861 could co-regulate the Runx2 during osteoblast differentiation via a regulatory loop.
During the course of generating functional miRNA, a mature miRNA is processed from a primary transcript (pri-miRNA) by RNA polymerase II. Sequential processing events by the ribonuclease III enzymes Drosha and Dicer yield the mature miRNA species (29–32). Recently, Marson et al. (24) predicted promoter sites and TSS for miRNAs by use of the genomic coordinates of the H3K4me3-enriched loci derived from multiple cell types and bioinformatics analysis. The authors created a library of candidate promoters and TSS for miRNA in both the human and mouse genomes. In the present study, we predicted the promoter and TSS of miR-3960 and miR-2861 based on these published data (24). We then examined the promoter region using the TF-Search prediction program and found a potential binding site (5′-ACCACA-3′) (33) for Runx2 just upstream of the miR-3960/miR-2861 cluster. In our studies, EMSA and ChIP experiments confirmed that Runx2 induced miR-3960 and miR-2861 expression through binding to the promoter regions of the miR-3960/miR-2861 cluster. Furthermore, overexpression of Runx2 induced miR-3960/miR-2861 transcription, and block of Runx2 expression attenuated BMP2-induced miR-3960/miR-2861 transcription. We drew the conclusion that Runx2 binds to the miR-3960/miR-2861 cluster promoter to transcriptionally induce the expression of miR-3960/miR-2861.
These data allowed us to develop a model for miR-3960/miR-2861 function (Fig. 8). Osteoblast-differentiation signals lead to the activation of Runx2 transcription factor in stromal cells. In addition to induction of genes essential for osteoblast differentiation, Runx2 transactivates miR-3960/miR-2861. In turn, miR-3960 and miR-2861 maintain the levels of Runx2 mRNA and protein via repressing Hoxa2 and histone deacetylase 5 and stabilizing the osteoblast differentiation. Thus, we propose that a unique autoregulatory feedback loop exists between Runx2 and the miR-3960/miR-2861 cluster.
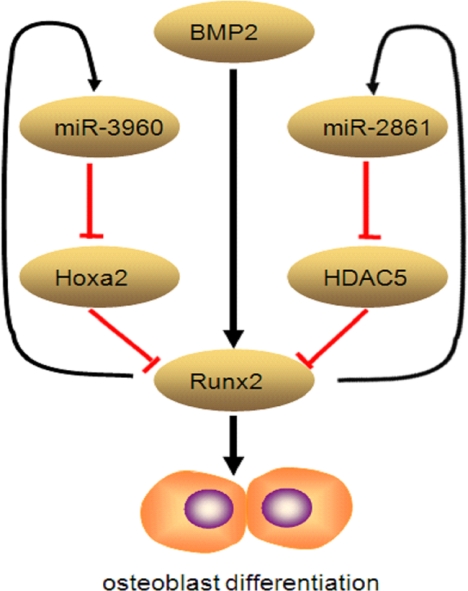
Graphic representation of autoregulatory feedback loop between Runx2 and miR-3960/miR-2861 cluster. Osteoblast differentiation signals lead to the activation of Runx2 transcription factor in stromal cells. In addition to induction of genes essential for osteoblast differentiation, Runx2 transactivates miR-3960/miR-2861. In turn, miR-3960 and miR-2861 maintain the levels of Runx2 mRNA and protein via repressing Hoxa2 and histone deacetylase 5 (HDAC5) and stabilizing the osteoblast differentiation.
In conclusion, the present study provides evidence that the newly described miR-3960 plays an important role in osteoblast differentiation through a novel Runx2/miR-3960/miR-2861 regulatory feedback loop. Our findings also provide new insights into the role of miRNAs in osteoblast differentiation.
Acknowledgments
We thank ELIXIGEN Corp. for native English speaking proofreading of the manuscript. We thank Dr. Genqing Xie and Peng Chen for helpful discussions and comments on the manuscript. We are grateful to Dr. Jicai Meng for the plasmid constructs and technical advice regarding the ChIP technique. We also thank Dr. Dan Wang for advice on quantitative PCR analysis.
*This work was supported by National Natural Science Foundation Grants 30870925, 81070696, and 81000122 from China.
3The abbreviations used are:
- miRNA
- microRNA
- ALP
- alkaline phosphatase
- BMP2
- bone morphogenic protein 2
- CDS
- coding sequence
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- pre-miRNA
- miRNA precursor
- Hoxa2
- Homeobox A2
- H3K4me3
- trimethylated Lys-4 of histone 3
- Runx2
- runt-related transcription factor 2
- TSS
- transcription start sites
- BLAST
- Basic Local Alignment Search Tool
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m110.176099
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/content/286/14/12328.full.pdf
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/286/14/12328.pdf
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/286/14/12328
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/286/14/12328
Citations & impact
Impact metrics
Citations of article over time
Article citations
CRISPR activation identifies a novel miR-2861 binding site that facilitates the osteogenesis of human mesenchymal stem cells.
J Orthop Surg Res, 19(1):730, 06 Nov 2024
Cited by: 0 articles | PMID: 39506798 | PMCID: PMC11542479
Streptococcus gordonii Supragingival Bacterium Oral Infection-Induced Periodontitis and Robust miRNA Expression Kinetics.
Int J Mol Sci, 25(11):6217, 05 Jun 2024
Cited by: 0 articles | PMID: 38892405 | PMCID: PMC11172800
Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities.
Int J Mol Sci, 25(10):5291, 13 May 2024
Cited by: 2 articles | PMID: 38791330 | PMCID: PMC11121608
Review Free full text in Europe PMC
MicroRNAs as potential biopredictors for premenopausal osteoporosis: a biochemical and molecular study.
BMC Womens Health, 23(1):481, 09 Sep 2023
Cited by: 3 articles | PMID: 37689658 | PMCID: PMC10493018
Aberrant expression of miR-33a-3p/IGF2 in postmenopausal osteoporosis patients and its role and mechanism in osteoporosis.
J Orthop Surg Res, 18(1):487, 06 Jul 2023
Cited by: 0 articles | PMID: 37415192 | PMCID: PMC10326950
Go to all (152) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NT_039206.7
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans.
J Clin Invest, 119(12):3666-3677, 16 Nov 2009
Cited by: 303 articles | PMID: 19920351 | PMCID: PMC2786801
A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program.
Proc Natl Acad Sci U S A, 107(46):19879-19884, 27 Oct 2010
Cited by: 235 articles | PMID: 20980664 | PMCID: PMC2993380
Runx2/miR-3960/miR-2861 Positive Feedback Loop Is Responsible for Osteogenic Transdifferentiation of Vascular Smooth Muscle Cells.
Biomed Res Int, 2015:624037, 28 Jun 2015
Cited by: 34 articles | PMID: 26221600 | PMCID: PMC4499372
Regulation of Runx2 by MicroRNAs in osteoblast differentiation.
Life Sci, 232:116676, 21 Jul 2019
Cited by: 53 articles | PMID: 31340165
Review




