Abstract
Free full text

What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer
Abstract
Mutations in metabolic enzymes, including isocitrate dehydrogenase 1 (IDH1) and IDH2, in cancer strongly implicate altered metabolism in tumorigenesis. IDH1 and IDH2 catalyze the interconversion of isocitrate and 2-oxoglutarate (2OG). 2OG is a TCA cycle intermediate and an essential cofactor for many enzymes, including JmjC domain-containing histone demethylases, TET 5-methylcytosine hydroxylases, and EglN prolyl-4-hydroxylases. Cancer-associated IDH mutations alter the enzymes such that they reduce 2OG to the structurally similar metabolite (R)-2-hydroxyglutarate [(R)-2HG]. Here we review what is known about the molecular mechanisms of transformation by mutant IDH and discuss their implications for the development of targeted therapies to treat IDH mutant malignancies.
Cellular metabolism has been hypothesized to play a central role in cancer since the observation made almost a century ago by Otto Warburg (Warburg 1956) that cancer cells preferentially generate energy by metabolizing glucose to lactate. Even in the presence of oxygen, cancer cells switch from generating ATP by the highly energy-efficient process of oxidative phosphorylation to the much less efficient process of glycolysis (Vander Heiden et al. 2009; Dang 2012). Why this switch occurs has long been a mystery, although the observation that normal cells use “aerobic glycolysis” during periods of increased proliferation supports the hypothesis that this metabolic switch is an important feature of rapidly dividing cells (Locasale and Cantley 2011; Lunt and Vander Heiden 2011). Recent work suggests that aerobic glycolysis facilitates cellular transformation by producing the high levels of glycolytic intermediates that proliferating cells need for the biosynthesis of lipids, amino acids, and nucleic acids. Moreover, metabolic reprogramming appears to be sufficient to mediate tumorigenesis in some systems (Lyssiotis and Cantley 2012; Sebastian et al. 2012). Nevertheless, whether altered cellular metabolism is a cause of cancer or merely an adaptive response of cancer cells in the face of accelerated cell proliferation is still a topic of some debate.
The recent identification of cancer-associated mutations in three metabolic enzymes suggests that altered cellular metabolism can indeed be a cause of some cancers (Pollard et al. 2003; King et al. 2006; Raimundo et al. 2011). Two of these enzymes, fumarate hydratase (FH) and succinate dehydrogenase (SDH), are bone fide tumor suppressors, and loss-of-function mutations in FH and SDH have been identified in various cancers, including renal cell carcinomas and paragangliomas. The third mutated enzyme, isocitrate dehydrogenase (IDH), is a more complicated case. Mutations in two isoforms of IDH, IDH1 and IDH2, are common in a diverse array of cancers, including gliomas and acute myelogenous leukemia (AML) (Dang et al. 2010). The mutant enzymes are not catalytically inactive. Rather, the cancer-associated mutations alter the catalytic activity of the enzymes such that they produce high levels of a metabolite, (R)-2-hydroxyglutarate [(R)-2HG], which is normally found at very low levels in cells (Dang et al. 2009; Ward et al. 2010). How this aberrant enzymatic activity contributes to cellular transformation is an area of intense interest and some controversy, and several hypotheses have been proposed for how IDH mutations promote tumorigenesis. It has been suggested that (R)-2HG functions as an “oncometabolite” to promote cellular transformation, perhaps by altering the redox state of cells or modulating the activity of metabolic and epigenetic tumor suppressor enzymes that use the structurally similar metabolite α-ketoglutarate as a cosubstrate (Reitman and Yan 2010; Yen et al. 2010; Kaelin 2011). Others have posited that loss of IDH activity from either loss of the wild-type allele or gain of a dominant-negative mutant allele alters normal mitochondrial function and promotes the metabolic switch of cancer cells to glycolysis (Leonardi et al. 2012; Oermann et al. 2012).
Here, we review the spectrum of tumors that harbor IDH mutations and discuss what has been learned to date about how mutant IDH and (R)-2HG contribute to tumorigenesis. We also discuss the potential of mutant IDH as a therapeutic target in cancer and discuss some of the outstanding questions about mutant IDH biology that warrant further investigation.
Normal functions of the IDH family of metabolic enzymes
Eukaryotic cells express three different isoforms of IDH (Fig. 1; Dalziel 1980). IDH1 and IDH2 are homodimeric NADP+-dependent enzymes that catalyze the oxidative decarboxylation of isocitrate to produce α-ketoglutarate (also known as 2-oxoglutarate [2OG]), NADPH, and CO2 (Fig. 2). IDH3 is a structurally unrelated heterotetrameric NAD+-dependent enzyme that similarly decarboxylates isocitrate and produces 2OG, NADH, and CO2. The reactions catalyzed by IDH1 and IDH2 are reversible, and the directionality of the reactions therefore depends in large part on the relative Km values of the forward and reverse reactions and the relative levels of isocitrate and 2OG in the cell (Lemons et al. 2010). The reaction catalyzed by IDH3, on the other hand, is irreversible under physiologic conditions and is principally regulated by substrate availability and positive and negative allosteric effectors. Calcium, ADP, and citrate activate IDH3, whereas ATP, NADH, and NADPH inhibit this enzyme (Gabriel et al. 1986).

The role of the IDH family of enzymes in the TCA cycle. Shown here are the mitochondrial and cytoplasmic reactions that involve IDH; also shown are the principal mechanisms of entry of glucose and glutamine-derived carbon molecules into the TCA cycle. IDH1 reversibly catalyzes the NADP+-dependent decarboxylation of isocitrate to 2OG in the cytoplasm, whereas IDH2 mediates the same reaction in the mitochondria. IDH3 catalyzes the NAD+-dependent conversion of isocitrate to 2OG in the mitochondria in a reaction that is irreversible under physiologic conditions. (ACO) Aconitase.

Reactions catalyzed by wild-type and mutant IDH1 and IDH2. Wild-type IDH1 and IDH2 catalyze a two-step reaction (indicated by blue arrows). The first step involves the oxidation of isocitrate to the unstable intermediate oxalosuccinate. In this reaction, NADP+ acts as the hydrogen acceptor and is reduced to NADPH. In the second step of the reaction, the β-carboxyl group is released as carbon dioxide (CO2), resulting in production of 2OG. Mutant IDH1 and IDH2 catalyze a single-step reaction (indicated by the purple arrow). In this reaction, NADPH is oxidized to NADP+, with concomitant reduction of 2OG to (R)-2HG. The mutant enzymes are unable to catalyze the carboxylation of (R)-2HG and therefore cannot generate 2OG. 2OG and 2HG are structurally very similar. They differ only in the replacement of the ketone group in 2OG with a hydroxyl group in 2HG.
The different IDH isoforms have overlapping, but nonredundant, roles in cellular metabolism (Fig. 1; Mailloux et al. 2007; Reitman and Yan 2010). IDH1 localizes to the cytoplasm and peroxisomes and plays a role in promoting the activity of the numerous cytoplasmic and nuclear dioxygenases that require 2OG as a cosubstrate (Hausinger 2004). Another important function of IDH1 is the generation of nonmitochondrial NADPH. NADPH provides critical reducing equivalents needed for lipid biosynthesis and is also an essential antioxidant that protects cells from oxidative stress and radiation damage (Jo et al. 2002; Lee et al. 2002; Kim et al. 2007). Finally, as the reaction catalyzed by IDH1 is reversible, IDH1 is capable of catalyzing the reductive carboxylation of 2OG to isocitrate, which can then be further metabolized to acetyl-CoA to support lipid biosynthesis (Koh et al. 2004; Filipp et al. 2012; Metallo et al. 2012).
IDH2 and IDH3, in contrast, localize to the mitochondrial matrix (Fig. 1). IDH3 plays a central role in mitochondrial respiration by catalyzing one of the rate-limiting steps of the TCA cycle. The 2OG produced by IDH3 is further metabolized to succinate, and the NADH is used by the electron transport chain to generate ATP (Barnes et al. 1971). It is not clear whether IDH2 likewise contributes to flux through the TCA cycle. Rather, IDH2 is thought to regulate energy metabolism by modulating the relative abundance of isocitrate and 2OG in the mitochondria (DeBerardinis et al. 2008). Under conditions where levels of mitochondrial glutamate are high or under hypoxic conditions when glucose-dependent citrate production is compromised, glutamate dehydrogenase (GDH) converts glutamate to 2OG (Sazanov and Jackson 1994; Comte et al. 2002). The resultant increase in the 2OG:isocitrate ratio favors the reverse IDH2 reaction, which results in conversion of 2OG and NADPH to isocitrate and NADP+. This isocitrate can then be fed into the TCA cycle by IDH3, which converts the isocitrate back to 2OG and produces additional NADH, or isomerized by aconitase to produce citrate. Citrate, in addition to being a TCA cycle intermediate, is a critical metabolite required for fatty acid biosynthesis. Accordingly, IDH2-mediated reductive carboxylation of 2OG plays an important role in sustaining energy production and promoting cell proliferation during periods of hypoxia (Wise et al. 2011; Filipp et al. 2012). IDH2, as an important source of mitochondrial NADPH, also plays a critical role in the protection of cells from mitochondrial-specific oxidative stress, such as that generated by the electron transport chain (Lee et al. 2004, 2007).
IDH mutations in human cancer
Glioblastoma multiforme (GBM) is a highly invasive brain tumor that is notoriously refractory to chemotherapy and radiation therapy. In 2008, in an effort to identify novel therapeutic targets in GBM, Parsons et al. (2008) undertook a landmark genomic sequencing effort of 22 adult primary and secondary GBM tumors. They identified recurrent somatic mutations at Arg132 of IDH1 that were present in five of the six secondary GBMs but none of the 16 primary GBMs included in the study. Several follow-up sequencing studies of malignant gliomas confirmed that IDH1 Arg132 mutations are very common in specific types of adult brain tumors, occurring in >70% of adult grade II and grade III gliomas and >80% of adult secondary GBMs (Balss et al. 2008; Bleeker et al. 2009). Moreover, many of the grade II/III gliomas and secondary GBMs that are IDH1 wild type harbor mutations at the analogous codon, Arg172, of IDH2 (Hartmann et al. 2009; Yan et al. 2009). Overall, 80%–90% of adult grade II/III gliomas and secondary GBMs harbor mutations at either Arg132 of IDH1 or Arg172 of IDH2 (Table 1; Chang et al. 2011). Conversely, <10% of primary GBMs and pediatric GBMs harbor IDH mutations, and no IDH mutations have been identified to date in brain tumors of nonglial subtypes (Balss et al. 2008; Yan et al. 2009; Capper et al. 2010; Byeon et al. 2012).
Table 1.
Frequency of IDH1 and IDH2 mutant alleles in human cancers
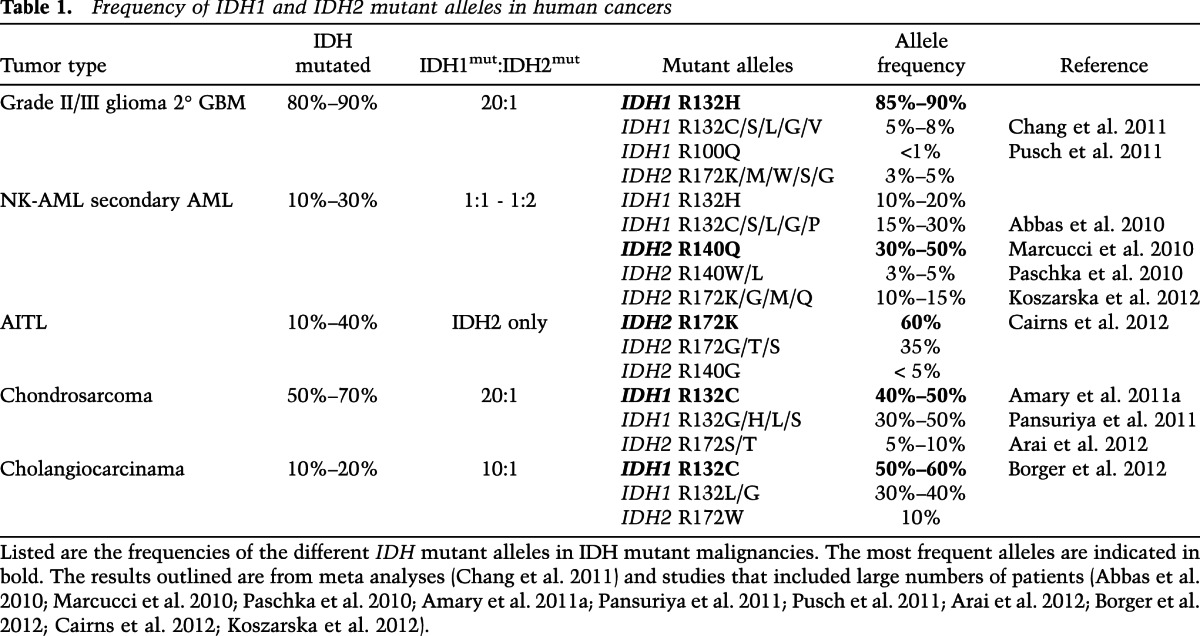
In 2009, Mardis et al. (2009) performed whole-genome sequencing of a case of normal karyotype AML (NK-AML) and identified a mutation at Arg132 of IDH1. Further sequencing efforts confirmed that IDH mutations are highly recurrent in clonal myeloid disorders. Mutations in IDH1 and IDH2 are present in 5%–20% of cases of de novo NK-AML and in 10%–20% of cases of secondary AML that result from leukemic transformation of “premalignant” myelodysplastic syndrome (MDS) and myeloproliferative neoplasm (MPN) (Kosmider et al. 2010; Pardanani et al. 2010; Tefferi et al. 2010; Patnaik et al. 2012; Rakheja et al. 2012; Zhou et al. 2012). IDH mutations are also present, albeit at a lower frequency (5%–10%), in chronic-phase MDS and MPN but are rare in translocation-positive AML (Mardis et al. 2009; Abbas et al. 2010). IDH2 mutations are also present in 10%–40% of cases of angioimmunoblastic T-cell lymphoma (AITL) but are uncommon in other T- or B-cell lymphoid malignancies (Cairns et al. 2012; Zhang et al. 2012).
IDH mutations have been found in a number of other solid tumors besides gliomas. Over 50% of chondrosarcomas harbor IDH mutations (Amary et al. 2011a; Arai et al. 2012; Meijer et al. 2012), and IDH mutations have been linked to the pathogenesis of the enchondromatosis syndromes Ollier disease and Maffucci syndrome (Pansuriya et al. 2010). Ollier disease and Maffucci syndrome are rare nonhereditary pediatric cancer predisposition syndromes in which affected children develop cartilaginous tumors as their developing bones undergo endochondral ossification. Recently, sequencing analysis has determined that 40%–90% of the tumors in these patients harbor mutations at Arg132 of IDH1 or Arg172 of IDH2 (Amary et al. 2011b; Pansuriya et al. 2011). Interestingly, many of these patients develop multiple independent IDH mutant tumors but express wild-type IDH1 and IDH2 in their normal tissues. This suggests that their IDH mutations are occurring during embryonic development and that their tumors are a consequence of somatic mosaicism of mutant IDH-expressing cells. Interestingly, Ollier disease and Maffucci syndrome appear to be associated with an increased risk of secondary neoplasms; in particular, gliomas and AML. IDH mutations have also been found in 10%–20% of cholangiocarcinomas as well as a few rare cases of paraganglioma, colon cancer, prostate cancer, and lung cancer (Sjoblom et al. 2006; Bleeker et al. 2009; Kang et al. 2009; Gaal et al. 2010; Sequist et al. 2011; Borger et al. 2012).
Among the cancers associated with IDH mutations, the mutations appear to have a predilection for certain tumor subtypes or locations. In cholangiocarcinoma, the frequency of IDH mutations in intrahepatic tumors is 28%, whereas the frequency is only 7% in extrahepatic tumors (Borger et al. 2012). In chondrosarcomas, IDH mutations appear to be exclusive to central, periosteal, and dedifferentiated tumors, with no IDH mutations having been identified to date in peripheral chondrosarcomas or other cartilaginous tumors (Amary et al. 2011a; Arai et al. 2012).
IDH1 and IDH2 mutations occur at different frequencies in different tumor types (Table 1). In gliomas, >80% of IDH mutations occur at Arg132 of IDH1 and result in replacement of the arginine residue with a histidine (IDH1 R132H) (Balss et al. 2008; Yan et al. 2009; Chang et al. 2011). In cholangiocarcinomas and chondrosarcomas, IDH1 Arg132 mutations are also far more common than IDH2 Arg172 mutations, although the IDH1 mutations in these tumors most often result in an arginine-to-cysteine substitution (IDH1 R132C) (Amary et al. 2011a; Arai et al. 2012; Borger et al. 2012). This is in contrast to AML, where IDH1 and IDH2 mutations occur at similar frequencies (Abbas et al. 2010; Marcucci et al. 2010; Paschka et al. 2010; Koszarska et al. 2012). The single most common IDH mutation in AML is the replacement of Arg140 of IDH2 with a glutamine (IDH2 R140Q). Interestingly, IDH2 Arg140 mutations have not been found in gliomas, cholangiocarcinomas, or chondrosarcomas. However, an IDH1 mutation analogous to Arg140 of IDH2, IDH1 Arg100, has been described in rare cases of glioma (Pusch et al. 2011). In AITL, all of the IDH mutations reported to date are IDH2 mutations, with >90% occurring at IDH2 Arg172 (Cairns et al. 2012). Why the frequency of these different amino acid substitutions is so variable in different tumors is not known. It is also worth noting that IDH1 and IDH2 mutations appear to be mutually exclusive. There are no reported cases of brain tumors harboring both mutations, and although rare cases of IDH1/IDH2 double mutant AML have been reported, it is not clear whether these mutations occur in the same or different leukemic subclones (Paschka et al. 2010).
The prognostic significance of IDH mutations is not consistent across all tumor types. In the case of GBM, IDH mutations appear to confer a more favorable prognosis, with a median survival for patients with IDH mutant GBM of 31 mo, as compared with a median survival of 15 mo for patients with IDH wild-type GBM (Yan et al. 2009; SongTao et al. 2012). Whether this difference is driven by the IDH mutational status of the tumors or reflects other fundamental biological differences between primary and secondary GBM is as yet unclear. In AML, the prognostic significance of IDH mutations is still somewhat ambiguous. Several studies have reported that IDH mutational status does not impact prognosis in AML, whereas other studies have found that IDH mutations are associated with an increased or decreased risk of disease relapse when compared with IDH wild-type disease, depending on the specific patient population examined (Abdel-Wahab et al. 2011; Rakheja et al. 2012; Zhou et al. 2012). In MDS and MPN, IDH mutations have been consistently found to be markers of poor prognosis. Furthermore, the observation that the frequency of IDH mutations is higher in late stage MPN and high-risk MDS (20%) than in early stage and low-risk disease (<4%), coupled with the observation that IDH mutations are frequently present in secondary AML, has lead to speculation that IDH mutations are involved in the progression of chronic MDS and MPN to full-blown leukemia. (Tefferi et al. 2010, 2012; Thol et al. 2010; Patnaik et al. 2012).
Catalytic activity of mutant IDH
When IDH mutations were first identified in cancer, they were hypothesized to contribute to tumorigenesis by causing loss of function and dominant-negative inhibition of wild-type IDH activity (Yan et al. 2009; Zhao et al. 2009). However, subsequent studies found that, on the contrary, tumor-associated IDH mutations cause a gain of function (Dang et al. 2009; Ward et al. 2010). The mutations are all located in the active sites of the enzymes. Arg100 and Arg132 in IDH1 and Arg140 and Arg172 in IDH2 form hydrogen bonds with the α-carboxyl and β-carboxyl groups of isocitrate and mediate isocitrate binding (Xu et al. 2004). Mutations at these residues decrease the binding affinity of the enzyme active sites for isocitrate and increase their binding affinity for NADPH, which significantly abrogates the normal oxidative decarboxylation activity of the enzymes (Dang et al. 2009). In addition to abrogating the “forward” catalytic activity of IDH, the mutations change the conformation of the enzyme active site such that the “reverse” reaction is only a partial reaction, one in which 2OG is reduced but not carboxylated. This results in conversion of 2OG to (R)-2HG rather than isocitrate (Fig. 2). In short, IDH mutations cause the reverse IDH reaction to be favored but partially disrupted, leading to high-level production of (R)-2HG.
Interestingly, although IDH1 and IDH2 mutants both produce (R)-2HG, they appear to have somewhat distinct enzymatic properties. Cells that express mutant IDH1 accumulate less (R)-2HG than do mutant IDH2-expressing cells (Ward et al. 2013). Moreover, (R)-2HG production by mutant IDH1 is enhanced by coexpression of wild-type IDH1, whereas mutant IDH2-expressing cells accumulate high levels of (R)-2HG independent of wild-type IDH2 activity. It has been hypothesized that the lower production of (R)-2HG by mutant IDH1 is due to the fact that cytoplasmic 2OG is limiting in cells, whereas mitochondrial 2OG is not. Consistent with this model, the addition of a mitochondrial localization signal to mutant IDH1 results in enhanced (R)-2HG production. Interestingly, mutant IDH1 forms heterodimers with wild-type IDH1, but mutant IDH2 does not bind wild-type IDH2 (Dang et al. 2009; Ward et al. 2013). It has been hypothesized that heterodimerization of wild-type and mutant IDH1 in the cytoplasm provides the mutant subunit with a local source of 2OG that can then be metabolized to (R)-2HG. Consistent with this model, it has recently been reported that loss of the wild-type IDH1 allele in IDH1 mutant gliomas is associated with significantly lower tumor levels of (R)-2HG than are seen in heterozygous IDH1 mutant tumors (Jin et al. 2013).
Mutant IDH is an oncogene
In a number of different in vitro transformation assays, expression of mutant IDH promotes the proliferation and inhibits the differentiation of cells. Expression of a tumor-derived IDH1 mutant (IDH1 R132H), but not a catalytically inactive form of the mutant enzyme, enhances the proliferation and soft agar colony formation of immortalized human astrocytes and confers growth factor independence and blocks the erythropoietin-induced differentiation of TF-1 cells, a human erythroleukemia cell line (Koivunen et al. 2012; Losman et al. 2013). Expression of mutant IDH is also able to inhibit the differentiation of murine 3T3-L1 fibroblasts and the differentiation of primary murine hematopoietic stem cells and immortalized murine myeloid progenitor cells (Figueroa et al. 2010; Lu et al. 2012; Losman et al. 2013).
Although mutant IDH is able to promote cellular transformation in vitro, the mutant enzyme does not appear to be sufficient to transform primary cells in vivo. Sasaki et al. (2012a,b) have characterized the phenotype of a conditional knock-in mouse model in which they inserted IDH1 R132H into the endogenous IDH1 locus and expressed mutant IDH in either the brains or the hematopoietic systems of mice. Brain-specific expression of IDH1 R132H during embryogenesis results in perinatal lethality of the mice from cerebral hemorrhage without evidence of underlying malignancy (Sasaki et al. 2012a). Although it is possible that the mice would develop brain tumors if they lived longer, the hematopoietic-specific IDH1 R132H knock-in mice, which have a normal life span, similarly do not develop malignancy (Sasaki et al. 2012b).
Despite the lack of a cancer phenotype in the IDH1 R132H knock-in mice, the hematopoietic stem cell phenotype of the mice does provide some insights into how mutant IDH contributes to tumorigenesis. Although IDH1 R132H-expressing bone marrow cells are not immortalized, the mice do show selective expansion of their hematopoietic stem and early progenitor compartments but not their committed myeloid and lymphoid progenitor compartments. This observation is quite interesting given the distinct self-renewal properties of these different cell types. Normal committed progenitors are unable to self-renew and are consigned to undergo further differentiation, whereas stem cells and early progenitors have inherent self-renewal capabilities (Chao et al. 2008; Orford and Scadden 2008; Seita and Weissman 2010). When stem cells and early progenitors divide, they make the decision to undergo either symmetric division, in which both daughter cells are identical to the original cell, or asymmetric division, in which one daughter cell is identical to the original cell and the other is committed to undergoing further differentiation. The balance between symmetric and asymmetric cell division is crucial to cellular homeostasis. Excessive symmetric division causes inappropriate expansion of stem cell compartments, whereas excessive asymmetric division results in stem cell exhaustion (Jacob and Osato 2009; Marciniak-Czochra et al. 2009; Matsumoto and Nakayama 2013). The observation that only stem and early progenitor cells are expanded in the IDH1 R132H knock-in mice suggests that mutant IDH promotes leukemogenesis by increasing symmetric division within a population of cells that have inherent self-renewal capacity but that mutant IDH is not, by itself, able to immortalize self-renewing cells or bestow self-renewal upon more differentiated cells. This observation has important biological implications. The limited ability of mutant IDH to promote the proliferation of primary cells suggests that the specific cell lineage in which the IDH mutation occurs significantly influences the ability of the mutation to contribute to transformation. An IDH mutation in a committed progenitor cell would likely not confer a proliferative advantage to that cell, whereas the same mutation in a more primitive cell would contribute to clonal expansion.
It is also notable that hematopoietic stem cells isolated from IDH1 R132H knock-in mice are able to differentiate normally in vitro and are able to competitively repopulate the bone marrow of transplanted recipient mice. This suggests that expression of mutant IDH has little if any effect on cellular differentiation in otherwise normal hematopoietic cells. This limited ability of mutant IDH to block the differentiation of normal primary cells suggests that other specific mutations that cooperate with mutant IDH to block cellular differentiation are likely to be required to bring about the maturation arrest that is characteristic of leukemia cells.
Mutant IDH-derived (R)-2HG is an oncometabolite
2HG is a five-carbon dicarboxylic acid with a chiral center at the second carbon atom (Fig. 2). There are therefore two possible enantiomers of 2HG: (R)-2HG [otherwise known as (D)-2HG] and (S)-2HG [otherwise known as (L)-2HG]. (R)-2HG and (S)-2HG are both byproducts of normal mitochondrial metabolism (Kranendijk et al. 2012). The principal source of (R)-2HG in IDH wild-type cells is a reaction catalyzed by hydroxyacid–oxoacid transhydrogenase (HOT) (Struys et al. 2005b). HOT converts γ-hydroxybutyrate (GHB) to succinic semialdehyde (SSA), with concomitant reduction of 2OG to (R)-2HG. (S)-2HG is generated during conversion of oxaloacetate to (L)-malate by (L)-malate dehydrogenase, an enzyme of the TCA cycle (Rzem et al. 2007).
(R)-2HG and (S)-2HG are believed to be unwanted byproducts of cellular metabolism, and their intracellular levels in normal cells are maintained at <0.1 mM. 2HG is prevented from accumulating in cells by the actions of two enzymes—(D)-2HG and (L)-2HG dehydrogenase (2HGDH)—that convert (R)-2HG and (S)-2HG, respectively, back to 2OG (Struys et al. 2005a; Steenweg et al. 2010). There is currently no known physiologic role for either enantiomer in normal metabolism.
IDH mutants exclusively produce the (R) enantiomer of 2HG, and the levels of (R)-2HG in IDH mutant tumors can be extremely elevated, ranging from 1 mM to as high as 30 mM (Dang et al. 2009; Gross et al. 2010; Choi et al. 2012). These high levels of (R)-2HG appear to be a consequence of a significant imbalance between (R)-2HG production and metabolism in tumor cells. IDH1 R132H has an estimated catalytic rate (Kcat) of 1.0 × 103 sec−1 (Dang et al. 2009), whereas recombinant D2HGDH has an estimated Kcat of 0.8 sec−1 (Engqvist et al. 2009). It is therefore likely that mutant IDH overwhelms the capacity of D2HGDH to oxidize the excess (R)-2HG back to 2OG. This model is supported by the observation that in patients with type I (D)-2-hydroxyglutaric aciduria (D2HGA), an inborn neurometabolic disorder caused by germline mutations in D2HGDH, levels of (R)-2HG are elevated but to a much lesser extent than is observed in IDH mutant tumors (Wickenhagen et al. 2009). Type II D2HGA, which is caused by germline IDH2 R140Q and IDH2 R140G mutations, is associated with higher levels of (R)-2HG and a more severe clinical course (Kranendijk et al. 2010, 2011). Interestingly, D2HGA is not associated with an increased incidence of cancer. It is important to note, however, that many patients with severe D2HGA die in infancy and early childhood, and it is therefore difficult to make definitive conclusions about an association between high levels of (R)-2HG and long-term susceptibility to cancer.
High intracellular concentrations of (R)-2HG are sufficient to mediate the in vitro transforming effects of mutant IDH. Treatment of TF-1 leukemia cells with concentrations of a cell-permeable form of (R)-2HG that achieve tumor-relevant intracellular levels is able to recapitulate the effects of mutant IDH expression (Losman et al. 2013), and treatment of cells with cell-permeable (R)-2HG is able to inhibit the differentiation of murine 3T3-L1 fibroblasts and immortalized murine myeloid progenitor cells (Lu et al. 2012; Losman et al. 2013). Furthermore, tumor cells that harbor IDH mutations and produce high levels of (R)-2HG appear to require continuous (R)-2HG to remain transformed. Withdrawal of (R)-2HG from TF-1 leukemia cells transformed by cell-permeable (R)-2HG reverses their growth factor independence and restores their ability to differentiate (Losman et al. 2013), and inhibition of mutant IDH1 expression in glioma cell lines that harbor naturally occurring IDH1 R132H mutations impairs in vitro colony formation and in vivo tumor engraftment by the cells (Rohle et al. 2013). Finally, highly potent and specific inhibitors of mutant IDH have been developed that are able to reverse the transformation of TF-1 cells expressing IDH1 R132H and IDH2 R140Q (Popovici-Muller et al. 2012; Losman et al. 2013; Wang et al. 2013). These inhibitors also induce the differentiation of primary IDH mutant human leukemia cells in vitro and recapitulate the effects of suppression of IDH1 R132H expression in IDH mutant glioma cell lines in vitro and in vivo (Rohle et al. 2013; Wang et al. 2013). Taken together, these findings suggest that the transforming activity of mutant IDH is mediated by (R)-2HG and that (R)-2HG is necessary to maintain the transformed phenotype of tumor cells that harbor IDH mutations.
Mechanisms of transformation by (R)-2HG
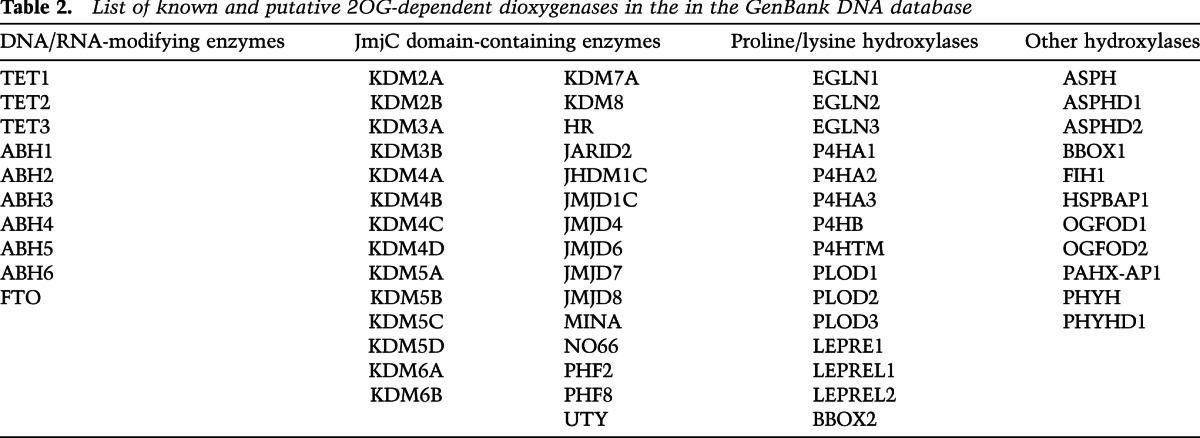
Several mechanisms of mutant IDH-mediated transformation have been proposed. One model for how mutant IDH contributes to tumorigenesis, and the one that has perhaps gained the most traction since the discovery of (R)-2HG, is transformation by 2OG antagonism. (R)-2HG is structurally and chemically very similar to 2OG (Fig. 2), and it has been proposed that (R)-2HG transforms cells by competitively inhibiting 2OG-dependent enzymes that function as tumor suppressors (Chowdhury et al. 2011; Xu et al. 2011; Koivunen et al. 2012; Lu et al. 2012). There are ~70 known and putative 2OG-dependent dioxygenases in the GenBank DNA database that could be relevant targets of (R)-2HG in mutant IDH-mediated transformation (Table 2).
Table 2.
List of known and putative 2OG-dependent dioxygenases in the in the GenBank DNA database
TET2
The myeloid tumor suppressor TET2 is a particularly intriguing candidate to be a pathogenically relevant target of (R)-2HG in IDH mutant tumors. TET1, TET2, and TET3 are members of a family of 2OG-dependent DNA-modifying enzymes that hydroxylate 5-methylcytosine (5mC) to generate 5-hydroxymethycytosine (5hmC) (Tahiliani et al. 2009; Ito et al. 2010). The TET enzymes are also able to further oxidize 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (He et al. 2011; Ito et al. 2011). Somatic mutations in TET2, including chromosomal microdeletions and missense, nonsense, and frameshift mutations, are common in clonal myeloid disorders, occurring in 10%–40% of cases of AML, MDS, MPN, and CMML (chronic myelomonocytic leukemia) (Abdel-Wahab et al. 2009; Delhommeau et al. 2009; Tefferi et al. 2009; Couronne et al. 2010; Chou et al. 2011). Interestingly, TET2 appears to be a haploinsufficient tumor suppressor. Although a subset of cases of MDS and AML present with biallelic TET2 loss, the majority of cases present with heterozygous TET2 mutations (Abdel-Wahab et al. 2009; Bejar et al. 2011; Gaidzik et al. 2012). The other TET family members are not recurrently mutated in hematopoietic malignancies, although TET1 is an infrequent MLL fusion partner in rare cases of MLL translocation-positive AML (Lorsbach et al. 2003).
TET enzymes are thought to play an important role in the epigenetic regulation of gene expression by mediating the demethylation of DNA. The conversion of 5mC to 5hmC and then to 5fC and 5caC by TET enzymes results in the generation of guanine:oxidized cytosine base-pair mismatches. It has been hypothesized that these mismatches are substrates for base excision repair by thymine–DNA glycosylase (TDG) (He et al. 2011; Raiber et al. 2012). It has also been suggested that 5caC residues are substrates for an as yet unidentified enzyme that can decarboxylate 5caC to generate unmethylated cytosine (Ito et al. 2011). In either case, oxidation of 5mC by TET enzymes, followed by TDG-mediated base excision or 5caC decarboxylation, would result in DNA demethylation. Indeed, overexpression of TET2 in HEK293 cells results in decreased 5mC levels and increased 5hmC, 5fC, and 5caC levels, whereas knockdown of TET1 in mouse embryonic stem cells results in decreased 5hmC, 5fC, and 5caC levels (Ito et al. 2011).
Given the putative role for TET2 in DNA demethylation, it would be expected that loss of TET2 activity should impair DNA demethylation and result in DNA hypermethylation. However, different studies have found inconsistent and even contradictory associations between TET2 mutational status and DNA methylation status in myeloid diseases. One study reported that TET2 mutant AML has a DNA hypermethylation signature (Figueroa et al. 2010). Two other studies found that genomic DNA is hypermethylated in TET2 wild-type myeloid diseases and is hypomethylated in TET2 mutant myeloid diseases (Ko et al. 2010; Perez et al. 2012). In yet another study, 5hmC levels were reported to be lower in TET2 mutant CMML than in TET2 wild-type disease, but global DNA methylation was the same in both cohorts (Yamazaki et al. 2012). In this last study, when locus-specific CpG island methylation was analyzed, only 11 of 27,578 CpG sites were differentially methylated between 5hmC-low and 5hmC-high patient samples. In the 5hmC-low patient samples, of the differentially methylated CpG sites, two were hypermethylated and the other nine were hypomethylated, suggesting that 5hmC levels do not correlate with 5mC levels at all. Some of these inconsistencies may be due to technical differences in experimental approach. It is also possible that TET2 mutations alter the methylation state of DNA at specific loci and only variably alter global DNA methylation.
Although 5mC and 5hmC are the only known substrates of TET2, TET2 may have other hydroxylation substrates besides DNA that mediate its tumor suppressor functions. Moreover, the hydroxylation of 5mC and the oxidation of 5hmC by TET2 might have other functions besides being intermediate steps in DNA demethylation. It has been observed that, in some cells, 5hmC modifications are copied to daughter strands during DNA replication (Szulwach et al. 2011). This requires a considerable expenditure of energy, as the daughter strands would first need to be methylated and then hydroxylated in order to maintain parental DNA 5hmC status. It seems unlikely that there would be significant selective pressure to maintain a nonfunctional transitional state of DNA demethylation. Rather, it seems more likely that 5hmC modifications and perhaps also 5fC and 5caC modifications play specific roles in the regulation of 5mC in the regulation of gene expression, perhaps by serving as recognition sites for the binding of distinct epigenetic and transcriptional regulators. Consistent with this hypothesis, specific “readers” of 5mC, 5hmC, and oxidized forms of 5hmC have recently been identified (Mellen et al. 2012; Spruijt et al. 2013). These include the transcriptional repressor MeCP2, which preferentially binds 5mC and 5hmC; the cell cycle regulator p53, which preferentially binds 5fC; and the DNA methyltransferase DNMT1, which preferentially binds 5caC. How the binding of these “readers” to modified cytosine residues contributes to the regulation of gene expression is not well understood, but it is likely that any perturbation of these interactions through imbalances in hydroxylation and oxidation of cytosine residues would have profound effects on cellular homeostasis.
There are several lines of evidence to suggest that TET2 is an important target of (R)-2HG in mutant IDH-mediated transformation. The catalytic activity of TET2 is potently inhibited by (R)-2HG in vitro (Xu et al. 2011; Koivunen et al. 2012). Furthermore, IDH mutant brain tumors and leukemias frequently display global DNA hypermethylation signatures (Figueroa et al. 2010; Ko et al. 2010; Turcan et al. 2012). Perhaps the most persuasive evidence for a link between IDH and TET2 mutations is the observation that IDH mutations are mutually exclusive with TET2 mutations in AML (Figueroa et al. 2010; Gaidzik et al. 2012). This suggests that the two mutations act on the same leukemogenic pathways and have redundant effects on cellular transformation. Indeed, (R)-2HG enhances the transforming effects of partial inhibition of TET2. In TF-1 cells, depletion of TET2 induces growth factor independence and blocks cellular differentiation similarly to expression of mutant IDH, and this transformation by TET2 depletion is potentiated by cell-permeable (R)-2HG (Losman et al. 2013).
It is unclear whether TET2 and IDH are similarly linked in other tumors. Depletion of TET2 in immortalized human astrocytes phenocopies the transforming effects of mutant IDH expression, but TET2 mutations have not been described in brain tumors (Koivunen et al. 2012). Interestingly, TET2 promoter methylation is increased in some cases of IDH wild-type grade II/III glioma (Kim et al. 2011). Given that promoter methylation is frequently associated with transcriptional silencing, this suggests that TET2 expression and activity are lost in these IDH wild-type gliomas. Taken as a whole, these observations suggest that loss of TET2 activity by either a loss-of-function mutational event, epigenetic silencing, or inhibition of TET2 activity by (R)-2HG is a frequent and important pathogenic event in brain tumors and myeloid diseases.
The evidence that IDH and TET2 mutations function in the same oncogenic pathway notwithstanding, it is worth noting that the phenotype of the conditional TET2 knockout mouse is quite distinct from that of the IDH1 R132H knock-in mouse described above (Moran-Crusio et al. 2011; Sasaki et al. 2012b). Homozygous deletion of TET2 in the hematopoietic compartment of mice does not cause leukemia but does result in a more significant expansion of hematopoietic stem and early progenitor cells than is seen in IDH1 R132H knock-in mice. Furthermore, TET2-null hematopoietic stem cells have a marked competitive repopulating advantage in transplanted recipient mice when compared with TET2 wild-type stem cells. This increased repopulating activity is not seen with IDH1 R132H knock-in stem cells, suggesting that self-renewal is more markedly enhanced by loss of TET2 than by expression of mutant IDH. TET2-null mice also show granulocyte and monocyte precursor lineage expansion and a skewing of myeloid differentiation toward monocyte/macrophage lineages, phenotypes that are not observed in IDH1 R132H knock-in mice.
There appears to be a dose-dependent effect of loss of TET2 activity on murine hematopoiesis. Mice with heterozygous loss of TET2 show an intermediate hematologic phenotype, with a less dramatic increase in stem cell self-renewal and a less dramatic dysregulation of myeloid differentiation than is seen in the TET2-null mice. This observation raises an interesting point. If transformation by TET2 suppression is indeed gene dosage-dependent, then it is reasonable to assume that selective pressure exists for heterozygous TET2 mutant cells and IDH mutant cells to further inhibit TET2 activity. A heterozygous TET2 mutant clone could do this by mutating the remaining wild-type TET2 allele or acquiring an IDH mutation. An IDH mutant clone could do this by acquiring a TET2 mutation. Why, then, are IDH mutations and heterozygous TET2 mutations mutually exclusive? One possible explanation is that the clonal advantage conferred by loss of TET2 is less dose-dependent in human myeloid cells than it is in mice. A cell that has already partially inhibited TET2 activity by either losing one allele of TET2 or acquiring an IDH mutation might not benefit significantly from further TET2 inhibition. With this in mind, it is interesting to note that, although biallelic TET2 loss is relatively common in MDS, occurring in >25% of cases, complete loss of TET2 is not associated with a more aggressive clinical disease phenotype (Jankowska et al. 2009; Mohamedali et al. 2009; Bejar et al. 2011).
Another possible explanation for the mutual exclusivity of TET2 and IDH mutations in leukemia is that acquisition of an IDH mutation confers a clonal disadvantage to TET2 mutant cells. Cells expressing mutant IDH grow more slowly under optimal culture conditions than their parental counterparts (Seltzer et al. 2010; Bralten et al. 2011; Losman et al. 2013). This suggests that mutant IDH and (R)-2HG have growth-suppressive effects that coexist with their transforming, growth-promoting effects. Perhaps in cells that do not require mutant IDH for cell growth and survival (for example, cells in which TET2 is already mutated), the negative effects of (R)-2HG on proliferation overwhelm the positive effects of (R)-2HG on transformation. This explanation could also account for the less pronounced phenotype of the IDH1 R132H knock-in mice as compared with the TET2 conditional knockout mice. Perhaps the mild phenotype of the IDH1 R132H knock-in mice is not the result of inadequate inhibition of TET2 but rather is a reflection of the balance between the protransforming effects of (R)-2HG inhibition of TET2 and the anti-transforming effects of (R)-2HG inhibition of other, growth-promoting enzymes.
JmjC histone demethylases
The JmjC family of histone lysine demethylases is another family of enzymes that may be a pathogenically relevant target of (R)-2HG. Histone demethylases play an important role in the epigenetic regulation of gene expression (Klose et al. 2006; Shilatifard 2006). Histone methylation defines the structure of chromatin, and methylation of some histone residues (H3K4, H3K36, and H3K79) is often associated with transcriptionally active euchromatin, whereas other histone methylation marks (H3K9, H3K27, and H4K20) are usually associated with transcriptionally silent heterochromatin. The patterns of histone methylation at different genetic loci are a function of the balance between the activities of histone methyltransferases and histone demethylases, each of which binds and alters the methylation of specific histone lysine residues. Disruption of histone methylation or demethylation can have profound effects on gene expression, and JmjC histone demethylases have been linked to the pathogenesis of a number of different cancers (Cloos et al. 2008; Varier and Timmers 2011).
Several JmjC histone demethylases appear to function as tumor suppressors. The H3K36 JmjC demethylase KDM2B (JHDM1B/FBX10) is a transcriptional repressor that regulates ribosomal gene expression and represses c-Jun-mediated transcription (Frescas et al. 2007; Koyama-Nasu et al. 2007). Consistent with its putative role as a tumor suppressor, KDM2B expression is significantly lower in brain tumors than in normal brain tissue. The H3K9 JmjC demethylase KDM3B (JMJD1B) is frequently deleted in 5q− MDS and AML (Hu et al. 2001), and the H3K4 JmjC demethylase KDM5C (JARID1C) is occasionally mutated and inactivated in clear cell renal carcinoma (Dalgliesh et al. 2010; Hakimi et al. 2013). The H3K27 JmjC demethylase KDM6A (UTX) and its paralog, KDM6B (JMJD3), can also function as tumor suppressors. KDM6A cooperates with the Retinoblastoma tumor suppressor protein to enforce cell cycle blockade and also negatively regulates the activity of NOTCH (Herz et al. 2010; Terashima et al. 2010; Wang et al. 2010). KDM6B is involved in the regulation of p53 and has been shown to promote the terminal differentiation of glioblastoma cells (Barradas et al. 2009; Sola et al. 2011).
It is possible that inhibition of tumor suppressor histone demethylases by (R)-2HG contributes to transformation by mutant IDH. In fact, (R)-2HG has been shown to inhibit the activity of many 2OG-dependent dioxygenases in vitro, including the JmjC histone demethylases KDM2A, KDM4A, KDM4C, and KDM7A (Chowdhury et al. 2011; Xu et al. 2011; Lu et al. 2012). Although the IC50 values for inhibition of 2OG-dependent enzymes by (R)-2HG vary considerably (ranging from 25 μM to >10 mM), it is important to remember that (R)-2HG can accumulate to millimolar levels in tumors. Moreover, overexpression of mutant IDH and treatment of cells with cell-permeable (R)-2HG has been reported to increase H3K4, H3K9, H3K27, H3K36, and H3K79 histone methylation marks in cell culture models (Lu et al. 2012).
The biological significance of the histone methylation changes induced in vitro by (R)-2HG is not clear, and it is not yet known whether (R)-2HG affects histone methylation in primary human IDH mutant tumors. H3K9 trimethylation is frequently increased in IDH mutant gliomas (Vanneti et al. 2013). However, many IDH wild-type gliomas also display H3K9 hypermethylation, suggesting that aberrant histone methylation is a common finding in brain tumors irrespective of IDH mutational status. In the hematopoietic-specific IDH1 R132H knock-in mice, bone marrow stem cells show a modest increase in methylated H3K4 levels but no change in other histone methylation marks (Sasaki et al. 2012b). Furthermore, no differences in global histone methylation are apparent in the brains of the brain-specific IDH1 R132H knock-in mice (Sasaki et al. 2012a). Although this could be interpreted as evidence that (R)-2HG does not significantly affect histone methylation in vivo, it is also possible that (R)-2HG induces biologically important changes in histone methylation only at specific genetic loci that promote tumor formation. Histone methylation changes that are beneficial could be preferentially retained in mutant IDH-expressing cells, whereas deleterious changes could be reversed by compensatory changes such as down-regulation of specific histone methyltransferases.
As described above, several JmjC histone demethylases can function as tumor suppressors. However, some histone demethylases are also known to promote tumor growth. Moreover, there are multiple examples of chromatin-modifying enzymes—including histone demethylases and their opposing methyltransferases—that can act as either oncoproteins or tumor suppressors in different cellular contexts (Cloos et al. 2008). It is interesting to speculate that the different functions of the JmjC histone demethylases in different tissues could explain, at least in part, the tissue specificity of IDH mutations in cancer. If the proliferation and survival of a particular cell were dependent on the activity of JmjC histone demethylases that are potently inhibited by (R)-2HG, that cell would likely be relatively resistant to transformation by mutant IDH. Conversely, a cell in which those same JmjC histone demethylases are dispensable for proliferation or have tumor suppressor functions might be particularly vulnerable to transformation by mutant IDH.
EglN prolyl-4-hydroxylases
EglN1, EglN2, and EglN3 are members of a family of 2OG-dependent dioxygenases that regulate the activity of HIF, a heterodimeric transcription factor that mediates the cellular response to hypoxia (Kaelin and Ratcliffe 2008; Semenza 2012). In the presence of oxygen, EglN hydroxylates specific proline residues on HIFα, and these hydroxylated proline residues are then recognized by the von Hippel-Lindau (VHL) E3–ubiquitin ligase complex. The VHL complex polyubiquitylates HIFα and targets it for proteasomal degradation. Under hypoxic conditions, when intracellular levels of oxygen are low, the activity of EglN is inhibited. This permits accumulation of HIFα, which can then heterodimerize with HIFβ, translocate to the nucleus, and activate the transcriptional response of cells to hypoxia. EglN1 is believed to be the principal HIF prolyl-hydroxylase, with EglN2 and EglN3 playing variable compensatory roles in the regulation of HIF in different tissues under specific cellular conditions.
EglN enzymes were initially reported to be inhibited by (R)-2HG (Zhao et al. 2009). However, further study has shown that, on the contrary, (R)-2HG potentiates EglN activity in vitro and in vivo and blunts the induction of HIFα in response to hypoxia in cell culture models (Koivunen et al. 2012; Losman et al. 2013). Consistent with this observation, IDH mutant brain tumors display decreased HIF activation compared with their wild-type counterparts (Williams et al. 2011; Koivunen et al. 2012). It should be noted that HIFα levels are elevated in the brains of the brain-specific IDH1 R132H knock-in mice (Sasaki et al. 2012a). However, these mice succumb at birth to cerebral hemorrhage, suggesting that their brain perfusion during development is abnormal. It is possible that tissue hypoxia, rather than a direct effect of (R)-2HG on EglN activity, is the cause of increased brain levels of HIFα in these mice.
Likewise, there is no evidence that HIF is activated in human IDH mutant leukemias. On the contrary, there is evidence to suggest that activation of HIF inhibits myeloid leukemia. A number of human myeloid leukemia cell lines have been reported to proliferate more slowly and undergo terminal differentiation and apoptosis upon exposure to hypoxia and upon overexpression of HIFα (Huang et al. 2003; di Giacomo et al. 2009; Zhang and Chen 2009; He et al. 2013), and inhibition of HIFα expression by RNAi blocks the differentiation of leukemia cells in response to all-trans retinoic acid (ATRA) (Zhang et al. 2008; Zhang and Chen 2009). Moreover, in a mouse model of acute promyelocytic leukemia, exposure of diseased mice to intermittent hypoxia induces leukemia cell differentiation and prolongs survival of the mice (Liu et al. 2006). These observations suggest that HIF may be a tumor suppressor in myeloid leukemia. However, it is not known whether the HIF response to hypoxia is truly blunted in IDH mutant tumors in vivo, as one would predict given that (R)-2HG can serve as an EglN agonist.
The ability of (R)-2HG to function as an EglN cosubstrate appears to play an important role in mutant IDH-mediated cellular transformation. In immortalized human astrocytes, either overexpression of EglN1 or depletion of HIF1α is sufficient to promote soft agar colony formation (Koivunen et al. 2012). Conversely, depletion of EglN1 in mutant IDH-transformed astrocytes inhibits the growth and soft agar colony formation of the cells. Intriguingly, (S)-2HG, which potently inhibits EglN in vitro, is unable to promote transformation of TF-1 leukemia cells. In fact, (S)-2HG antagonizes TF-1 cell transformation induced by TET2 loss (Losman et al. 2013). This is despite the fact that (S)-2HG is a more potent inhibitor of TET2 than is (R)-2HG (Xu et al. 2011; Koivunen et al. 2012). Depletion of EglN1 in transformed TF-1 cells reverses transformation induced by expression of mutant IDH or by depletion of TET2, suggesting that the anti-leukemic properties of (S)-2HG relate to its ability to inhibit EglN1 (Losman et al. 2013). In short, inhibition of EglN1 by (S)-2HG reverses the leukemic transformation that would otherwise ensue as a result of its inhibition of TET2 activity.
Other candidate targets of (R)-2HG
A number of other 2OG-dependent enzymes might be inhibited by (R)-2HG in IDH mutant tumors (Table 2). Three types of collagen hydroxylases—the Leprecan family of prolyl-3-hydroxylases, the P4HA family of prolyl-4-hydroxylases, and the PLOD family of lysyl-5-hydroxylases—all require 2OG for activity, and P4HA1 has been found to be inhibited by (R)-2HG in vitro (Koivunen et al. 2012). The collagen prolyl-hydroxylases mediate the hydroxylation of proline residues required for formation of the collagen triple helix (Gorres and Raines 2010), and the collagen lysyl-hydroxylases mediate the hydroxylation of lysine residues required for collagen cross-linking into stable fibrils (Bank et al. 1999). Interestingly, collagen maturation has been found to be impaired in the brain-specific IDH1 R132H knock-in mice, suggesting that these enzymes are targets of inhibition by mutant IDH in vivo (Sasaki et al. 2012a). Although a role for these enzymes in cancer has not been established, the observation that expression of collagen prolyl-3-hydroxylases and prolyl-4-hydroxylases is down-regulated in many B-cell lymphomas suggests that these enzymes function as tumor suppressors in some tissues (Teodoro et al. 2006; Hatzimichael et al. 2012).
Other potential 2OG-dependent targets of (R)-2HG include FIH1 (factor inhibiting hypoxia-inducible factor 1), an asparaginyl hydroxylase that regulates the transcriptional activity of HIF (Mahon et al. 2001); the ABH family of DNA demethylases that are involved in DNA damage repair (Lee et al. 2005); and the RNA demethylase FTO (fat mass and obesity-associated), which is believed to be important for the regulation of cellular metabolism (Jia et al. 2008; Berulava et al. 2013).
Other possible mechanisms of transformation by mutant IDH
Another proposed mechanism of transformation by mutant IDH harkens back to Warburg's mitochondrial model of oncogenesis. Warburg (1956) hypothesized that the primary defect in cancer cells is impaired mitochondrial function, which causes cells to switch from oxidative phosphorylation to aerobic glycolysis. Warburg's model has been somewhat discredited by the observation that mitochondrial function is not impaired in most cancer cells (Wallace 2012). However, there are data to suggest that (R)-2HG directly interferes with normal mitochondrial function. Some D2HGA patients have elevated levels of urinary lactate, suggesting that they have some degree of mitochondrial dysfunction (Kranendijk et al. 2012). Furthermore, the activity of two enzyme complexes in the electron transport chain, complex IV (cytochrome c oxidase) and complex V (ATP synthase), can be inhibited by (R)-2HG in vitro (Kolker et al. 2002; Latini et al. 2005). Inhibition of ATP synthase expression and activity has been observed in some cancers and is believed to directly promote cellular transformation by causing the accumulation of excess electrons that are then transferred to molecular oxygen to generate superoxide (O2−) (Willers and Cuezva 2011).
Mutant IDH-expressing cells may have an impaired ability to neutralize reactive oxygen species (ROS) (Latini et al. 2003). Conversion of 2OG to (R)-2HG by mutant IDH consumes NADPH, and the high catalytic rate of the mutant IDH reaction could conceivably result in depletion of cellular NADPH (Yan et al. 2009; Bleeker et al. 2010). NADPH is the electron donor used by glutathione reductase to regenerate reduced glutathione, which is the principal cellular and mitochondrial antioxidant that regulates cellular redox (Kirsch and De Groot 2001; Block and Gorin 2012). Depletion of NADPH might therefore decrease the ability of cells to neutralize DNA-damaging oxygen-free radicals. NADPH also has other regulatory functions in cells that could contribute to cellular transformation. NADPH plays a role in nutrient sensing and participates in the regulation of the cellular metabolic rate (Newsholme et al. 2010; Newsholme and Krause 2012), and NADPH is also involved in the regulation of thioredoxins, enzymes that regulate the thiol-disulphide redox state and activity of several transcription factors, including NF-κB and AP1 (Arner and Holmgren 2000; Kabe et al. 2005).
Although dysregulation of cellular redox has been shown to contribute to transformation in other types of cancer, it is not clear whether NADPH and ROS are in fact dysregulated in IDH mutant tumors. Consistent with the notion that mutant IDH can perturb cellular NADPH levels, the embryonic brains of the brain-specific IDH1 R132H knock-in mice have a slight increase in their NADP+:NADPH ratio and slightly decreased levels of reduced glutathione (Sasaki et al. 2012a). However, the brains of the IDH1 R132H mice have lower, not higher, levels of ROS than control mice. In the hematopoietic-specific IDH1 R132H knock-in mice, the NADP+:NADPH ratio and the levels of ROS are normal in the bone marrow (Sasaki et al. 2012b). Despite the apparent lack of increased ROS in the IDH1 R132H knock-in mice, it is nonetheless possible that cellular redox dysregulation plays a role in transformation by mutant IDH. The mice may have subtle or compartmentalized, and yet pathophysiologically relevant, perturbations in their brain and bone marrow redox states that only become apparent under conditions of oxidative stress. It is also possible that otherwise normal cells have compensatory mechanisms that largely maintain redox homeostasis in the presence of mutant IDH and that these mechanisms are defective in cancer cells.
Another potential contributor to mutant IDH-mediated transformation is deficiency of wild-type IDH activity. Although most tumor-associated mutations in IDH result in (R)-2HG production, a few rare IDH1 and IDH2 mutants that do not produce (R)-2HG have recently been identified in lymphoid and thyroid tumors (Ward et al. 2012). These mutations are monoallelic and result in either decreased enzymatic activity or loss of expression of the mutant enzyme. It has not yet been determined whether these loss-of-function mutations contribute to cellular transformation or are simply passenger mutations, but they do raise the possibility that IDH haploinsufficiency in IDH mutant cells could be tumorigenic.
Therapeutic targeting of IDH mutant tumors
The observation that (R)-2HG is sufficient to promote cellular transformation and that transformation by (R)-2HG is reversible suggests that inhibiting (R)-2HG production by mutant IDH would have efficacy in the treatment of IDH mutant cancers. In this regard, first-generation mutant IDH inhibitors capable of suppressing the production of (R)-2HG in vitro and in vivo have recently been reported (Popovici-Muller et al. 2012; Losman et al. 2013; Rohle et al. 2013; Wang et al. 2013).
As described above, EglN activation plays a causal role in the transformation of astrocytes by mutant IDH in cell culture models and appears to play a permissive role in the transformation of TF-1 leukemia cells by mutant IDH. These findings, if proven robust, warrant further exploration of the potential of EglN inhibitors for the treatment of IDH mutant neoplasms. A number of EglN inhibitors have been developed for the treatment of anemia and tissue ischemia, some of which have advanced to human clinical trials (Robinson et al. 2008; Miyata et al. 2011; Sen Banerjee et al. 2012; Forristal et al. 2013).
Finally, it seems likely that the millimolar levels of (R)-2HG that accumulate in IDH mutant tumors also create certain liabilities for tumor cells. For example, inhibition of certain enzymes by (R)-2HG, whether they are pathogenic targets or “bystanders,” might decrease cellular fitness and hence decrease the resistance of cells to certain forms of therapeutic attack. There is some indirect evidence to support this idea. For example, introduction of mutant IDH into TF-1 cells confers a growth disadvantage to the cells under cytokine-rich conditions even though mutant IDH confers a growth advantage to the cells under cytokine-poor conditions (Losman et al. 2013). Moreover, stable expression of IDH1 R132H impedes the growth and migration of human IDH1 wild-type GBM cell lines both in vitro and in mouse xenografts models (Bralten et al. 2011). The relatively good prognosis and enhanced sensitivity to chemotherapy and radiation therapy of IDH mutant brain tumors might also reflect deleterious effects of (R)-2HG in tumor cells (Houillier et al. 2010; SongTao et al. 2012; Li et al. 2013). Therefore, another strategy for tackling IDH mutant tumors could be to systematically identify and target the unique dependencies that are created in the presence of millimolar concentrations of (R)-2HG, thereby converting the oncometabolite into an Achilles' heel.
Acknowledgments
We thank Dr. Matthew Vander Heiden, Dr. Rafael Bejar, and Dr. Samuel McBrayer for critical reading of the manuscript. We apologize to colleagues whose work was not cited due to space limitations or oversight. Please bring errors and egregious omissions to our attention. W.G.K. consults for, and owns equity in, Fibrogen, which is developing EglN inhibitors, and Agios, which is targeting cancer metabolism. Supported by grants from NIH, HHMI, and the Breast Cancer Research Foundation.
References
- Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, van Putten WJ, Rijneveld AW, Lowenberg B, Valk PJ 2010. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood 116: 2122–2126 [Abstract] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. 2009. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114: 144–147 [Europe PMC free article] [Abstract] [Google Scholar]
- Abdel-Wahab O, Patel J, Levine RL 2011. Clinical implications of novel mutations in epigenetic modifiers in AML. Hematol Oncol Clin North Am 25: 1119–1133 [Abstract] [Google Scholar]
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. 2011a. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 [Abstract] [Google Scholar]
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. 2011b. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet 43: 1262–1265 [Abstract] [Google Scholar]
- Arai M, Nobusawa S, Ikota H, Takemura S, Nakazato Y 2012. Frequent IDH1/2 mutations in intracranial chondrosarcoma: A possible diagnostic clue for its differentiation from chordoma. Brain Tumor Pathol 29: 201–206 [Abstract] [Google Scholar]
- Arner ES, Holmgren A 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267: 6102–6109 [Abstract] [Google Scholar]
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A 2008. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116: 597–602 [Abstract] [Google Scholar]
- Bank RA, Robins SP, Wijmenga C, Breslau-Siderius LJ, Bardoel AF, van der Sluijs HA, Pruijs HE, TeKoppele JM 1999. Defective collagen crosslinking in bone, but not in ligament or cartilage, in Bruck syndrome: Indications for a bone-specific telopeptide lysyl hydroxylase on chromosome 17. Proc Natl Acad Sci 96: 1054–1058 [Europe PMC free article] [Abstract] [Google Scholar]
- Barnes LD, Kuehn GD, Atkinson DE 1971. Yeast diphosphopyridine nucleotide specific isocitrate dehydrogenase. Purification and some properties. Biochemistry 10: 3939–3944 [Abstract] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. 2009. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23: 1177–1182 [Europe PMC free article] [Abstract] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, et al. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364: 2496–2506 [Europe PMC free article] [Abstract] [Google Scholar]
- Berulava T, Ziehe M, Klein-Hitpass L, Mladenov E, Thomale J, Ruther U, Horsthemke B 2013. FTO levels affect RNA modification and the transcriptome. Eur J Hum Genet 21: 317–323 [Europe PMC free article] [Abstract] [Google Scholar]
- Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, et al. 2009. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat 30: 7–11 [Abstract] [Google Scholar]
- Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, Tigchelaar W, Troost D, Vandertop WP, Bardelli A, et al. 2010. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 119: 487–494 [Europe PMC free article] [Abstract] [Google Scholar]
- Block K, Gorin Y 2012. Aiding and abetting roles of NOX oxidases in cellular transformation. Nat Rev Cancer 12: 627–637 [Europe PMC free article] [Abstract] [Google Scholar]
- Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, et al. 2012. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17: 72–79 [Europe PMC free article] [Abstract] [Google Scholar]
- Bralten LB, Kloosterhof NK, Balvers R, Sacchetti A, Lapre L, Lamfers M, Leenstra S, de Jonge H, Kros JM, Jansen EE, et al. 2011. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol 69: 455–463 [Abstract] [Google Scholar]
- Byeon SJ, Myung JK, Kim SH, Kim SK, Phi JH, Park SH 2012. Distinct genetic alterations in pediatric glioblastomas. Childs Nerv Syst 28: 1025–1032 [Abstract] [Google Scholar]
- Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, Parrens M, Martin A, Xerri L, Brousset P, et al. 2012. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 119: 1901–1903 [Europe PMC free article] [Abstract] [Google Scholar]
- Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D, Ackermann U, Tessmer C, Korshunov A, Zentgraf H, et al. 2010. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol 20: 245–254 [Abstract] [Google Scholar]
- Chang C, Xu K, Shu H 2011. The role of isocitrate dehydrogenase mutations in glioma brain tumors. In Molecular targets of CNS tumors (ed. M Garami), pp. 413–436. InTechOpen, Atalanta, GA. [Google Scholar]
- Chao MP, Seita J, Weissman IL 2008. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol 73: 439–449 [Abstract] [Google Scholar]
- Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, et al. 2012. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 18: 624–629 [Europe PMC free article] [Abstract] [Google Scholar]
- Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY, Lee MC, Ko BS, Tang JL, Yao M, et al. 2011. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 118: 3803–3810 [Abstract] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [Europe PMC free article] [Abstract] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K 2008. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev 22: 1115–1140 [Europe PMC free article] [Abstract] [Google Scholar]
- Comte B, Vincent G, Bouchard B, Benderdour M, Des Rosiers C 2002. Reverse flux through cardiac NADP+-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol 283: H1505–H1514 [Abstract] [Google Scholar]
- Couronne L, Lippert E, Andrieux J, Kosmider O, Radford-Weiss I, Penther D, Dastugue N, Mugneret F, Lafage M, Gachard N, et al. 2010. Analyses of TET2 mutations in post-myeloproliferative neoplasm acute myeloid leukemias. Leukemia 24: 201–203 [Abstract] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363 [Europe PMC free article] [Abstract] [Google Scholar]
- Dalziel K 1980. Isocitrate dehydrogenase and related oxidative decarboxylases. FEBS Lett 117: K45–K55 [Abstract] [Google Scholar]
- Dang CV 2012. Links between metabolism and cancer. Genes Dev 26: 877–890 [Europe PMC free article] [Abstract] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [Europe PMC free article] [Abstract] [Google Scholar]
- Dang L, Jin S, Su SM 2010. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med 16: 387–397 [Abstract] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB 2008. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [Abstract] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301 [Abstract] [Google Scholar]
- di Giacomo V, Rapino M, Miscia S, Di Giulio C, Cataldi A 2009. Dual role of HIF-1α in delivering a survival or death signal in hypoxia exposed human K562 erythroleukemia cells. Cell Biol Int 33: 49–56 [Abstract] [Google Scholar]
- Engqvist M, Drincovich MF, Flugge UI, Maurino VG 2009. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and β-oxidation pathways. J Biol Chem 284: 25026–25037 [Europe PMC free article] [Abstract] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [Europe PMC free article] [Abstract] [Google Scholar]
- Filipp FV, Scott DA, Ronai ZA, Osterman AL, Smith JW 2012. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 25: 375–383 [Europe PMC free article] [Abstract] [Google Scholar]
- Forristal CE, Winkler IG, Nowlan B, Barbier V, Walkinshaw G, Levesque JP 2013. Pharmacologic stabilization of HIF-1α increases hematopoietic stem cell quiescence in vivo and accelerates blood recovery after severe irradiation. Blood 121: 759–769 [Abstract] [Google Scholar]
- Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M 2007. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450: 309–313 [Abstract] [Google Scholar]
- Gaal J, Burnichon N, Korpershoek E, Roncelin I, Bertherat J, Plouin PF, de Krijger RR, Gimenez-Roqueplo AP, Dinjens WN 2010. Isocitrate dehydrogenase mutations are rare in pheochromocytomas and paragangliomas. J Clin Endocrinol Metab 95: 1274–1278 [Abstract] [Google Scholar]
- Gabriel JL, Zervos PR, Plaut GW 1986. Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria. Metabolism 35: 661–667 [Abstract] [Google Scholar]
- Gaidzik VI, Paschka P, Spath D, Habdank M, Kohne CH, Germing U, von Lilienfeld-Toal M, Held G, Horst HA, Haase D, et al. 2012. TET2 mutations in acute myeloid leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol 30: 1350–1357 [Abstract] [Google Scholar]
- Gorres KL, Raines RT 2010. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol 45: 106–124 [Europe PMC free article] [Abstract] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. 2010. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339–344 [Europe PMC free article] [Abstract] [Google Scholar]
- Hakimi AA, Chen YB, Wren J, Gonen M, Abdel-Wahab O, Heguy A, Liu H, Takeda S, Tickoo SK, Reuter VE, et al. 2013. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur Urol 63: 848–854 [Europe PMC free article] [Abstract] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et al. 2009. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol 118: 469–474 [Abstract] [Google Scholar]
- Hatzimichael E, Lo Nigro C, Lattanzio L, Syed N, Shah R, Dasoula A, Janczar K, Vivenza D, Monteverde M, Merlano M, et al. 2012. The collagen prolyl hydroxylases are novel transcriptionally silenced genes in lymphoma. Br J Cancer 107: 1423–1432 [Europe PMC free article] [Abstract] [Google Scholar]
- Hausinger RP 2004. FeII/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39: 21–68 [Abstract] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [Europe PMC free article] [Abstract] [Google Scholar]
- He M, Wang QY, Yin QQ, Tang J, Lu Y, Zhou CX, Duan CW, Hong DL, Tanaka T, Chen GQ, et al. 2013. HIF-1α downregulates miR-17/20a directly targeting p21 and STAT3: A role in myeloid leukemic cell differentiation. Cell Death Differ 20: 408–418 [Europe PMC free article] [Abstract] [Google Scholar]
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A 2010. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 30: 2485–2497 [Europe PMC free article] [Abstract] [Google Scholar]
- Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, Paris S, Boisselier B, Idbaih A, Laigle-Donadey F, et al. 2010. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75: 1560–1566 [Abstract] [Google Scholar]
- Hu Z, Gomes I, Horrigan SK, Kravarusic J, Mar B, Arbieva Z, Chyna B, Fulton N, Edassery S, Raza A, et al. 2001. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31. Oncogene 20: 6946–6954 [Abstract] [Google Scholar]
- Huang Y, Du KM, Xue ZH, Yan H, Li D, Liu W, Chen Z, Zhao Q, Tong JH, Zhu YS, et al. 2003. Cobalt chloride and low oxygen tension trigger differentiation of acute myeloid leukemic cells: Possible mediation of hypoxia-inducible factor-1α. Leukemia 17: 2065–2073 [Abstract] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [Europe PMC free article] [Abstract] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [Europe PMC free article] [Abstract] [Google Scholar]
- Jacob B, Osato M 2009. Stem cell exhaustion and leukemogenesis. J Cell Biochem 107: 393–399 [Abstract] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O'Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP 2009. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 113: 6403–6410 [Europe PMC free article] [Abstract] [Google Scholar]
- Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C 2008. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett 582: 3313–3319 [Europe PMC free article] [Abstract] [Google Scholar]
- Jin G, Reitman ZJ, Duncan CG, Spasojevic I, Gooden DM, Rasheed BA, Yang R, Lopez GY, He Y, McLendon RE, et al. 2013. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res 73: 496–501 [Europe PMC free article] [Abstract] [Google Scholar]
- Jo SH, Lee SH, Chun HS, Lee SM, Koh HJ, Lee SE, Chun JS, Park JW, Huh TL 2002. Cellular defense against UVB-induced phototoxicity by cytosolic NADP(+)-dependent isocitrate dehydrogenase. Biochem Biophys Res Commun 292: 542–549 [Abstract] [Google Scholar]
- Kabe Y, Ando K, Hirao S, Yoshida M, Handa H 2005. Redox regulation of NF-κB activation: Distinct redox regulation between the cytoplasm and the nucleus. Antioxid Redox Signal 7: 395–403 [Abstract] [Google Scholar]
- Kaelin WG Jr 2011. Cancer and altered metabolism: Potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol 76: 335–345 [Europe PMC free article] [Abstract] [Google Scholar]
- Kaelin WG Jr, Ratcliffe PJ 2008. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell 30: 393–402 [Abstract] [Google Scholar]
- Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH 2009. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer 125: 353–355 [Abstract] [Google Scholar]
- Kim SY, Lee SM, Tak JK, Choi KS, Kwon TK, Park JW 2007. Regulation of singlet oxygen-induced apoptosis by cytosolic NADP+-dependent isocitrate dehydrogenase. Mol Cell Biochem 302: 27–34 [Abstract] [Google Scholar]
- Kim YH, Pierscianek D, Mittelbronn M, Vital A, Mariani L, Hasselblatt M, Ohgaki H 2011. TET2 promoter methylation in low-grade diffuse gliomas lacking IDH1/2 mutations. J Clin Pathol 64: 850–852 [Abstract] [Google Scholar]
- King A, Selak MA, Gottlieb E 2006. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 25: 4675–4682 [Abstract] [Google Scholar]
- Kirsch M, De Groot H 2001. NAD(P)H, a directly operating antioxidant? FASEB J 15: 1569–1574 [Abstract] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y 2006. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [Abstract] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843 [Europe PMC free article] [Abstract] [Google Scholar]
- Koh HJ, Lee SM, Son BG, Lee SH, Ryoo ZY, Chang KT, Park JW, Park DC, Song BJ, Veech RL, et al. 2004. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J Biol Chem 279: 39968–39974 [Abstract] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484–488 [Europe PMC free article] [Abstract] [Google Scholar]
- Kolker S, Pawlak V, Ahlemeyer B, Okun JG, Horster F, Mayatepek E, Krieglstein J, Hoffmann GF, Kohr G 2002. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci 16: 21–28 [Abstract] [Google Scholar]
- Kosmider O, Gelsi-Boyer V, Slama L, Dreyfus F, Beyne-Rauzy O, Quesnel B, Hunault-Berger M, Slama B, Vey N, Lacombe C, et al. 2010. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia 24: 1094–1096 [Abstract] [Google Scholar]
- Koszarska M, Bors A, Feczko A, Meggyesi N, Batai A, Csomor J, Adam E, Kozma A, Orban TI, Lovas N, et al. 2012. Type and location of isocitrate dehydrogenase mutations influence clinical characteristics and disease outcome of acute myeloid leukemia. Leuk Lymphoma 10.3109/10428194.2012.736981 [Abstract] [Google Scholar]
- Koyama-Nasu R, David G, Tanese N 2007. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol 9: 1074–1080 [Abstract] [Google Scholar]
- Kranendijk M, Struys EA, Gibson KM, Wickenhagen WV, Abdenur JE, Buechner J, Christensen E, de Kremer RD, Errami A, Gissen P, et al. 2010. Evidence for genetic heterogeneity in D-2-hydroxyglutaric aciduria. Hum Mutat 31: 279–283 [Abstract] [Google Scholar]
- Kranendijk M, Salomons GS, Gibson KM, Van Schaftingen E, Jakobs C, Struys EA 2011. A lymphoblast model for IDH2 gain-of-function activity in d-2-hydroxyglutaric aciduria type II: Novel avenues for biochemical and therapeutic studies. Biochim Biophys Acta 1812: 1380–1384 [Abstract] [Google Scholar]
- Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C 2012. Progress in understanding 2-hydroxyglutaric acidurias. J Inherit Metab Dis 35: 571–587 [Europe PMC free article] [Abstract] [Google Scholar]
- Latini A, Scussiato K, Rosa RB, Llesuy S, Bello-Klein A, Dutra-Filho CS, Wajner M 2003. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur J Neurosci 17: 2017–2022 [Abstract] [Google Scholar]
- Latini A, da Silva CG, Ferreira GC, Schuck PF, Scussiato K, Sarkis JJ, Dutra Filho CS, Wyse AT, Wannmacher CM, Wajner M 2005. Mitochondrial energy metabolism is markedly impaired by D-2-hydroxyglutaric acid in rat tissues. Mol Genet Metab 86: 188–199 [Abstract] [Google Scholar]
- Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW 2002. Cytosolic NADP+-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med 32: 1185–1196 [Abstract] [Google Scholar]
- Lee SH, Jo SH, Lee SM, Koh HJ, Song H, Park JW, Lee WH, Huh TL 2004. Role of NADP+-dependent isocitrate dehydrogenase (NADP+-ICDH) on cellular defence against oxidative injury by γ-rays. Int J Radiat Biol 80: 635–642 [Abstract] [Google Scholar]
- Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O'Connor TR 2005. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem 280: 39448–39459 [Abstract] [Google Scholar]
- Lee JH, Kim SY, Kil IS, Park JW 2007. Regulation of ionizing radiation-induced apoptosis by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem 282: 13385–13394 [Abstract] [Google Scholar]
- Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, Coller HA 2010. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol 8: e1000514. [Europe PMC free article] [Abstract] [Google Scholar]
- Leonardi R, Subramanian C, Jackowski S, Rock CO 2012. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem 287: 14615–14620 [Europe PMC free article] [Abstract] [Google Scholar]
- Li S, Chou AP, Chen W, Chen R, Deng Y, Phillips HS, Selfridge J, Zurayk M, Lou JJ, Everson RG, et al. 2013. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro-oncol 15: 57–68 [Europe PMC free article] [Abstract] [Google Scholar]
- Liu W, Guo M, Xu YB, Li D, Zhou ZN, Wu YL, Chen Z, Kogan SC, Chen GQ 2006. Induction of tumor arrest and differentiation with prolonged survival by intermittent hypoxia in a mouse model of acute myeloid leukemia. Blood 107: 698–707 [Abstract] [Google Scholar]
- Locasale JW, Cantley LC 2011. Metabolic flux and the regulation of mammalian cell growth. Cell Metab 14: 443–451 [Europe PMC free article] [Abstract] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR 2003. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 17: 637–641 [Abstract] [Google Scholar]
- Losman JA, Looper R, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley G, Root D, Ebert BL, Kaelin WG Jr 2013. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339: 1621–1625 [Europe PMC free article] [Abstract] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478 [Europe PMC free article] [Abstract] [Google Scholar]
- Lunt SY, Vander Heiden MG 2011. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27: 441–464 [Abstract] [Google Scholar]
- Lyssiotis CA, Cantley LC 2012. SIRT6 puts cancer metabolism in the driver's seat. Cell 151: 1155–1156 [Europe PMC free article] [Abstract] [Google Scholar]
- Mahon PC, Hirota K, Semenza GL 2001. FIH-1: A novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 15: 2675–2686 [Europe PMC free article] [Abstract] [Google Scholar]
- Mailloux RJ, Beriault R, Lemire J, Singh R, Chenier DR, Hamel RD, Appanna VD 2007. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE 2: e690. [Europe PMC free article] [Abstract] [Google Scholar]
- Marciniak-Czochra A, Stiehl T, Ho AD, Jager W, Wagner W 2009. Modeling of asymmetric cell division in hematopoietic stem cells–regulation of self-renewal is essential for efficient repopulation. Stem Cells Dev 18: 377–385 [Abstract] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, et al. 2010. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. J Clin Oncol 28: 2348–2355 [Europe PMC free article] [Abstract] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [Europe PMC free article] [Abstract] [Google Scholar]
- Matsumoto A, Nakayama KI 2013. Role of key regulators of the cell cycle in maintenance of hematopoietic stem cells. Biochim Biophys Acta 1830: 2335–2344 [Abstract] [Google Scholar]
- Meijer D, de Jong D, Pansuriya TC, van den Akker BE, Picci P, Szuhai K, Bovee JV 2012. Genetic characterization of mesenchymal, clear cell, and dedifferentiated chondrosarcoma. Genes Chromosomes Cancer 51: 899–909 [Abstract] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430 [Europe PMC free article] [Abstract] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. 2012. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481: 380–384 [Europe PMC free article] [Abstract] [Google Scholar]
- Miyata T, Takizawa S, van Ypersele de Strihou C 2011. Hypoxia. 1. Intracellular sensors for oxygen and oxidative stress: Novel therapeutic targets. Am J Physiol Cell Physiol 300: C226–C231 [Abstract] [Google Scholar]
- Mohamedali AM, Smith AE, Gaken J, Lea NC, Mian SA, Westwood NB, Strupp C, Gattermann N, Germing U, Mufti GJ 2009. Novel TET2 mutations associated with UPD4q24 in myelodysplastic syndrome. J Clin Oncol 27: 4002–4006 [Abstract] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [Europe PMC free article] [Abstract] [Google Scholar]
- Newsholme P, Krause M 2012. Nutritional regulation of insulin secretion: Implications for diabetes. Clin Biochem Rev 33: 35–47 [Europe PMC free article] [Abstract] [Google Scholar]
- Newsholme P, Gaudel C, McClenaghan NH 2010. Nutrient regulation of insulin secretion and β-cell functional integrity. Adv Exp Med Biol 654: 91–114 [Abstract] [Google Scholar]
- Oermann EK, Wu J, Guan KL, Xiong Y 2012. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol 23: 370–380 [Europe PMC free article] [Abstract] [Google Scholar]
- Orford KW, Scadden DT 2008. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet 9: 115–128 [Abstract] [Google Scholar]
- Pansuriya TC, Kroon HM, Bovee JV 2010. Enchondromatosis: Insights on the different subtypes. Int J Clin Exp Pathol. 3: 557–569 [Europe PMC free article] [Abstract] [Google Scholar]
- Pansuriya TC, van Eijk R, d'Adamo P, van Ruler MA, Kuijjer ML, Oosting J, Cleton-Jansen AM, van Oosterwijk JG, Verbeke SL, Meijer D, et al. 2011. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet 43: 1256–1261 [Europe PMC free article] [Abstract] [Google Scholar]
- Pardanani A, Lasho TL, Finke CM, Mai M, McClure RF, Tefferi A 2010. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia 24: 1146–1151 [Abstract] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [Europe PMC free article] [Abstract] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, et al. 2010. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 28: 3636–3643 [Abstract] [Google Scholar]
- Patnaik MM, Hanson CA, Hodnefield JM, Lasho TL, Finke CM, Knudson RA, Ketterling RP, Pardanani A, Tefferi A 2012. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: A Mayo Clinic study of 277 patients. Leukemia 26: 101–105 [Abstract] [Google Scholar]
- Perez C, Martinez-Calle N, Martin-Subero JI, Segura V, Delabesse E, Fernandez-Mercado M, Garate L, Alvarez S, Rifon J, Varea S, et al. 2012. TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS ONE 7: e31605. [Europe PMC free article] [Abstract] [Google Scholar]
- Pollard PJ, Wortham NC, Tomlinson IP 2003. The TCA cycle and tumorigenesis: The examples of fumarate hydratase and succinate dehydrogenase. Ann Med 35: 632–639 [Abstract] [Google Scholar]
- Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F, Gross S, Dang L, Yen KE, Yang H, et al. 2012. Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett 3: 850–855 [Europe PMC free article] [Abstract] [Google Scholar]
- Pusch S, Sahm F, Meyer J, Mittelbronn M, Hartmann C, von Deimling A 2011. Glioma IDH1 mutation patterns off the beaten track. Neuropathol Appl Neurobiol 37: 428–430 [Abstract] [Google Scholar]
- Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S 2012. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol 13: R69. [Europe PMC free article] [Abstract] [Google Scholar]
- Raimundo N, Baysal BE, Shadel GS 2011. Revisiting the TCA cycle: Signaling to tumor formation. Trends Mol Med 17: 641–649 [Europe PMC free article] [Abstract] [Google Scholar]
- Rakheja D, Konoplev S, Medeiros LJ, Chen W 2012. IDH mutations in acute myeloid leukemia. Hum Pathol 43: 1541–1551 [Abstract] [Google Scholar]
- Reitman ZJ, Yan H 2010. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J Natl Cancer Inst 102: 932–941 [Europe PMC free article] [Abstract] [Google Scholar]
- Robinson A, Keely S, Karhausen J, Gerich ME, Furuta GT, Colgan SP 2008. Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology 134: 145–155 [Europe PMC free article] [Abstract] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. 2013. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 10.1126/science.1236062 [Europe PMC free article] [Abstract] [Google Scholar]
- Rzem R, Vincent MF, Van Schaftingen E, Veiga-da-Cunha M 2007. L-2-hydroxyglutaric aciduria, a defect of metabolite repair. J Inherit Metab Dis 30: 681–689 [Abstract] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, Cairns RA, McCracken S, Wakeham A, Haight J, et al. 2012a. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 26: 2038–2049 [Europe PMC free article] [Abstract] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. 2012b. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488: 656–659 [Europe PMC free article] [Abstract] [Google Scholar]
- Sazanov LA, Jackson JB 1994. Proton-translocating transhydrogenase and NAD- and NADP-linked isocitrate dehydrogenases operate in a substrate cycle which contributes to fine regulation of the tricarboxylic acid cycle activity in mitochondria. FEBS Lett 344: 109–116 [Abstract] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. 2012. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151: 1185–1199 [Europe PMC free article] [Abstract] [Google Scholar]
- Seita J, Weissman IL 2010. Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2: 640–653 [Europe PMC free article] [Abstract] [Google Scholar]
- Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, et al. 2010. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 70: 8981–8987 [Europe PMC free article] [Abstract] [Google Scholar]
- Semenza GL 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148: 399–408 [Europe PMC free article] [Abstract] [Google Scholar]
- Sen Banerjee S, Thirunavukkarasu M, Tipu Rishi M, Sanchez JA, Maulik N, Maulik G 2012. HIF-prolyl hydroxylases and cardiovascular diseases. Toxicol Mech Methods 22: 347–358 [Abstract] [Google Scholar]
- Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, Lennes IT, Digumarthy S, Waltman BA, Bast E, et al. 2011. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 22: 2616–2624 [Europe PMC free article] [Abstract] [Google Scholar]
- Shilatifard A 2006. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [Abstract] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [Abstract] [Google Scholar]
- Sola S, Xavier JM, Santos DM, Aranha MM, Morgado AL, Jepsen K, Rodrigues CM 2011. p53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation. PLoS ONE 6: e18421. [Europe PMC free article] [Abstract] [Google Scholar]
- SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, LanXiao W, Fei Y 2012. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 103: 269–273 [Abstract] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. 2013. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152: 1146–1159 [Abstract] [Google Scholar]
- Steenweg ME, Jakobs C, Errami A, van Dooren SJ, Adeva Bartolome MT, Aerssens P, Augoustides-Savvapoulou P, Baric I, Baumann M, Bonafe L, et al. 2010. An overview of L-2-hydroxyglutarate dehydrogenase gene (L2HGDH) variants: A genotype-phenotype study. Hum Mutat 31: 380–390 [Abstract] [Google Scholar]
- Struys EA, Salomons GS, Achouri Y, Van Schaftingen E, Grosso S, Craigen WJ, Verhoeven NM, Jakobs C 2005a. Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am J Hum Genet 76: 358–360 [Europe PMC free article] [Abstract] [Google Scholar]
- Struys EA, Verhoeven NM, Ten Brink HJ, Wickenhagen WV, Gibson KM, Jakobs C 2005b. Kinetic characterization of human hydroxyacid–oxoacid transhydrogenase: Relevance to D-2-hydroxyglutaric and γ-hydroxybutyric acidurias. J Inherit Metab Dis 28: 921–930 [Abstract] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 2011. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci 14: 1607–1616 [Europe PMC free article] [Abstract] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [Europe PMC free article] [Abstract] [Google Scholar]
- Tefferi A, Pardanani A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Gangat N, Finke CM, Schwager S, Mullally A, et al. 2009. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23: 905–911 [Europe PMC free article] [Abstract] [Google Scholar]
- Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, Caramazza D, Pieri L, Finke CM, Kilpivaara O, Wadleigh M, et al. 2010. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 24: 1302–1309 [Europe PMC free article] [Abstract] [Google Scholar]
- Tefferi A, Jimma T, Sulai NH, Lasho TL, Finke CM, Knudson RA, McClure RF, Pardanani A 2012. IDH mutations in primary myelofibrosis predict leukemic transformation and shortened survival: Clinical evidence for leukemogenic collaboration with JAK2V617F. Leukemia 26: 475–480 [Europe PMC free article] [Abstract] [Google Scholar]
- Teodoro JG, Parker AE, Zhu X, Green MR 2006. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science 313: 968–971 [Abstract] [Google Scholar]
- Terashima M, Ishimura A, Yoshida M, Suzuki Y, Sugano S, Suzuki T 2010. The tumor suppressor Rb and its related Rbl2 genes are regulated by Utx histone demethylase. Biochem Biophys Res Commun 399: 238–244 [Abstract] [Google Scholar]
- Thol F, Weissinger EM, Krauter J, Wagner K, Damm F, Wichmann M, Gohring G, Schumann C, Bug G, Ottmann O, et al. 2010. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica 95: 1668–1674 [Europe PMC free article] [Abstract] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483 [Europe PMC free article] [Abstract] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [Europe PMC free article] [Abstract] [Google Scholar]
- Varier RA, Timmers HT 2011. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta 1815: 75–89 [Abstract] [Google Scholar]
- Venneti S, Felicella MM, Coyne T, Phillips JJ, Gorovets D, Huse JT, Kofler J, Lu C, Tihan T, Sullivan LM, et al. 2013. Histone 3 Lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J Neuropathol Exp Neurol 72: 298–306 [Europe PMC free article] [Abstract] [Google Scholar]
- Wallace DC 2012. Mitochondria and cancer. Nat Rev Cancer 12: 685–698 [Europe PMC free article] [Abstract] [Google Scholar]
- Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, Shi Y, Chang HY 2010. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev 24: 327–332 [Europe PMC free article] [Abstract] [Google Scholar]
- Wang F, Travins J, de la Barre B, Lacronique-Penard V, Schalm S, Hansen E, Straley KS, Kernytsky A, Liu W, Gliser C, et al. 2013. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 10.1126/science.1234769 [Abstract] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [Abstract] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [Europe PMC free article] [Abstract] [Google Scholar]
- Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB 2012. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene 31: 2491–2498 [Europe PMC free article] [Abstract] [Google Scholar]
- Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, Thompson CB 2013. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 288: 3804–3815 [Europe PMC free article] [Abstract] [Google Scholar]
- Wickenhagen WV, Salomons GS, Gibson KM, Jakobs C, Struys EA 2009. Measurement of D:-2-hydroxyglutarate dehydrogenase activity in cell homogenates derived from D:-2-hydroxyglutaric aciduria patients. J Inherit Metab Dis 32: 264–268 [Abstract] [Google Scholar]
- Willers IM, Cuezva JM 2011. Post-transcriptional regulation of the mitochondrial H+-ATP synthase: A key regulator of the metabolic phenotype in cancer. Biochim Biophys Acta 1807: 543–551 [Abstract] [Google Scholar]
- Williams SC, Karajannis MA, Chiriboga L, Golfinos JG, von Deimling A, Zagzag D 2011. R132H-mutation of isocitrate dehydrogenase-1 is not sufficient for HIF-1α upregulation in adult glioma. Acta Neuropathol 121: 279–281 [Europe PMC free article] [Abstract] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB 2011. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci 108: 19611–19616 [Europe PMC free article] [Abstract] [Google Scholar]
- Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J 2004. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem 279: 33946–33957 [Abstract] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [Europe PMC free article] [Abstract] [Google Scholar]
- Yamazaki J, Taby R, Vasanthakumar A, Macrae T, Ostler KR, Shen L, Kantarjian HM, Estecio MR, Jelinek J, Godley LA, et al. 2012. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics 7: 201–207 [Europe PMC free article] [Abstract] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [Europe PMC free article] [Abstract] [Google Scholar]
- Yen KE, Bittinger MA, Su SM, Fantin VR 2010. Cancer-associated IDH mutations: Biomarker and therapeutic opportunities. Oncogene 29: 6409–6417 [Abstract] [Google Scholar]
- Zhang J, Chen GQ 2009. Hypoxia-HIF-1α-C/EBPα/Runx1 signaling in leukemic cell differentiation. Pathophysiology 16: 297–303 [Abstract] [Google Scholar]
- Zhang J, Song LP, Huang Y, Zhao Q, Zhao KW, Chen GQ 2008. Accumulation of hypoxia-inducible factor-1 α protein and its role in the differentiation of myeloid leukemic cells induced by all-trans retinoic acid. Haematologica 93: 1480–1487 [Abstract] [Google Scholar]
- Zhang Y, Wei H, Tang K, Lin D, Zhang C, Mi Y, Wang L, Wang C, Wang M, Wang J 2012. Mutation analysis of isocitrate dehydrogenase in acute lymphoblastic leukemia. Genet Test Mol Biomarkers. 16: 991–995 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. 2009. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324: 261–265 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou KG, Jiang LJ, Shang Z, Wang J, Huang L, Zhou JF 2012. Potential application of IDH1 and IDH2 mutations as prognostic indicators in non-promyelocytic acute myeloid leukemia: A meta-analysis. Leuk Lymphoma 53: 2423–2429 [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.217406.113
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/27/8/836.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gad.217406.113
Article citations
IDH2 mutation accelerates TPO-induced myelofibrosis with enhanced S100a8/a9 and NFκB signaling in vivo.
EJHaem, 5(4):738-748, 28 Jul 2024
Cited by: 0 articles | PMID: 39157630 | PMCID: PMC11327712
Targeting Isocitrate Dehydrogenase (IDH) in Solid Tumors: Current Evidence and Future Perspectives.
Cancers (Basel), 16(15):2752, 02 Aug 2024
Cited by: 0 articles | PMID: 39123479 | PMCID: PMC11311780
Review Free full text in Europe PMC
DNA damage response in brain tumors: A Society for Neuro-Oncology consensus review on mechanisms and translational efforts in neuro-oncology.
Neuro Oncol, 26(8):1367-1387, 01 Aug 2024
Cited by: 1 article | PMID: 38770568
Review
Treatment of IDH-mutant glioma in the INDIGO era.
NPJ Precis Oncol, 8(1):149, 19 Jul 2024
Cited by: 0 articles | PMID: 39025958 | PMCID: PMC11258219
Review Free full text in Europe PMC
GLS and GLS2 Glutaminase Isoenzymes in the Antioxidant System of Cancer Cells.
Antioxidants (Basel), 13(6):745, 20 Jun 2024
Cited by: 0 articles | PMID: 38929183
Review
Go to all (382) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Isocitrate Dehydrogenase Mutations in Cancer: Mechanisms of Transformation and Metabolic Liability.
Cold Spring Harb Perspect Med, 14(5):a041537, 02 May 2024
Cited by: 2 articles | PMID: 38191174
Review
(R)-2-Hydroxyglutarate Inhibits KDM5 Histone Lysine Demethylases to Drive Transformation in IDH-Mutant Cancers.
Cancer Discov, 13(6):1478-1497, 01 Jun 2023
Cited by: 10 articles | PMID: 36847506 | PMCID: PMC10238656
(R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible.
Science, 339(6127):1621-1625, 07 Feb 2013
Cited by: 463 articles | PMID: 23393090 | PMCID: PMC3836459
The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep, 12(5):463-469, 01 Apr 2011
Cited by: 636 articles | PMID: 21460794 | PMCID: PMC3090014
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (3)
Grant ID: R01 CA068490
Grant ID: F31 CA260794
Grant ID: R01 CA076120





