Abstract
Free full text

Signaling cross-talk between TGF-β/BMP and other pathways
Abstract
Transforming growth factor-beta (TGF-β)/bone morphogenic protein (BMP) signaling is involved in the vast majority of cellular processes and is fundamentally important during the entire life of all metazoans. Deregulation of TGF-β/BMP activity almost invariably leads to developmental defects and/or diseases, including cancer. The proper functioning of the TGF-β/BMP pathway depends on its constitutive and extensive communication with other signaling pathways, leading to synergistic or antagonistic effects and eventually desirable biological outcomes. The nature of such signaling cross-talk is overwhelmingly complex and highly context-dependent. Here we review the different modes of cross-talk between TGF-β/BMP and the signaling pathways of Mitogen-activated protein kinase, phosphatidylinositol-3 kinase/Akt, Wnt, Hedgehog, Notch, and the interleukin/interferon-gamma/tumor necrosis factor-alpha cytokines, with an emphasis on the underlying molecular mechanisms.
Introduction
The transforming growth factor-beta (TGF-β) family of cytokines, including TGF-β, bone morphogenic proteins (BMPs), and activin/inhibin, plays crucial roles in embryonic development, adult tissue homeostasis and the pathogenesis of a variety of diseases. The highly conserved core of the canonical TGF-β/BMP signaling is a simple linear cascade that involves the TGF-β/BMP ligands, two types of receptors (type I and II) and the signal transducers, Smads. On activation, the receptor complex phosphorylates the carboxy-terminus of receptor-regulated Smad proteins (R-Smads), including Smad1, 5 and 8 for BMP signaling and Smad2 and 3 for TGF-β signaling. Activated R-Smads interact with the common partner Smad, Smad4, and accumulate in the nucleus, where the Smad complex directly binds defined elements on the DNA and regulates target gene expression together with numerous other factors [1–3]. Simple as it is, the TGF-β/BMP pathway controls a myriad of events, including cell proliferation, differentiation, apoptosis, migration, extracellular matrix (ECM) remodeling, immune functions, and tumor invasion/metastasis [4–8]. On the other hand, the activity and the signaling outcomes of this pathway are also influenced by many intracellular and extracellular signals [1, 3, 9]. This interplay between TGF-β/BMP and other pathways, which is tightly regulated both spatially and temporally, gives rise to the remarkable complexity, diversity, flexibility, and delicacy of TGF-β/BMP functions that have been exemplified by a great number of studies.
Signaling cross-talk is a perennial theme of TGF-β research. In retrospect, the entire field of TGF-β research sprang from a few groundbreaking observations, which we now know as a typical form of signaling cross-talk (reviewed by [10]). In the early 1980s, Roberts and colleagues isolated two fractions from murine sarcoma cell extracts that could synergistically induce remarkable growth of normal fibroblasts (NRK cells) on soft agar, a hallmark of cellular transformation. These two components were therefore named transforming growth factor α and β (TGF-α and TGF-β) [11–13]. TGF-α alone had only limited transforming activity, and it was soon proven to be a ligand for the epithelial growth factor receptor (EGFR) and equivalent to EGF in promoting newborn mouse eyelid opening [14]. The other polypeptide, TGF-β, was shown to be a potent inducer of NRK cell transformation, but only in the presence of TGF-α or EGF. This was undoubtedly the first classic example of functional interaction between TGF-β and other signaling pathways, although the underlying mechanisms remain not completely understood even today.
At the organism level, TGF-β/BMP talks with other pathways at every stage of the life of a metazoan from birth to death. During embryonic development, the complex but delicate interactions between the TGF-β/BMP, Wnt/Wg, Hedgehog (Hh), Notch, mitogen-activated protein kinase (MAPK), and other pathways are crucial for stem cell maintenance, body patterning, cell fate determination, organogenesis, and so on. These signals are also instrumental for the proper growth and functioning of cells and tissues in adult animals (homeostasis), whereas concurrent alterations of these pathways are commonly found in aged or diseased animals, as repetitively seen during the development of cancer [7, 15–20].
In the past three decades since its discovery, the downstream signaling cascade of TGF-β has been clearly delineated. The biochemical basis for the extensive cross-talk between TGF-β and other pathways is essentially two-fold: (1) multiple components of the TGF-β pathway (mainly Smads) make direct and dynamic contacts with numerous other proteins; and (2) TGF-β has a great many targets, both transcriptional and non-transcriptional (Figure 1). Many of these binding partners and target molecules are essential constituents of other pathways, naturally integrating TGF-β with other signals to produce highly regulated cellular responses. In this review, we focus on these features and discuss the different modes of TGF-β signaling cross-talk as well as the ensuing context-dependent outcomes.
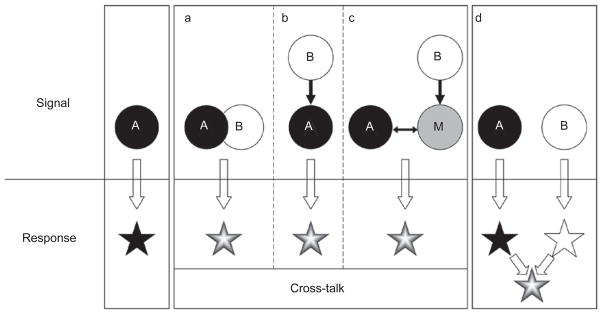
Basic modes of signaling cross-talk. A cross-talk exists between pathways A and B when both of the following criteria are met. Functionally, the combinatorial signal from A and B must produce a different response than that triggered by A or B alone. Mechanistically, the A and B pathways must be connected in at least one of the three depicted ways: (a) components of the two pathways physically interact; (b) components of one pathway are enzymatic or transcriptional targets of the other; and (c) one signal modulates or competes for a key modulator or mediator (“M”) of the other. In this scheme, A and B are interchangeable, and the arrows may represent either positive or negative regulations. Note that an altered response can arise from independent (non-cross-talk) inputs (d).
TGF-β and the MAPK pathway
MAPKs, including Erk1/2, JNK1/2/3, and p38/MAPKs, are evolutionarily conserved regulators essential for a variety of cellular events. Multiple extracellular stimuli can initiate a cascade of serial phosphorylation activation from MAP kinase kinase kinase (MAPKKK) to MAP kinase kinase (MAPKK) and finally MAPK [21]. Activated MAPKs phosphorylate a battery of proteins (primarily nuclear transcription factors) with diverse functions in regulating proliferation, survival, migration, and so on. One of the best characterized trigger for this MAPK pathway is Ras activation, which propagates signals from a number of ligand- or self-activated receptor tyrosine kinases (RTKs), such as EGFR (including HER2/Neu/ErbB2), FGFR, IGFR, PDGFR, and insulin receptor [21–23]. In addition, MAPKs can also be regulated by TGF-β/BMP stimulation [24, 25], which represents an important mechanism for non-Smad TGF-β signaling and is discussed in a separate review. Here, we mainly focus on how MAPK activity modulates the function of TGF-β/BMP.
A series of studies have shown that HER2/Neu/ErbB2 signaling, which activates both the MAPK and phosphati-dylinositol-3 kinase (PI3K)/Akt pathways, communicates intimately with TGF-β/Smad in controlling mammary epithelial cell biology and breast cancer development [6,26–31]. A general notion emerging from these studies is that HER2/Ras can antagonize TGF-β-induced apoptosis and cell cycle arrest, while allowing for the pro-migratory and pro-invasive functions of TGF-β. Therefore, both positive and negative regulations exist between the two pathways.
The synergy between the TGF-β and HER2/Ras/MAPK pathways often leads to the secretion of additional growth factors and cytokines, including TGF-β itself, which in turn promote epithelial-to-mesenchymal transition (EMT) and cell invasion [32–34], whereas JNK kinases seem to negatively regulate the autocrine expression of TGF-β1 [35]. MEK/Erk has been reported to positively regulate SMAD3 gene transcription in epithelial and smooth muscle cells [36]. On the other hand, TGF-β/Smad induces the expression of platelet-derived growth factor (PDGF) in liver cancer and glioma, which is required for fulfilling the pro-oncogenic and pro-metastatic functions of TGF-β [37, 38].
Numerous studies have revealed that the linker region of Smad proteins is a critical platform for integrating RTK/MAPK signals with the TGF-β/BMP pathway. The Smad linker region is loosely organized and highly flexible in structure, rendering it readily accessible for a number of kinases. This region is also rich in serine, threonine as well as proline residues, favoring phosphorylation by proline-directed kinases such as MAPKs and glycogen synthase kinase 3-beta (GSK3-β). Human cancer cells harboring oncogenic Ras are often resistant to TGF-β-induced cytostasis, which was thought to result from Erk-mediated Smad2/3 linker phosphorylation and Smad nuclear exclusion [39]. However, this effect is debatable and may be cell context-dependent [40, 41]. Several reports showed that Erk or JNK activation by RTKs leads to strong phosphorylation of endogenous Smad2/3 in mammalian cells without affecting their nuclear accumulation and transcriptional activity [42–44]. More recently, three residues in the linker region of Smad3 (Thr178, Ser203, and Ser207) were identified as Erk1/2 phosphorylation sites both in vitro and in vivo. Erk-mediated phosphorylation of these sites inhibits Smad3 transcriptional activity but does not prevent Smad3 from entering the nucleus [45], suggesting the existence of a yet unknown mechanism for Smad3 inhibition by the linker phosphorylation. Adding more complexity, the same serine residues (203 and 207) of Smad3 are also targets for other kinases. In human breast cancer MCF10CA1h cells, both Rho-dependent kinase (ROCK) and p38 MAPK phosphorylate Ser203/207 and facilitate, rather than inhibit, TGF-β-induced growth inhibition [46]. Our recent study indicates that GSK3-β, which is structurally similar to MAPKs, selectively phosphorylates Ser203 of Smad3 in vivo [47]. We also noticed that, unlike Erk, which primarily phosphorylates Smad3 linker in the nucleus ([45] and our unpublished result), GSK3-β mainly phosphorylates the cytoplasmic Smad3 (our unpublished result). The linker phosphorylation by GSK3-β does not seem to affect Smad3 localization or activity, and its functional role in TGF-β signaling is unknown. Together, these findings suggest that linker phosphorylation of Smad2/3 can yield distinct outcomes depending on the identity of the kinase, the specific intracellular localization where the phosphorylation occurs, the collateral events caused by MAPK activation, and other cell type-specific factors (Figure 2).
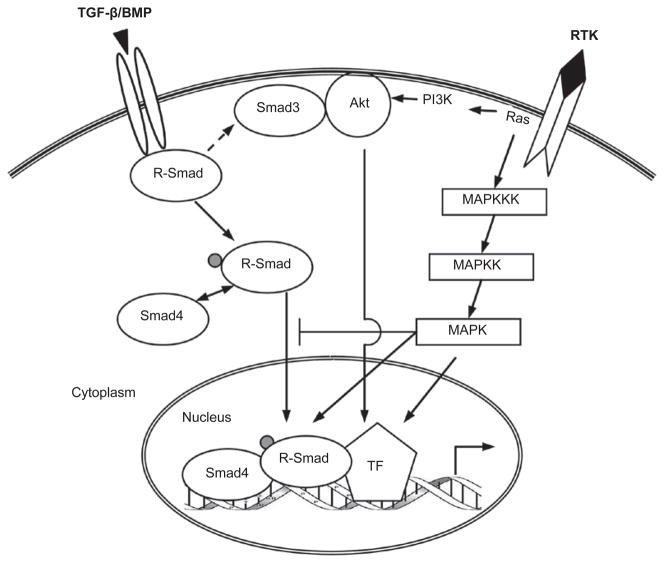
TGF-β/BMP and RTK/Ras-activated MAPK and PI3K/Akt pathways. The MAPK and PI3K/Akt pathways impinge on TGF-β/BMP signaling primarily by modulating Smad functions. MAPKs and Akt bind and/or phosphorylate R-Smads to control their intracellular distribution and transcriptional activity. MAPKs and Akt also phosphorylate and regulate a variety of Smad binding partners in the nucleus, indirectly affecting the Smads.
MAPKs (especially Erk1/2) also phosphorylate the linker of Smad1/5, which almost always blocks Smad1/5 nuclear translocation. As a result, BMP function can be suppressed by several signals that activate RTK/MAPK, including EGF, fibroblast growth factor (FGF) and insulin-like growth factor (IGF) [48–50]. Multiple Ser/Thr residues in Smad1 linker can be sequentially phosphorylated by Erk and then GSK3-β, creating a docking site for the Smad1/5-specific E3 ubiquitin ligase, Smurf1. Smurf1 binding not only causes ubiquitination and degradation of the Smads but also occludes their interaction with the nuclear pore complex, thereby preventing Smad nuclear translocation [50]. As a functional consequence, FGF/MAPK relieves BMP-mediated repression to induce neural differentiation of Xenopus embryonic cells and rat neural precursor cells [51, 52]. Importantly, Wnt signaling, which is known to inactivate GSK3-β, reduces Smad1 ubiquitination and stabilizes the protein [53]. Together, these studies have provided a compelling molecular mechanism for the long-known Wnt—| FGF —| BMP axis during embryonic patterning and cell differentiation [54]. It is interesting to note that MAPK- and GSK3-β-mediated linker phosphorylation has not been shown to regulate the protein stability of Smad2/3. Such difference between Smad1/5 and Smad2/3 could be because of the variation in amino-acid sequences of their respective linker regions. A different mode of regulation has been observed in prostate cells, in which Erk-mediated linker phosphorylation allows BMP-activated Smad1 to physically interact with the androgen receptor (AR) and act as a co-repressor. This induced binding of Smad1 and AR culminates in an antagonism of androgen-stimulated prostate cell growth by BMP [55]. Additionally, MH1 domain phosphorylation of Mad (Drosophila homolog of Smad1/5) by the MAPK-like kinase Nemo also leads to Mad nuclear exclusion [56].
In addition to R-Smads, MAPKs also phosphorylate and regulate the Co-Smad, Smad4, and the inhibitory Smad, Smad7. For example, oncogenic Ras decreases Smad4 protein stability in an MEK/Erk-dependent manner [57]. JNK and p38 seem to preferentially phosphorylate tumor-derived mutant Smad4 and promote its proteasomal degradation [58]. Erk, JNK, and p38 have all been implicated in the transcriptional regulation of Smad7, therefore indirectly regulating TGF-β signaling [59–61].
Although FGF often suppresses BMP activity as described earlier, these pathways can have synergistic functions. In chicken liver, hepatoblast differentiation into biliary epithelial cells is induced by BMP-4 and also requires an active FGF/MAPK pathway. This collaboration of BMP and FGF occurs specifically at certain stages of liver development and results in the expression of several cell lineage-restricted genes [62]. On the other hand, FGF/MAPK activity can also be opposed by BMPs. Mice lacking the receptor BMPR1a and/or BMPR1b exhibit defects in cartilage development, partly because of an elevation in FGF signaling that suppresses chondrogenesis. In the growth plate of these mutant mice, both FGFR1 protein level and Erk1/2 activity are higher than in wild-type animals, suggesting inhibition of the FGF/MAPK pathway by BMP [63]. In early mouse embryos, limb-bud outgrowth is promoted by Shh and FGFs, whereas termination of this growth requires BMP-mediated inhibition of FGFs. Interestingly, high-level FGF downregulates Gremlin1, an antagonist of BMP. Thus, a negative feed-back loop is established to tune down FGF signals by indirectly activating BMP, which is an ideal way to prevent limb-bud overgrowth ([64] and references therein).
MAPKs phosphorylate a number of nuclear transcription factors, many of which can physically interact with Smads and regulate TGF-β/BMP responses. The best-characterized ones in this category are the AP-1 proteins, including members of the Jun, Fos, Maf, and ATF subfamilies [65]. Functional interaction between Smad and the Jun/Fos family proteins has been widely studied, and their relationship can be synergistic or antagonistic depending on their target genes and other binding partners [66–72]. Recently, inhibition of MafA-dependent transcription by a TGF-β-activated Smad complex was reported [73]. The ATF proteins (ATF1, ATF2, and ATF3), which are activated by p38 MAPK, have also been shown to bind Smads and participate in a variety of TGF-β-regulated activities [74–78]. It is noteworthy that several of the Jun and ATF sub-family members are themselves Smad target genes, thus establishing the so-called “self-enabling” or “self-disabling” TGF-β responses [75,79–81].
TGF-β and the PI3K/Akt pathway
The most widely studied PI3K, p110α/p85, converts phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 is a potent signaling molecule that recruits and regulates a number of downstream effectors, the most important one being the serine/threonine kinase Akt (also known as PKB). RTK/Ras, integrin, and several other signals can activate the PI3K/Akt pathway, which usually promotes cell survival, growth, and motility through Akt-mediated phosphorylation of a slew of relevant proteins. Consistent with these physiological functions, oncogenic mutations and protein overproduction of PI3K and Akt are commonly found in human cancers. The activity of PI3K is counteracted by the tumor suppressor protein PTEN (phosphatase and tensin homolog deleted on chromosome 10), which is a lipid phosphatase that removes the phosphate group from the 3′ position of the inositol ring of PIP3, thereby blocking Akt activation. Loss-of-function mutations of PTEN also occur at a high frequency in human cancers (reviewed by [23, 82–84]).
The PI3K/Akt activity is known to alleviate TGF-β-induced apoptosis and/or cell cycle arrest in multiple types of cells in response to insulin, IGF, interleukin (IL)-6, and viral proteins [85–89]. Interestingly, Smad3, but not Smad2, seems to be the primary target of inhibition by PI3K/Akt, consistent with the indispensable function of Smad3 in mediating the pro-apoptotic effects of TGF-β [6].
Several mechanisms have been proposed by which PI3K/Akt restricts Smad3 activity (Figure 2). Studies using pharmacological inhibitors of PI3K or the downstream kinase mammalian target of rapamycin (mTOR) have implicated PI3K/Akt in regulating Smad3 activation by the TGF-β receptors, although conflicting results have been observed such that PI3K/Akt could either enhance or attenuate TGF-β responses [90, 91]. Exactly how PI3K/Akt modulates Smad3 activation remains a question unanswered. Other reports have suggested a “Smad3-trap” model for the anti-TGF-β effects of Akt in protecting liver cancer cells from cell death [92, 93]. PI3K-activated, plasma membrane-anchored Akt can physically sequester Smad3 and block its nuclear translocation in a kinase-independent manner, without affecting the C-terminal phosphorylation (activation) of Smad3. However, this mechanism may be cell type-specific and dependent on the stoichiometry between the Akt and Smad3 proteins, as Akt can also facilitate Smad3 function (see below), and PI3K/Akt has been shown to be required for the nuclear accumulation of BMP-activated Smad1 [94]. A third way for PI3K/Akt to debilitate Smad3 is through the inactivation of certain nuclear factors that are necessary for Smad3 function. For instance, activated Smad3 directly interacts with the FoxO family of transcription factors and they are simultaneously recruited to the promoter of p21 as an integral step of the TGF-β-induced cytostatic program [95]. However, both extracellular (e.g. IGF) and intracellular (e.g. Bcr-Abl) signals can defy this activity of TGF-β by enhancing Akt-mediated phosphorylation of FoxO, which forces FoxO out of the nucleus and foils p21 induction [95,96].
On the flip side, the PI3K/Akt pathway is also subjected to TGF-β/BMP regulation. Akt activity increases in response to TGF-β treatment, which seems to be required for a variety of TGF-β-induced activities, such as cell migration of HER2-expressing breast cancer cells [97], EMT of normal mammary epithelial cells [98, 99], cell survival of mouse hippocampal neurons and mesenchymal cells [100, 101], as well as growth stimulation of certain fibroblasts [102]. BMP also activates Akt to induce osteoblast differentiation [94]. It is to be noted that Akt activation by TGF-β/BMP is cell type-dependent and very likely indirect, often requiring either MAPKs or autocrine actions of secreted molecules.
Alteration of PTEN function represents another route for TGF-β/BMP to influence Akt activity. TGF-β has been shown to transcriptionally downregulate PTEN in Smad4 null pancreatic cancer cells, which, again, seems to rely on the function of the Ras/MAPK pathway [103]. In the same cells, TGF-β elicits EMT by dislodging β-catenin from the adherence junctions, a process that involves TGF-β-dependent PTEN dissociation from β-catenin and Akt activation [104]. On the other hand, TGF-β/Smad can reduce Akt activity in hematopoietic cells by inducing the expression of SHIP (SH2 domain-containing 5′ inositol phosphatase), a lipid phosphatase that removes the 5 position phosphate from PIP3 [105].
BMP also regulates PTEN activity. In one report, BM-PR1a deletion in mouse intestine caused elevated PTEN phosphorylation (indicative of PTEN inactivation) and therefore Akt hyperactivation, suggesting a positive correlation between BMP signaling and PTEN activity. As a result, those mutant mice developed polyposis that was reminiscent of human diseases with perturbed functions of BMPR1a and PTEN [106,107]. In addition, BMP-2 treatment of MCF7 human breast cancer cells slightly increased the protein level and stability of PTEN [108]. In contrast, Beck and Carethers [109] noticed that long-term BMP-2 treatment of Smad4-null SW480 colon cancer cells could decrease the mRNA level of PTEN, which requires Ras/Erk activity. It is yet to be shown whether the above observations are cell type- and tissue-specific; and more work is needed to understand how TGF-β/BMP and PTEN are connected at the molecular level.
TGF-β and the Wnt pathway
The Wnt proteins are secreted, lipid-modified signaling molecules that have diverse roles in regulating cell proliferation, differentiation, migration, survival, and so on. The canonical Wnt signaling is mediated by the transcription co-factor β-catenin, which undergoes nucleocytoplasmic shuttling and is also essential for the formation of adherence junctions between cells through its interaction with the cadherins. In the absence of Wnt, the level of cytosolic β-catenin is controlled by the so-called “β-catenin destruction complex”, comprising Axin, adenomatous polyposis coli (APC), GSK3 and casein kinase 1 alpha (CKIα). In this complex, Axin scaffolds all the other members by direct binding to facilitate CKIα- and GSK3-mediated serial phosphorylation and subsequent polyubiquitination/degradation of β-catenin. This constitutive turnover of β-catenin keeps the Wnt pathway in an “OFF” state. When the Wnt ligand binds to its receptor Frizzled (Fz) and co-receptor LRP5/6, the signal is transduced to the β-catenin destruction complex through an intracellular protein Dishevelled (Dvl), leading to Axin downregulation, GSK3 inactivation and β-catenin stabilization. The cytosolic accumulation of β-catenin favors its translocation into the nucleus, where it binds the Lef/TCF (lymphocyte enhancer factor/T-cell transcription factor) family of transcription factors and turns the Wnt pathway on. Given the power of Wnt signaling to stimulate cell proliferation, hyperactivation of this pathway often contributes to carcinogenesis [110–113].
The cross-talk between TGF-β/BMP and Wnt pathways has been known for a long time and is probably the most extensively studied. The two pathways are intertwined throughout the life of an animal, and molecularly they interact at multiple levels. First, TGF-β/BMP and Wnt reciprocally regulate their ligand production, which is critical for establishing extracellular gradients of these morphogens during embryonic development. Second, the best-defined venue of TGF-β/Wnt cross-talk is in the nucleus, where the Smad/β-catenin/Lef protein complex regulates a host of shared target genes, often in a synergistic manner. Third, recent research has identified cytoplasmic interactions between components of these pathways as novel mechanisms for fine-tuning their respective signaling. In fact, this multi-level paradigm also holds true for the cross-talk between TGF-β and other pathways such as Hh, Notch, IL, and interferon-gamma (IFNγ), which will be discussed later (Figures 3 and and4).4). Moreover, the molecular details of such cross-talk are often conserved across species, further highlighting the biological relevance of integrated signaling.
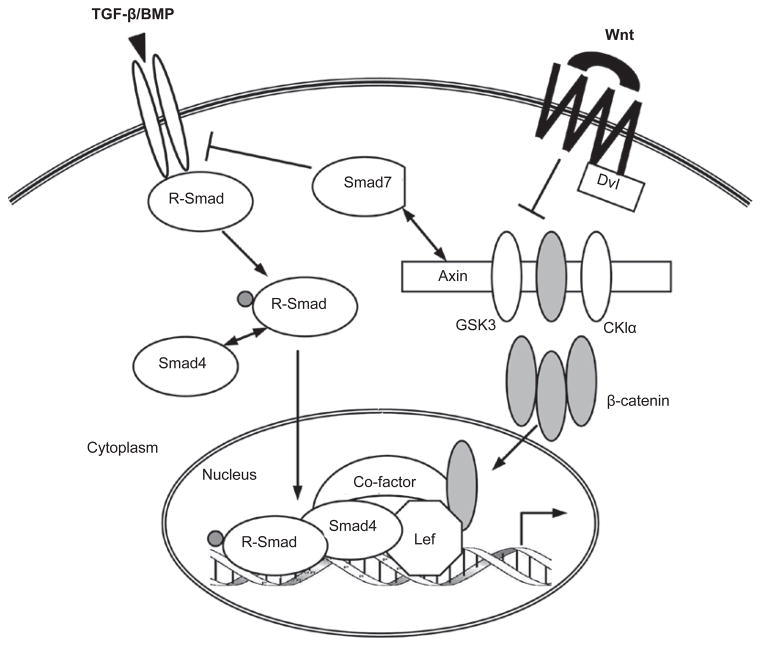
TGF-β/BMP and the Wnt pathway. The most common format of TGF-β/Wnt cross-talk occurs in the nucleus, where the Smad and Lef/β-catenin synergistically regulate a set of shared target genes. TGF-β/BMP and Wnt can determine the ligand production of each other (see text and Figure 4). In addition, protein interactions in the cytoplasm (such as Smad7-Axin binding) also link the two pathways in various settings.
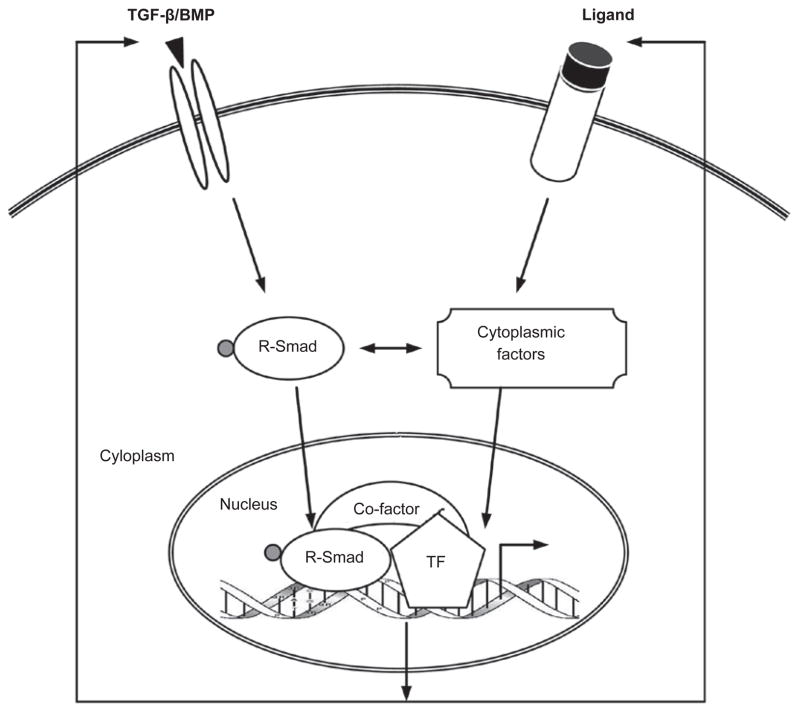
simplified and unified view of the multi-level cross-talk. TGF-β/BMP communicates with other signals at several levels. Physical interactions between pathway components, both in the cytoplasm and in the nucleus, can lead to protein redistribution and/or post-translational modifications, and eventually changes in target gene expression. Often the ligands themselves, as well as certain extracellular regulators of the ligands, are transcriptional targets of other pathways. All these events are context-dependent.
Mutual regulations between TGF-β/BMP and Wnt ligands are prevalent and important during early development, but are also seen in adult tissues. In chicken embryos, Wnt-8c induces the expression of Nodal (a TGF-β family member) in a β-catenin-dependent manner, which is required for the establishment of the left–right body axis [114]. In Xenopus, BMP2/4 regulate Wnt-8 expression and they cooperatively pattern the mesoderm [115], whereas BMP-2 downregulates Wnt-7a and β-catenin in chicken embryonic mesenchymal cells in a p38-dependent manner, leading to enhanced chondrogenesis [75]. Oncogenic β-catenin has been implicated in promoting BMP-4 production in human colon cancer cells [116]. However, Kosinski et al. [117] showed that Wnt activity overlaps with the expression of a number of BMP antagonists at the bottom of colon crypts, which is believed to be the niche for intestinal stem cells. This study suggests that one of the mechanisms by which Wnt signaling maintains the intestinal stem cell population is through inhibition of the opposing activity of BMP. In Xenopus embryos, Wnt and TGF-β co-regulate the expression of connective tissue growth factor (CTGF). Interestingly, CTGF directly interacts with the BMP-4 and TGF-β1 ligands in the extracellular space, which blocks the ligand–receptor binding of BMP but enhances that of TGF-β [118]. CTGF is also co-regulated by Wnt and BMP in mesenchymal stem cells, and its induction stunts the osteoblastic differentiation driven by BMP [119]. Another Wnt target, Cripto, is a TGF-β/Nodal co-receptor that plays important roles, both positive and negative, in transmitting TGF-β/Nodal signals during development and in cellular transformation as well as tumorigenesis (reviewed in [120]).
Many development-relevant genes in several model systems have been documented to be co-regulated by TGF-β/BMP and Wnt pathways at the transcription level, including, but not limited to, Ultrabithorax, Goosecoid, Siamois, Xnr, Chordin, Cerberus, Crescent, and Noggin [121–125]. During the early stage of vertebrate development, TGF-β(Nodal)/BMP and Wnt signaling play critical roles in the formation of the Spemann’s organizer, an embryonic dorsal signaling center that controls the movement and fate of neighboring cells [126]. In Xenopus, the organizer genes Xtwn and Xsia are synergistically controlled by both activin/Vg1-like (members of the TGF-β subfamily) and Wnt activity, and inactivation of either pathway leads to significant reduction in Xtwn transcription [127]. The two pathways were found to converge at the promoter of Xtwn, where Smad4, β-catenin, and Lef1 form a complex and bind to adjacent regulatory elements to co-regulate Xtwn expression. The interaction between Smad4 and β-catenin is mediated by Lef1, and their cooperation was soon corroborated in mammalian cells [128]. Interestingly, not all Wnt target genes in the same organizer cells are regulated by Smad4 [127], and neither Xtwn nor Xsia is affected by BMP, which also signals through Smad4 [129]. These findings suggest that cross-talk between the transcription (co)factors of different pathways is target gene-specific and is dependent on the context of target gene promoters as well as other relevant co-factors (Figure 3).
Besides Xtwn and Xsia, several other genes are jointly regulated by Smads and β-catenin/Lef in a similar manner as described above. During mouse embryonic development, Wnt and BMP collaborate to upregulate the Emx2 and Msx2 genes that play important roles in neural development, and critical elements in the promoters of both genes were found co-occupied by the Smad/β-catenin/Lef1 complex [130, 131]. In human embryonic carcinoma cells, Msx1, Msx2, and Id2 were synergistically induced by Wnt3-A and BMP-4 [132]. In mouse gastric cancer cells, the pro-tumorigenic protein gastrin is activated by both Wnt and TGF-β [133]. Interestingly, although the promoter of gastrin contains both Smad and Lef/Tcf binding sites, either site alone is sufficient to recruit the Smads/β-catenin/Tcf4 complex. Smads and β-catenin/Tcf4 can function as mutual co-factors, and the interaction between these proteins is thought to be stabilized by the p300 co-activator protein [133]. The implication of cooperative TGF-β and Wnt signaling in tumor progression has recently been examined by Labbé et al. [134], who identified several shared target genes of the two pathways in normal mouse epithelial cells, such as Ctgf, Robo1, Gpc1, and Inhba. Importantly, when analyzed in transgenic mouse models, many of these genes were found to be overexpressed in breast and colon tumors with active TGF-β and Wnt signaling. Controlled inactivation of the TGF-β pathway in these animals resulted in weakened expression of some of the above genes as well as delayed tumor formation, indicating that TGF-β and Wnt can synergistically promote tumorigenesis.
An unusual way of Smad3-β-catenin cross-talk has been observed in human mesenchymal stem cells (hMSCs). TGF-β and Wnt cooperatively stimulate the proliferation of these cells and inhibit their differentiation into the osteocytic and adipocytic lineages, thereby supporting hMSC self-renewal [135]. TGF-β stimulation of hMSCs leads to a rapid co-translocation of Smad3 and β-catenin into the nucleus, which is a unique feature of this cell type and is required for the above functions of TGF-β. Moreover, Smad3 and β-catenin co-regulate a cohort of genes in these cells that are otherwise not known to be TGF-β or Wnt targets, such as the Src family tyrosine kinase BLK. The functions of these genes in TGF-β/Wnt-regulated hMSC self-renewal remain to be determined [135].
In contrast to TGF-β, BMP impedes Wnt-induced β-catenin translocation and cell proliferation in mouse MSCs as a result of Smad1 interaction with Dvl-1 in the cytoplasm [136]. Axin also interacts with Smad proteins in the cytoplasm. Overexpression studies suggest that Axin can facilitate TGF-β signaling by presenting Smad3 to the type I TGF-β receptor [137]. However, our findings indicate that endogenous Axin negatively regulates TGF-β function by promoting Smad3 basal degradation in a GSK3-β-dependent and Wnt-independent fashion [47].
Several lines of evidence have suggested perplexing roles of Smad7 in connecting the TGF-β and Wnt pathways. As an inhibitory Smad, Smad7 primarily functions to downregulate the TGF-β/BMP receptors and hence R-Smad activation through the recruitment of E3 ubiquitin ligases, Smurf1 and Smurf2 [3]. Smad7 itself is induced by TGF-β/BMP and undergoes nucleocytoplasmic shuttling. Smad7 can directly bind β-catenin and promote β-catenin degradation by Smurf2-mediated ubiquitination, thereby reducing Wnt activity. As a result, mice carrying a Smad7 transgene experience abnormal epidermal development, which may be owing to inefficient Wnt signaling in the epidermal stem cells [138]. However, a recent report described a conflicting scenario that Smad7 could bind to Axin, disassemble the β-catenin destruction complex, prevent Smurf2 recruitment, and stabilize β-catenin as well as the adherence junctions [139]. Moreover, Smad7-Axin interaction was also reported to cause Smad7 ubiquitination and degradation through the Axin-assisted action of an E3 ubiquitin ligase called Arkadia [140]. Finally, Edlund et al. [141] observed that TGF-β could trigger β-catenin nuclear translocation in a Smad7-dependent manner in human prostate cancer PC-3U cells. The physical interaction between Smad7 and β-catenin was shown to be important for TGF-β-induced, β-catenin-regulated apoptotic responses in those cells. The most probable explanation for these seemingly opposite results is that they represent different aspects of Smad7, Axin, and β-catenin functions that are only visible in certain cell types under specific experimental conditions.
TGF-β and the Hh pathway
Like the Wnt proteins, Hedeghog (Hh) is a family of lipid-modified, secreted molecules that participates in a variety of cellular functions, functioning as a potent mitogen and morphogen. In flies, Hh binds the membrane-bound receptor Patched (Ptc) and relieves its suppression on another membrane protein called Smoothened (Smo). This leads to, through a series of intracellular events, activation of the signal transducer, Ci, which is otherwise processed to a repressor form (Cirep) by proteolysis in the cytoplasm of resting cells. Activated Ci (Ciact) joins with the co-activator protein CBP in the nucleus and controls Hh target genes. Three Hh proteins are produced in mammals: sonic, desert, and Indian hedgehog (Shh, Dhh, and Ihh), with Shh being the best characterized. The mammalian homologs of Ci are the Gli proteins (Gli1, Gli2, and Gli3). The Hh signal transduction is well conserved during evolution, and aberrant Hh signaling leads to developmental defects as well as tumorigenesis [142–144].
During development and oncogenesis, Hh and TGF-β/BMP pathways can directly regulate key components of each other. In the wing imaginal disc of flies, Ci can either suppress or induce the transcription of Dpp (Decapentaplegic, the fly version of BMP) depending on the availability of Hh. The different forms of Ci (activator or repressor) bind to the same regulatory elements of the Dpp promoter, yet with probably distinct co-factors [145–147]. Luciferase reporter analysis also revealed potential binding sites for the Gli proteins in the promoters of human BMP-4 and -7 genes [148]. Shh provokes an invasive phenotype of cultured gastric cancer cells, which is thought to be mediated by Shh-induced TGF-β ligand production as well as TβRI expression [149]. During bone development, Shh upregulates TGF-β2 to inhibit hypertrophic chondrocyte differentiation [150]; whereas Shh/Gli2-induced BMP-2 expression is responsible for osteoblast differentiation [151]. On the other hand, notochord-derived activin-βB (as well as FGF) represses Shh expression and permits proper development of the pancreas in chicken embryos [152]. Dorsalin-1, a BMP family member, has been shown to compete with Hh and repress the development of muscle pioneer cells in zebrafish [153].
Smads regulate the Gli genes and modulate Hh activity. In several types of cells, TGF-β/Smad3 directly induces Gli2 transcription, which in turn upregulates Gli1. Of particular relevance, in pancreatic cancer cells that are resistant to Hh inhibition, blocking TGF-β function could attenuate Gli-mediated Hh signaling and reduce cell growth [154]. In developing cerebellum, Shh stimulates the proliferation of granule cell precursors (GCPs) [155], whereas BMPs have the opposite functions [156]. BMP-2 and -4, but not BMP-7, are expressed in the same group of cells and antagonize the proliferative function of Shh specifically through Smad5. Interestingly, Shh pathway components such as Smo and Gli1 can be downregulated by BMP treatment, probably through direct or indirect transcriptional repression [157, 158]. In addition, the BMP-2 target gene TIEG-1 (TGF-β inducible early gene-1) blocks Gli-mediated transcription of N-myc, an oncogene and essential target of Shh in GCPs, thereby inhibiting cell proliferation and promoting cell differentiation [158].
Several Smad proteins were shown to bind to a C-terminally truncated form of Gli3, which is produced in human diseases and may resemble the endogenously expressed Glirep. Despite the unknown function of this Smad-Gli3 complex, TGF-β or BMP treatment could dissociate Smads from the trunctated Gli3, which may allow the Gli repressor to antagonize Shh signaling [159].
Our knowledge about the molecular nature of the TGF-β/Hh interaction is still limited. However, in view of the similarity between the Hh and Wnt pathways [143, 160], more sophisticated cross-talk between Hh and TGF-β is expected.
TGF-β and the Notch pathway
The Notch signaling plays integral roles in cell fate determination and is activated in a cell contact-dependent manner. The Notch protein (receptor) of one cell binds the transmembrane ligand, Jagged or Delta-like, that is expressed on the surface of an adjacent cell. Such ligand engagement triggers the shedding of the ectodomain of Notch proteins and further proteolytic cleavage that releases the Notch intracellular domain (NICD). This NICD fragment translocates into the nucleus and activates the CSL family of transcription factors. This NICD-CSL complex, in conjunction with other co-factor proteins, then drives the expression of Notch target genes, including Hes, Herp, and Hey, which encode the basic helix-loop-helix (bHLH) transcription factors necessary for mediating the downstream effects of Notch [161].
TGF-β can induce the expression of Notch ligands. In Ciona embryos, Nodal induces the local expression of Delta2 to specify the fate of notochord cells [162, 163]. Jagged has been shown to be a TGF-β target gene in multiple types of mammalian cells. Smad3-dependent expression of Jagged1 and Hey1 seems to be critical for TGF-β-induced EMT in cells derived from several organs [164]. Jagged1 upregulation also contributes to TGF-β-stimulated p21 expression and cytostasis in epithelial cells [165]. Induction of Jagged and Hes1 by TGF-β seems particularly evident in diabetic patients with nephropathy, which might be relevant to the pathogenic process [166].
TGF-β/BMP and Notch synergistically regulate their common target genes in many cell types. In chicken embryos and in mouse myofibroblasts with active TGF-β and Notch signaling, Smad3 and NICD directly interact and form a complex with CSL that binds to specific DNA sequences as found in the promoter of Hes-1 [167]. A similar cooperation is seen in mouse regulatory T cells (Treg) in that the Notch1 ICD not only interacts with activated Smad3 and facilitates its nuclear translocation [168], but also remains bound with pSmad3 in the nucleus, where they jointly upregulate the transcription factor Foxp3 [169]. Moreover, the membrane-bound form of TGF-β expressed in Treg cells activates the Notch pathway in the target cells, an event necessary for Treg/TGF-β-regulated immunosuppression [170]. BMP inhibits myogenic differentiation of C2C12 cells, and this function requires an intact Notch pathway as well as Smad1-regulated induction of Notch target genes Hes-1 and Hey-1 [171]. In mouse neuroepithelial cells, BMP-2-activated Smad1 joins with NICD with the aid of p300 to regulate the expression of Hes-5 and Hesr-1 [172]. Similarly, the Notch target gene Herp2 is positively regulated by BMP in endothelial cells. However, Herp2 in turn suppresses the expression of the BMP target gene, Id1, which is required for BMP-stimulated endothelial cell migration. Consequently, the transcriptional synergy of the two pathways in fact leads to functional antagonism [173].
Notch can also act against TGF-β/BMP. Activated Notch1 has been shown to inhibit the anti-proliferative function of TGF-β by sequestering the p300 co-factor from Smads [174]. Notch4 ICD can interact with Smad2/3/4 and inhibit TGF-β-regulated cytostasis, although it does not interfere with Smad2/3 activation [175]. Very recently, Carlson et al. [176] reported an interesting antagonism between TGF-β and Notch in the muscles. In old muscles with failing ability to regenerate, high levels of TGF-β ligand and activated Smad3 were observed in association with decreased activity of Notch. Re-activation of Notch could rejuvenate the muscle cells by preventing Smad3 binding to the promoters of p15, p16, p21, and p27, key regulators of cell cycle and senescence that contribute to the aging of muscle [176]. A mutual inhibition of Notch3 and TGF-β was also seen during the differentiation of 10T1/2 fibroblasts into smooth muscle-like cells [177]. BMP and Notch signaling can also have opposing effects on prostate development in mice. Deletion of BMP7 in mouse urogenital cells led to excessive branching morphogenesis and elevated Notch activity in vivo, whereas BMP7 treatment decreased Hes1 expression in vitro. Therefore, BMP and Notch are antagonistic in this setting, although the underlying mechanism is not clear [178].
TGF-β and the IL, TNFβ, and IFN-γ pathways
ILs, tumor necrosis factor-alpha (TNFα), and IFNγ are key regulators of immune functions, inflammatory responses, and many other physiological/pathological activities. These cytokines collectively signal through the Jak/STAT (signal transducers and activators of transcription) pathway and the NF-κB pathway to regulate multiple aspects of cell survival, proliferation, and differentiation. When ILs and IFNγ bind their cognate receptors, the Jak kinase becomes activated and phosphorylates the STAT proteins, allowing for STAT dimerization and nuclear translocation. The NF-κB proteins, including p50, p52, RelA/p65, and RelB, exist as dimers and are normally kept inactive by the IκB protein. Extracellular stimuli such as TNFα can trigger the degradation of IκB thereby freeing the NF-κB complexes. Activated NF-κB proteins enter the nucleus and, like the STATs, function as transcription factors to regulate a wide range of target genes involved in the aforementioned processes [179–183]. TGF-β, with its diverse and crucial functions in the immune system, inevitably overlaps with ILs, TNFα, and IFN-γ in many signaling events (reviewed by [8, 184]). TGF-β regulates the bioavailability of these cytokines as well as their signal transduction. In turn, TGF-β activity is modulated by these factors in various ways.
TGF-β potently suppresses T cell proliferation by inhibiting IL-2 production via Smad3 [185]. TGF-β/Smad3 also selectively blocks the expression of IL-2 target genes involved in cell proliferation, without affecting other IL-2 targets that support cell survival [186]. However, TGF-β/Smad3 also induces the expression of IL-2 receptor [187]. In addition, TGF-β probably synergizes with IL-2/STAT5 signals to upregulate Foxp3, an essential transcription factor for the differentiation of induced Treg (iTreg) cells [188–191]. These results therefore suggest a two-faced relationship between TGF-β and IL-2 in T cells, depending on the specific developmental and functional status of the cells.
IL-11 has been identified as a TGF-β/Smad target gene, and it plays a unique part in mediating the facilitatory effects of TGF-β during breast cancer bone metastasis [2, 192, 193]. A related finding is that osteoblast-derived TGF-β, probably in conjunction with AP-1 and NF-κB, stimulates the expression of IL-8 in human cancer cells [194].
TGF-β and IL-6 signaling cooperate in promoting the differentiation of Th17 cells, a new type of T helper cell, although the molecular mechanisms for such cooperation are still elusive ([8] and references therein). IL-6 treatment of human renal proximal tubular epithelial cells promotes TGF-β receptor internalization via the endosomal (signaling) pathway as opposed to the caveolar (degradation) pathway, thereby stabilizing the receptor and augmenting Smad3 activity [195]. In contrast, Smad2-mediated inhibition of STAT3 activation by IL-6 has been observed in human intestinal epithelial cells [196].
In mice with T-cell-specific deletion of TβRII, a higher amount of IFNγ is produced by the CD4+ and CD8+ T cells, which correlates with an altered pattern of T-cell differentiation and suggests an inhibition of IFNγ expression by TGF-β [197, 198]. In line with this, TGF-β secreted from tumor cells directly inhibits the production of IFNγ and several other factors by cytotoxic T cells, providing an important mechanism for the escape of tumor cells from immune surveillance [77].
Conversely, IFN-γ antagonizes TGF-β activity in many cellular responses, for which Smad7 induction by Jak1/STAT1 seems to be a major mechanism [199]. On skin injury, loss of IFN-γ correlates with a reduction in the Smad7 level and an increase in TGF-β ligand production and Smad2 activation, which favors the wound-healing process [200]. Smad7 is also a target of ILs and TNFα (see below). In mouse gastric epithelial cells, hyperactive gp130 (the receptor for IL-6) upregulates Smad7 through activation of STAT3, and subsequent inhibition of TGF-β by Smad7 leads to gastric adenoma growth in the animals [201]. IL-7 signaling in pulmonary fibrosis fibroblasts also induces Smad7 expression via Jak1/STAT1 and inhibits the pro-fibrotic functions of TGF-β [202].
TGF-β/Smad and NF-κB antagonize each other in inflammatory and adaptive immune responses [203]. NF-κB/RelA-mediated Smad7 induction inhibits Smad2/3 activation and allows cells to respond to a variety of pro-inflammatory stimuli, including TNF-α and IL-1β [204]. IL-1β has also been shown to alleviate TGF-β-induced growth arrest and transcriptional responses in epithelial cancer cells and hematopoietic cells without conscripting Smad7. Instead, IL-1β activates TAK1, an MAPKKK known to be activated by TGF-β and hence the name. TAK1 directly interacts with Smad3 in response to IL-1β and modulates Smad3 activity (probably by phosphorylation) without affecting its C-terminal phosphorylation or nuclear translocation [205].
NF-κB is also subject to TGF-β regulation. For example, TGF-β treatment of mouse intestinal epithelial cells decreases NF-κB activity by downregulation of the Toll-like receptor 2 (TLR2) protein and subsequently lowers IL-6 production. In IL-10 null mice, owing to severely impaired TGF-β signaling, bacterial infection leads to persistent NF-κB activation, which does not involve Smad7 induction [206,207]. On the other hand, an additive effect of Smad and NF-κB/RelA on the expression of type VII collagen (COL7A1) has been reported [208]. Smads have also been shown to bind and cooperate with NF-κB/p52 to upregulate JunB [209].
In Xenopus embryos, the leukemia inhibitory factor (LIF), which signals through gp130 and STAT3, inhibits activin/Smad2 activity [210]. On the other hand, BMP2 and LIF (or IL-6) synergistically induce astrocyte formation from mouse neural progenitor cells. In response to these ligands, Smad1 and STAT3 form a complex with p300 on the promoter of astrocyte-specific genes such as GFAP and co-direct cell differentiation [211]. In addition, BMP-induced Id expression provides a permissive environment for LIF to support self-renewal of the ES cells [212].
Concluding remarks
With a growing list of new regulatory factors and targets being identified, the TGF-β pathway has been interwoven into the vast network of cell signaling. Countless experiments performed in distinct systems and/or with different approaches have provided all sorts of answers as to how the TGF-β/BMP pathway interacts with the rest of this signaling network. Sometimes these studies show contradictory results that are not easily reconcilable. Although such discrepancies could have arisen from the variations of experimental conditions, they may also reflect the true adaptability that an organism must possess to survive in a constantly changing environment. In this sense, signaling cross-talk between different pathways creates a comprehensive view of the outside world, so that the cell can orchestrate these pieces of information and respond in an accurate, efficient, and balanced manner.
The core concept of TGF-β signaling cross-talk is the context dependency. It is clear from all the above discussions that no simple rule can be easily generalized to describe how TGF-β interacts with any other signaling cascade. All experimental data should be interpreted with specific confinement parameters, including cell type, developmental stage, physiological/pathological status, protein intracellular localization, nature of modifying enzymes, co-factors, identity of targets, and so forth.
It should also be noted that cross-talk between pathways can be direct or indirect, unidirectional or bidirectional, and often occurs as part of a feed-back loop. On the other hand, synergy or antagonism can also result from independent inputs that do not cross-talk. Therefore, in-depth mechanistic studies are necessary to distinguish the cause from the consequence and to identify the specific convergence point of the pathways (Figure 4).
The outcome of any signaling cross-talk is an integrated and quantitative reflection of all individual input signals, which should be kept within a physiologically relevant range. Otherwise, distorted artifacts may occur. With the availability of techniques such as RNAi and gene targeting, some of the earlier conclusions drawn from experiments solely dependent on overexpression and dominant-negative strategies warrant re-evaluation.
Owing to space limit, several other pathways that functionally interact with TGF-β signaling are not covered in this review, including the nuclear receptor and apoptosis pathways. On the other hand, recent literatures have suggested some new areas that are interconnected with TGF-β activity, such as energy metabolism (glucose uptake/consumption, AMPK and mTOR signaling) and NO (nitric oxide) signaling. In addition, the recent discovery that TGF-β/BMP/Smad signaling also regulates microRNA expression has pointed out a path toward yet another unexplored territory of TGF-β research [213]. It can be envisioned that macro-scale screening/modeling in combination with current techniques in molecular biology, biochemistry, genetics, structural biology, and bioinformatics will reveal a good number of new signaling partners of TGF-β and greatly assist the elucidation of many fundamental mechanisms regarding TGF-β function and regulation.
Acknowledgments
We apologize to the researchers whose work was not included in this review owing to space limit. Xiao-Fan Wang was supported by NIH grants DK064113 and GM083000.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/cr.2008.302
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/cr2008302.pdf
Citations & impact
Impact metrics
Article citations
The role of epithelial-mesenchymal transition in pulmonary fibrosis: lessons from idiopathic pulmonary fibrosis and COVID-19.
Cell Commun Signal, 22(1):542, 13 Nov 2024
Cited by: 0 articles | PMID: 39538298 | PMCID: PMC11558984
Review Free full text in Europe PMC
Repulsive guidance molecules b (RGMb): molecular mechanism, function and role in diseases.
Expert Rev Mol Med, 26:e24, 08 Oct 2024
Cited by: 0 articles | PMID: 39375839 | PMCID: PMC11488336
Review Free full text in Europe PMC
Regulation of Bone Morphogenetic Protein Receptor Type II Expression by <i>FMR1</i>/Fragile X Mental Retardation Protein in Human Granulosa Cells in the Context of Poor Ovarian Response.
Int J Mol Sci, 25(19):10643, 03 Oct 2024
Cited by: 0 articles | PMID: 39408972 | PMCID: PMC11477111
TGFβ in Pancreas and Colorectal Cancer: Opportunities to Overcome Therapeutic Resistance.
Clin Cancer Res, 30(17):3676-3687, 01 Sep 2024
Cited by: 1 article | PMID: 38916900
Review
In vitro evidence for the potential of EGFR inhibitors to decrease the TGF-β1-induced dispersal of circulating tumour cell clusters mediated by EGFR overexpression.
Sci Rep, 14(1):19980, 28 Aug 2024
Cited by: 0 articles | PMID: 39198539 | PMCID: PMC11358385
Go to all (633) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
TGF-β and BMP signaling in osteoblast differentiation and bone formation.
Int J Biol Sci, 8(2):272-288, 21 Jan 2012
Cited by: 927 articles | PMID: 22298955 | PMCID: PMC3269610
Review Free full text in Europe PMC
Signaling Cross Talk between TGF-β/Smad and Other Signaling Pathways.
Cold Spring Harb Perspect Biol, 9(1):a022137, 03 Jan 2017
Cited by: 290 articles | PMID: 27836834 | PMCID: PMC5204325
Review Free full text in Europe PMC
Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis.
Am J Respir Crit Care Med, 177(3):321-329, 01 Nov 2007
Cited by: 120 articles | PMID: 17975199 | PMCID: PMC2218851
Vasohibin-1 expression is regulated by transforming growth factor-β/bone morphogenic protein signaling pathway between tumor-associated macrophages and pancreatic cancer cells.
J Interferon Cytokine Res, 33(8):428-433, 07 May 2013
Cited by: 16 articles | PMID: 23651239 | PMCID: PMC3741417
Funding
Funders who supported this work.
NIDDK NIH HHS (2)
Grant ID: DK064113
Grant ID: R01 DK064113
NIGMS NIH HHS (2)
Grant ID: R01 GM083000
Grant ID: GM083000





