Abstract
Free full text

Mechanism of action of chromogranin A on catecholamine release: molecular modeling of the catestatin region reveals a β-strand/loop/β-strand structure secured by hydrophobic interactions and predictive of activity
Abstract
A novel fragment of chromogranin A, known as ‘catestatin’ (bovine chromogranin A344–364), inhibits catecholamine release from chromaffin cells and noradrenergic neurons by acting as a non-competitive nicotinic cholinergic antagonist, and may therefore constitute an endogenous autocrine feedback regulator of sympathoadrenal activity. To characterize how this activity depends on the peptide’s structure, we searched for common 3-dimensional motifs for this primary structure or its homologs. Catestatin’s primary structure bore significant (29–35.5% identity, general alignment score 44–57) sequence homology to fragment sequences within three homologs of known 3-dimensional structures, based on solved X-ray crystals: 8FAB, 1PKM, and 2IG2. Each of these sequences exists in nature as a β-strand/loop/β-strand structure, stabilized by hydrophobic interactions between the β-strands. The catestatin structure was stable during molecular dynamics simulations. The catestatin loop contains three Arg residues, whose electropositive side chains form the terminus of the structure, and give rise to substantial uncompensated charge asymmetry in the molecule. A hydrophobic moment plot revealed that catestatin is the only segment of chromogranin A predicted to contain amphiphilic β-strand. Circular dichroism in the far ultraviolet showed substantial (63%) β-sheet structure, especially in a hydrophobic environment. Alanine-substitution mutants of catestatin established a crucial role for the three central arginine residues in the loop (Arg351, Arg353, and Arg358), though not for two arginine residues in the strand region toward the amino-terminus. [125I]Catestatin bound to Torpedo membranes at a site other than the nicotinic agonist binding site. When the catestatin structure was ‘docked’ with the extracellular domain of the Torpedo nicotinic cholinergic receptor, it interacted principally with the β and δ subunits, in a relatively hydrophobic region of the cation pore extracellular orifice, and the complex of ligand and receptor largely occluded the cation pore, providing a structural basis for the non-competitive nicotinic cholinergic antagonist properties of the peptide. We conclude that a homology model of catestatin correctly predicts actual features of the peptide, both physical and biological. The model suggests particular spatial and charge features of the peptide which may serve as starting points in the development of non-peptide mimetics of this endogenous nicotinic cholinergic antagonist.
1. Introduction
Chromogratlin A, a 48-kilodalton acidic, soluble protein [36], is found in the core of catecholamine storage vesicles of chromaffin cells and sympathetic nerves [3,29], from which it is co-released, by exocytosis, with epinephrine and norepinephrine [30]. Chromogranin A is a prohormone, subject to proteolytic cleavage during and after secretion [2,10], yielding several biologically active peptides, including the mid-molecule fragment pancreastatin (porcine chromogranin A240–288), which inhibits glucose-stimulated insulin release from pancreatic islet β-cells [32] and parathyroid hormone release from chief cells [11], and the amino-terminal fragment vasostatin (human chromogranin A1–76; [1]), which relaxes vascular smooth muscle [1] and inhibits parathyroid hormone release [7]).
The recently described chromogranin A mid-molecule proteolytic fragment catestatin (bovine chromogranin A344–364), formed within chromaffin granules, is a potent inhibitor of catecholamine release [20], acting as a non-competitive antagonist at the nicotinic cholinergic receptor, the physiologic trigger for catecholamine release from chromaffin cells and sympathetic axons.
Little is known about the 3-dimensional structure of the 431 amino acid full-length chromogranin A molecule. Although chromogranin A is the best-characterized member of the chromogranin/secretogranin protein family, it seems to have an overall extended structure [23,28], and no proteins with known structure have a high level of homology to chromogranin A as a whole.
To better understand the basis of catestatin’s catecholamine release-inhibitory effect, we investigated whether its primary structure was predictive of particular 3-dimensional conformations. We found several primary structural homologs of catestatin whose 3-dimensional structure was a consistent β-sheet/loop/β-sheet motif. Such a model for catestatin correctly predicted several of its structural and pharmacological features.
2. Materials and methods
2.1. Molecular modeling and computational methods
2.1.1. Homology modeling
To perform sequence alignments, we used the programs BLAST (for preliminary screening) and FASTA [24] for the following pairwise alignments. The program Homology (Molecular Simulations Inc., San Diego, CA; 1997) was used to build a catestatin model based on template (homolog) structures found using multi-template correlations. After homology modeling, the structure was energy-minimized for 2000 iterations in a water shell (thickness, 10 Å) using the steepest-descent method. Then, 300 ps molecular dynamics simulations in the same water shell were performed on the peptide structure, using the following strategy at T = 300 K. Initially the peptide was fixed and a new water shell was introduced. Molecular dynamics (MD) for 10 ps followed by a 1000 iteration energy minimization were then performed on water alone to ensure the homogeneous distribution of water molecules. After that, MD of the entire system were studied. Snap-shots of trajectories were saved every 0.5 ps.
2.1.2. Electrostatic energy calculations
Models then underwent calculation of total electrostatic energy, including Coulombic and solvent energy components. The electrostatic energies were calculated by solving the linearized Poisson—Boltzmann equation using the program DelPhi 2.5 [12], as distributed by Molecular Simulations Inc. The dielectric constant of the solvent was set to 80, and that of the protein interior was set to 4; the ionic strength was equivalent to 0.145 M sodium chloride. The Poisson—Boltzmann equation was solved numerically on a 0.9 Å grid. The ionizable residues Glu, Arg, Lys, and Asp were assigned full charge, as expected at pH 7. The Amber partial charge distribution [35] was used to place partial charges on all atoms in the system.
2.1.3. Hydrophobic moment plot
A hydrophobic moment plot (MacVector; Oxford Molecular, Oxford, UK) searched chromogranin A’s entire primary structure for regions suggesting amphiphilic β-sheet, by testing for alternating polar and apolar amino acid side chains at 160° to 180° intervals [8,9]. At each residue, results were averaged over an 11-amino-acid window.
2.1.4. Docking
Since catestatin is a non-competitive nicotinic cholinergic antagonist, we evaluated its interaction with its likely target on the nicotinic receptor: the extracellular domain in the region of the cation pore. We have recently homology-modeled the structure of the extracellular domain of the Torpedo nicotinic receptor [34]. Calculations were done using the Grid-Grid docking program in Insight-II (Molecular Simulations Inc.). The grid unit size was 1 Å on a 128 × 128 × 128 matrix for the pentameric nicotinic cholinergic receptor as the stationary target, and catestatin as the movable ligand. The program used van der Waals and electrostatic energies as parameters to minimize docking energy.
2.2. Experimental procedures
2.2.1. Cell culture and catecholamine secretion
PC12 rat pheochromocytoma cells [13] were cultured (at passage 8–20) as previously described [20]. PC12 cells’ chromaffin granules were pre-loaded with [3H]-L-norepinephrine, and catecholamine secretion was monitored over a 30-min time course, in the presence or absence of secretagogues (see below), by efflux of [3H]-L-norepinephrine. Tritium counts in release medium and cell lysate were determined by liquid scintillation, and norepinephrine secretion was expressed as % of cell total norepinephrine released.
2.2.2. Pharmacology and synthetic peptides
The nicotinic cholinergic secretory pathway was triggered by nicotine (Sigma, St. Louis, MO), 1–1000 μM; peak secretory responses typically occur at 60–100 μM nicotine. Synthetic peptides (20–100 μmol scale) were prepared by the solid phase method, using f-moc or t-boc protection chemistry. Purification was by C-18 RP-HPLC. Authenticity of the resulting peptides was confirmed by mass spectrometry, using either MALDI (matrix-assisted laser-desorption ionization) or electrospray ionization. Some peptides were prepared by small (1 μmol) scale simultaneous synthesis on pins (PepSet; Chiron, Melbourne, Australia), then cleaved and solvent-washed prior to use.
2.2.3. Circular dichroism
Circular dichroism [37] tested peptide secondary structure by circularly polarized light rotation in the far ultraviolet (195–270 nm), on a Cary instrument, at 0.54 mg/ml peptide concentration. Percent secondary structure (α-helix, β-sheet, or random coil) was estimated as described [4].
2.2.4. Equilibrium radioligand binding of [125I]catestatin to Torpedo nicotinic receptor-containing membranes
[Cys0]-human catestatin (C0-SSMKLSFRARAYGFRGPGPQL) was iodinated on its endogenous tyrosine using the chloramine T method [31] to a specific activity of ~300 Ci/mmol. The [125I]catestatin was purified on a Sep-Pak C-18 silica column with a CH3CN/H2O gradient. The solvent system consisted of 0.1% trifluoroacetic acid in water (solvent A) and 0.1% trifluoroacetic acid in 80% CH3CN (solvent B).
Nicotinic cholinergic receptors were partially purified from Torpedo californica electroplax membranes [18]; the final preparation contained nicotinic receptors as ~20% of total protein, with 1.32 nmol acetylcholine binding sites/mg protein, at 8.4 μM concentration.
Receptor-enriched membranes (10 nM receptor final concentration) were incubated at room temperature for 90 min in 600 μl of assay buffer (10 mM sodium phosphate pH 7.4, 5 mM EDTA, 0.1% Triton X-100, 0.03 mg/ml bovine albumin, 3 mM NaN3, 100 μM phenylmethyl-sulfonyl fluoride) containing [125I]catestatin (120 nCi/tube), in the presence or the absence of unlabelled catestatin or the nicotinic agonist carbachol. The incubation was terminated by rapid filtration under vacuum through Whitman DE-81 cation exchange filter paper previously equilibrated with wash buffer (10 mM sodium phosphate buffer pH 7.4, 0.1% Triton X-100, 0.03 mg/ml bovine albumin). Filters were then washed twice with 3 ml of ice-cold wash buffer, and counted in an LKR-Wallac Ria-gamma 1274 counter.
2.3. Statistics
Results are shown as mean values ± 1 standard error. Significance was inferred at P < 0.05. Peptide IC50 values for nicotinic cholinergic inhibition were determined by interpolation (Kaleidagraph) on dose-response curves at 0.1, 1, and 10 μM peptide.
3. Results and discussion
3.1. Sequence alignments and homology-modeled tertiary structure
Fig. 1 shows the multiple sequence alignments built on the basis of pairwise alignments of the 8FAB (a pepsindigest Fab fragment of a human myeloma IgG1 immunoglobulin [26]) 1PKM (cat muscle pyruvate kinase M1 [22]) and 2IG2 (human monoclonal IgGl immunoglobulin Kol [21]) primary structures with the catestatin region of chromogranin A. One can see that the overall sequence identity of each of these homologs to catestatin is no less than 29% and, in the case of 8FAB, 35.5%. Sequence identity between template proteins in these regions is similarly congruent, with a high degree of 3-dimensional structural similarity (Fig. 2): two anti-parallel β-strands about 10 Å from each other, linked by a loop. This structure is similar to a motif in neuraminidase [5] created from four antiparallel β-strands joined by hairpin loops. Such antiparallel motifs frequently contain hydrophobic residues within the hairpin regions [5], and the hydrophobic residues’ side chains may stabilize the overall structure of the motif by ‘hydrophobic collapse’. The catestatin region of chromogranin A is thus predicted to have such an anti-parallel structure.

See lengthwise print.
Alignment of the catestatin region of chromogranin A with and homologous regions of proteins with partial amino acid sequence identity and known three-dimensional structure, as determined by xray crystallography: 8FAB (a human myeloma immunogobulin), 1PKM (cat muscle pyruvate kinase), and 2IG2 (a monoclonal human immunogobulin). Sequence homologies were detected by the algorithms BLAST and FASTA, while three-dimensional structures were found in the Protein Data Bank (PDB). Homology modeling was performed using the program HOMOLOGY (Molecular Simulations, Inc., San Diego, CA). Columns in bold contain residues found not only in the majority of catestatin regions but also in at least one of the homologous proteins, in the FASTA alignment, the gap penalty was -12/-2. Human chromogranin A sequence (1): Konecki, D.S., U.M. Benedum, H.H. Gerdes, and W.B. Huttner. 1987. The primary structure of human chromogranin A and pancreastatin. J. Biol. Chem. 262:17026-17030. Human chromogranin A sequence (2): Helman, L.J., T.G. Ahn, M.A. Levine, A. Allison, P.S. Cohen, M.J. Cooper, D.V. Cohn, M.A. Israel. 1988. Molecular cloning and primary structure of human chromogranin A (secretory protein I) cDNA. J. Biol. Chem. 263:11559-11563. For other chromogranin A sequences in the catestatin region, see Mahata et al, 1997.
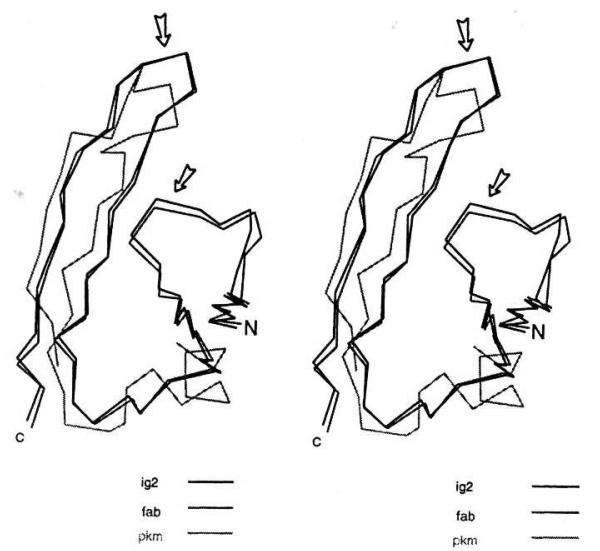
Three-dimensional (stereo) view of the tertiary structures of the homologous regions of proteins 8FAB, 1PKM and 2IG2, in the region of partial sequence identity to catestatin (bovine chromogranin A344–364). N, amino-terminus; C, carboxy-terminus. Arrows denote the beginning and end of the β-strand/loop/β-strand regions of homology with catestatin.
The portion of chromogranin A corresponding to the biologically active catestatin (bovine chromogranin A344–364) is thus framed by a loop joining two anti-parallel β-sheets, as shown in Fig. 3a, to display the orientation of the strands about the connecting loop in the modeled region, bovine chromogranin A342–370 (Pro342 → Pro370; PDRSMRLSFRARGYGFRGPGLQLRRGWRP). Two predictions might be considered here. Firstly, before catestatin is cleaved from chromogranin A, its constraint within the overall chromogranin A molecule may prevent its biological activity. Indeed, both we [20] and Simon et al. [27] have found that the nicotinic cholinergic antagonist (i.e. catestatin) activity in chromogranin A is liberated by proteolysis. Secondly, if the β-strands flanking the catestatin region (Fig. 2) are removed, the force constraining the loop from unfolding consists of hydrophobic interactions especially among apolar side chains of Leu348, Phe350, Tyr355, Phe360, Pro370, Leu362, and Leu364; the inter-carbon distances between these side chains are sufficiently proximate ( ≤4.5 Å) to form a hydrophobic ‘node’ (Fig. 3a). Residue pairs whose side chains may so associate are (in order from the loop tip): Tyr355 with Pro360; Tyr355 with Leu362; Pro360 with Leu362; Phe357 with Leu362; Phe350 with Leu362; Phe350 with Leu364; Leu364 with Trp368; and Leu348 with Leu364.
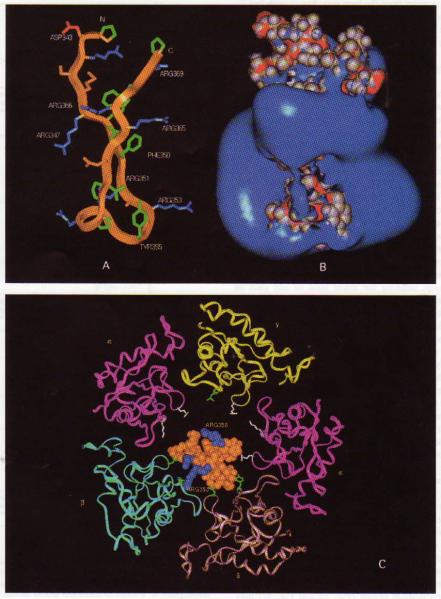
Models of catestatin structure, (a) Structure of catestatin: results of homology modeling. The region shown is bovine chromogranin A342–370, PDRSMRLSFRARGYGFRGPGLQLRRGWRP, thus extending from Pro342 (at its amino-terminus) to Pro370 (at its carboxy-terminus). N, amino-terminus; C, carboxy-terminus. Red, electronegative side chain. Blue, electropositive side chain. Green, hydrophobic side chain. Brown, other hydrophilic side chain. (b) Isopotential surfaces of catestatin (same primary structure, length, and orientation as in panel 3a), as determined by electrostatic energy calculations. Potentials: blue surface, + 2.5 kT/e; red surface, − 2.5 kT/e. Gray spheres: CPK models of atoms. (c) ‘Docking’ of the homology-modeled structure of bovine catestatin (same primary structure and length as in panel 3a) with the extracellular domain of the Torpedo nicotinic cholinergic receptor [34], in the region of the cation pore vestibule. Extracellular portions of the α, β, γ, and δ subunits of the Torpedo nicotinic receptor are shown. Blue CPK atoms in catestatin: basic residues (Arg353 and Arg358). White stick side chains (basic residues) in nicotinic receptor subunits: Arg87 and Lys104 in α subunit; Arg87 in γ subunit. Green stick side chains (hydrophobic residues) in nicotinic receptor subunits: Leu87 and Leu104 in β subunit; Leu104 in γ subunit; Leu89 and Tyr106 in δ subunit.
3.2. Stability of the structure
To determine whether this hydrophobic core provides sufficient stability to maintain the β-strand/loop/β-strand conformation, we conducted molecular dynamics simulations, giving the molecule an opportunity to reconform in a water environment under physiological conditions (T = 300 K). We simulated two length variations of the catestatin region: a longer 29-amino-acid peptide (bovine chromogranin A342–370; PDRSMRLSFRARGYGFRGPGLQLRRGWRP), and a shorter 21-amino-acid peptide (bovine chromogranin A344–364; RSMRLSFRARGYGFRGPGLQL), chosen because of extensive knowledge of its biological activity [20]. Fig. 4a through c reports inter-residue interatomic distances during the simulations. The distance between Cα atoms of Arg358 and Arg353 represents the largest distance between backbone atoms in the tip of the loop, defining the lower limit of any cleft or cavity which might be accessed by catestatin; this distance has a stable value of ~ 12 Å for both the longer and shorter peptides (Fig. 4a). The distance between Cα atoms of Arg344 and Pro360 defines the long axis of the catestatin region; this distance is also stable at ~27–30 Å during molecular dynamics (Fig. 4b). The distance between the terminal side chain carbon (Cζ) atoms of Arg358 and Arg353 represents to some extent the maximum width of this region; this distance (Fig. 4c) changes quite sharply during the molecular dynamics run, from an initial distance of ~ 11 Å to ~ 18 Å for the longer peptide and ~ 24 Å for the shorter peptide. These changes may result from electrostatic repulsion between the side chains of the Arg358 and Arg353. The initial ~11 Å Cζ side chain distance after molecular mechanics energy minimization of the homology-derived catestatin structure (Fig. 4c) apparently resulted from a local minimum of potential energy, a circumstance which can be overcome by molecular dynamics. Molecular dynamics of catestatin in water also showed that the region from Leu348 Cα to Leu364 Cα was stable throughout a 300-ps molecular dynamics simulation at 300 K (data not shown).
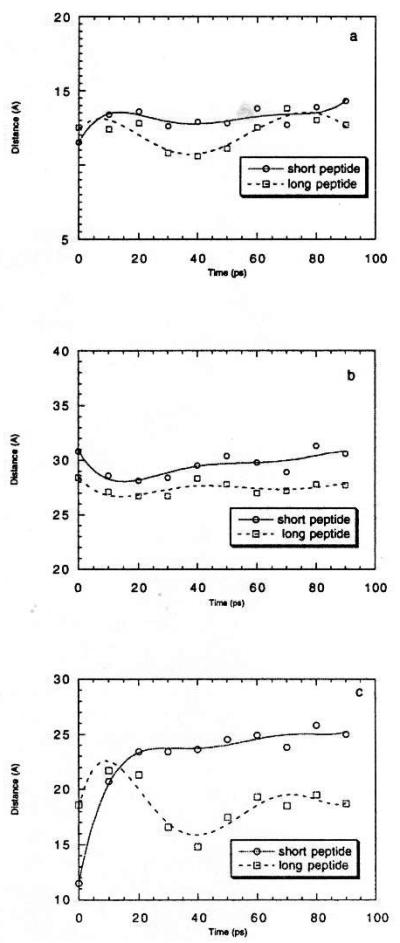
Stability of the catestatin 3-dimensional structure during sustained molecular dynamics simulations in water. Simulations (100 ps, T = 300 K) were conducted for two length versions of the catestatin region of chromogranin A: a longer 29-amino-acid peptide (bovine chromogranin A342–370; PDRSMRLSFRARGYGFRGPGLQLRRGWRP), and a shorter 21-amino-acid peptide (bovine chromogranin A344–364; RSMRLSFRARGYGFRGPGLQL). Distances between specified atoms of catestatin are shown as a function of time: (a) Cα (α-carbon) atoms of Arg358 and Arg353; this distance defines the longest backbone Cα—Cα distance in the tip of the loop, and hence the lower limit of any cleft or cavity which might be accessed by catestatin. (b) Cα (α-carbon) atoms of Arg344 and Pro360; this distance defines the long axis of catestatin. (c) Cζ (terminal carbon in the side chain) atoms of Arg358 and Arg353; this distance defines the maximum strand-to-strand width of catestatin.
A Cζ inter-carbon distance of ~24–25 Å as a maximum width of the catestatin region (Fig. 3a and Fig. 4c) may have implications for the peptide’s mechanism of action as a non-competitive nicotinic cholinergic antagonist [20], since this value corresponds to a value of ~ 24 Å for the width of the cation pore vestibule of the Torpedo nicotinic acetylcholine receptor [34], in turn suggesting that catestatin might completely occlude the pore vestibule. By contrast, the ionic radius of Na+, the primary cation moving through the nicotinic cation pore, is only 0.95 Å (plus its hydralion shell) [17].
3.3. Prediction of secondary structure: hydrophobia moment plot
The entire 431 amino acid primary structure of the mature bovine chromogranin A protein was subjected to a hydrophobic moment plot [8,9], looking for regions suggestive of amphiphilic β-pleated sheet. Only chromogranin A345–356 (SMRLSFRARGYG) which is towards the N-terminus of the catestatin region (bovine chromogranin A344–364) showed a region suggesting amphiphilic β-sheet, bounded by residues Ser345 and Gly356, peaking at and residue Arg351 (Fig. 5). The cationic amphiphilic character of this region (alternating cationic and hydrophobic residues) is clear upon visual inspection: SMRLSFRARGYG. By contrast, there were no regions of predicted amphiphilic α-helix.
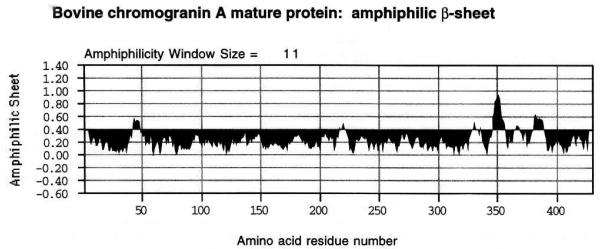
Hydrophobic moment plot of bovine chromogranin A (431 amino acid mature protein, minus signal peptide), to detect regions of likely amphiphilic β-sheet (alternating polar and apolar residues). The catestatin region is bovine chromogranin A344–364 (RSMRLSFRARGYGFRGPGLQL). The amphiphilic peak is bounded by residues Ser345 and Gly356, and is maximally amphiphilic at residue Arg351. At each residue, results were averaged over an 11-amino-acid window.
3.4. Experimental evidence for global secondary structure: circular dichroism with solvent titration
When synthetic bovine chromogranin A344–364 was studied for rotation of circularly polarized light in the far ultraviolet, the circular dichroism spectrum in water showed a deep minimum at 200 nm, which did not suggest substantial secondary structure for the peptide (Fig. 6). In organic solvent (100% trifluoroethanol), the minimum shifted to 206 nm, with an inflection at 221 nm. When even as little as 25% organic solvent (trifluoroethanol) was included in the medium, the calculated global secondary structure assumed substantial (63%) β-sheet character, and the amount of calculated β-sheet did not increase with further addition of organic solvent, even up to 100% trifluoroethanol. Thus, the peptide had a propensity to assume β-sheet conformation with only modest reduction in solvent dielectric constant.
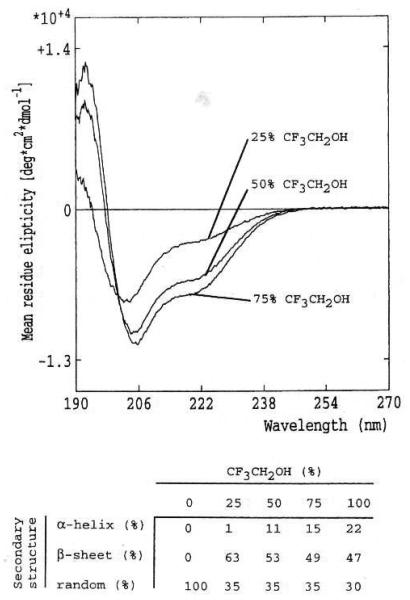
Circular dichroism spectra of catestatin (bovine chromogranin A334–364; RSMRLSFRARGYGFRGPGLQL; 0.54 mg/ml), as a function of dielectric constant of the solvent, as varied by increasing % composition with an apolar solvent (trifluoroethanol).
By contrast, an α-helical conformation could be provoked only by inclusion of very high concentrations of organic solvent (22% α-helix at 100% trifluoroethanol).
3.5. Predictions of the catestatin model: functional results of point mutagenesis
Catestatin (bovine chromogranin A344–364) inhibits nicotinic cholinergic-stimulated PC12 cell catecholamine release, with IC50 ~ 200–500 nM peptide [20]. Selective alanine substitution (‘alanine scanning’) mutagenesis was accomplished by replacement of individual amino acid residues by alanine (methylglycine), the smallest amino acid which retains chirality (in this case, [L]-alanine). Alanine scanning revealed important functional roles for the three central arginine residues in the tip or the catestatin loop (Fig. 3a): Arg351, Arg353, or Arg358. Replacement of any of these three central arginine residues by alanine (to R351A, R353A, or R358A) resulted in substantial increases in catestatm’s IC50 values (that is, decreases in its molar potency) (Fig. 7). By contrast, alanine replacement of the arginine residues in the aminoterminal strand of catestatin (Fig. 3a), Arg344 (in R344A) or Arg387 (in R347A), had little effect on catestatin’s IC50; nor did replacement of Pro360 by alanine (in P360A) (Fig. 7). Thus, particular amino acids contribute specifically to catestatin’s function, and residues in the loop tip (Fig. 3a) are especially crucial. Lack of effect of the P360A mutation suggests that Pro360 is not absolutely required in formation of the turn in the catestatin loop (Fig. 7).
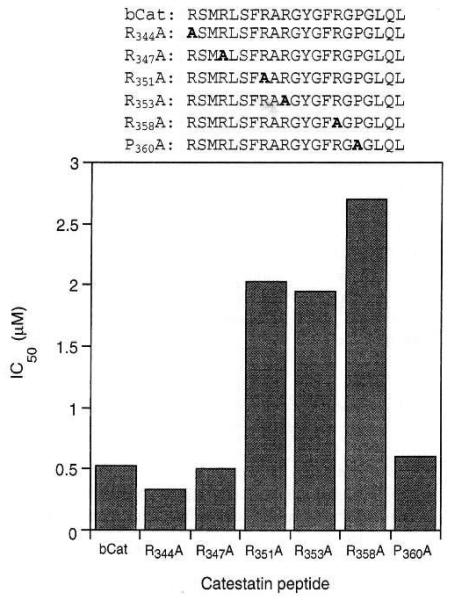
Point mutants of catestatin: functional results of selective alanine substitutions (alanine scan) on nicotinic cholinergic-stimulated catecholamine secretion from PC12 pheochromocytoma cells. IC50, the concentration of peptide required to inhibit 60 μM nicotine-stimulated catecholamine secretion by 50% (derived from dose-response curves determined at 0.1, 1, and 10 μM peptide). Mutant alanine residues are in bold.
3.6. Binding of catestatin to Torpedo membranes
[125I]Catestatin bound to Torpedo membranes, and an excess (50 μM) of unlabeled catestatin displaced the tracer from the membranes, defining specific binding (Fig. 8). By contrast, the nicotinic agonist carbachol, even at 100 μM, did not displace [125I]catestatin. Thus, catestatin seems to bind to a site other than the agonist binding site. For such non-competitive nicotinic cholinergic antagonists [20], that binding site is typically in the cation channel vestibule or pore [6].
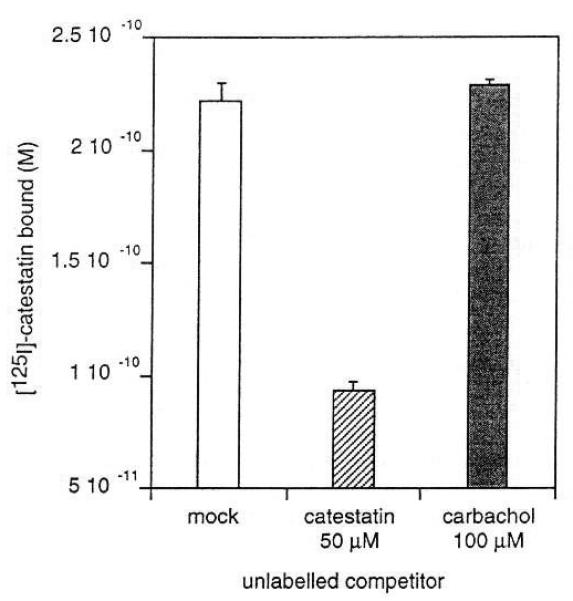
Effect of unlabeled catestatin itself or the nicotinic cholinergic agonist carbachol on equilibrium binding of [125I]catestatin to nicotinic receptor-rich Torpedo membranes. Torpedo californica membranes were incubated for 90 min with [125I]catestatin in the presence of vehicle (mock), unlabeled catestatin (50 μM) or carbachol (carbamylcholine chloride, 100 μM). Values are given as the means of triplicate determinations±S.E. of one representative experiment.
3.7. Electrostatic potential
Electrostatic field calculations (Fig. 3b) show that a strong maximum of positive potential is located at the tip of the catestatin loop between two β-strands, largely the result of three arginines: Arg351, Arg353 and Arg358. This positive potential suggests a directional orientation of the peptide’s binding to the highly electronegative extracellular domain of the acetylcholine nicotinic receptor [34].
3.8. Docking of catestatin with the extracellular domain of the nicotinic cholinergic receptor
The directional positive potential of catestatin (Fig. 3b) also suggested that catestatin might interact with the electronegative extracellular domain or the nicotinic receptor [34].
When catestatin was ‘docked’ to the extracellular face of the Torpedo nicotinic receptor, the energy of interaction was minimized as catestatin approached the extracellular orifice of the cation pore. In the final state, catestatin most closely approached the β and δ subunits of the receptor, with deep minima of both electrostatic energy and van der Waals energy (Fig. 3c). In this configuration of minimal energy, it can be seen that catestatin almost completely occludes the ~24–25 Å extracellular orifice of the nicotinic cation pore (Fig. 3c).
The final docked position of catestatin on the nicotinic receptor may be influenced by two major charge factors on the receptor.
Overall charge: the very negative electrostatic energy potential of the extracellular domain of the Torpedo nicotinic pentamer, which is unevenly distributed in its cation pore [34], attracts the electropositive (Fig. 3b) catestatin.
Specific charged sites: in the Torpedo nicotinic α subunit, cationic side chains (cationic side chains shown as white sticks in Fig. 3c) of α subunit residues Arg87 and Lys104 and γ subunit residue Arg87 are directed towards the cation pore [34]. By contrast, in the nicotinic β and δ subunits, leucine residues are found at positions equivalent to those α or γ subunit basic residues (hydrophobic side chains shown as green sticks in Fig. 3c): Leu87 and Leu104 in the β subunit, and Leu89 in the δ subunit. Thus, the three arginine residues towards the tip of the catestatin loop (Figs. (Figs.33 and and7;7; Arg351, Arg353, and Arg358) are likely to find a position of minimal energy away from the arginine and lysine side chains in the cation porefacing domains of the nicotinic α and γ subunits.
4. Conclusions
An oligopeptide domain within chromogranin A is a potent, selective, non-competitive inhibitor at the nicotinic cholinergic receptor on chromaffin cells, the receptor which controls catecholamine secretion from such cells [20]. This ‘catestatin’ domain bears partial (up to 35.5%) sequence identity to regions of three proteins with known tertiary structure, as previously determined hy X-ray crystallography: 8FAB (a human myeloma immunoglobulin), 1PKM (cat muscle pyruvate kinase), and 2IG2 (a human monoclonal immunoglobulin). Proteins with regions of sequence homology down to as little as 30% amino acid sequence identity tend to assume generally similar tertiary structures [14,15,25]. Similar empirically determined tertiary structures for 8FAB, 1PKM, and 2IG2, despite at most 40% sequence identity, reinforce the validity of such homology modeling as an approach to the determination of the tertiary structure of catestatin.
Several independent lines of evidence, both computational and experimental, give support to features of this homology model-derived 3-dimensional structure of catestatin.
Firstly, the model predicts two β-strand regions in catestatin, especially from residues chromogranin A343–353 (DRSMRLSFRAR) and chromogranin A361–369 (GLQLRRGWR). Substantial β-strand character for the catestatin region is also predicted by the hydrophobic moment algorithm [8,9], which recognizes regions of alternating polar and apolar residues as predictive of amphiphilic β-sheet; indeed, a portion of the catestatin region (chromogranin A345–356) is the only such region in the 431-amino-acid primary structure of bovine chromogranin A (Fig. 5). In addition, circular dichroism spectra (Fig. 6) of chromogranin A indicated substantial (~63%) β-strand character, especially when even small amounts of organic solvent (25% trifluoroethanol) were included in the solvent. By contrast, substantial α-helix was not apparent under any conditions, and even inclusion of very large fractions of organic solvent (up to 100%) seemed to result in only ~22% of apparent α-helical character in the peptide. Lack of clearcut global structure for the peptide in water alone (Fig. 6) is not unusual for small biologically active peptides [19], which may yet interact specifically with their cell surface receptors; perhaps cell surface receptors provide a specific conformation-inducing milieu for their cognate ligands, small biologically active peptides [16].
Secondly, the electropositive loop structure (Fig. 3a and b), with three directionally displayed positive arginine side chains (Fig. 3a), suggests a domain crucial for the peptide’s secretory action. Indeed, replacement of any of the loop’s three arginine residues Arg351, Arg353, or Arg358) by uncharged alanine (Fig. 7: mutants R351A, R353A, or R358A) resulted in a marked fall in the peptide’s potency on nicotinic-stimulated secretion (rise in IC50), while replacement of arginine residues outside the loop (in mutants R344A or R347A) was without effect on activity.
Substitution of an alanine for Pro360 (in mutant P360A) did not affect the peptide’s biological activity (Fig. 7); perhaps the turn typically induced by a proline residue is not essential to the ability of the peptide to form its central cationic loop structure, especially as the peptide approaches its receptor, where ‘induced fit’ might prevail [16].
The electropositive nature of the catestatin loop structure corresponds to an overall directionality of charge of the catestatin molecule, as evidenced by the electrostatic potential calculations (Fig. 3b). Several features suggested a model for the electrostatic interaction of catestatin with the extracellular orifice of the nicotinic receptor: (1) catestatin’s pharmacological action as a non-competitive nicotinic cholinergic antagonist to limit cation flux through the nicotinic ligand-gated cation pore [20], (2) the lack of competition of a nicotinic agonist for catestatin binding to nicotinic receptors (Fig. (Fig.8),8), and ((3)3) the marked electronegativity of the extracellular domain of the nicotinic receptor in the region of its cation pore orifice [34]. ‘Docking’ of catestatin to the Torpedo nicotinic receptor extracellular domain, with both electrostatic and van der Waals energy minima, revealed a complex of catestatin at the orifice of the cation pore, with catestatin’s electropositive loop closely abutting the β and δ subunits of the nicotinic receptor, away from those positively charged receptor subunit side chains in the vestibule (Fig. 3c); indeed, the complex virtually occluded the extracellular orifice of the cation pore, thereby providing a structural basis for the non-competitive nicotinic antagonist properties of the peptide [20].
In conclusion, we developed a homology model for the endogenous nicotinic cholinergic antagonist catestatin. The model predicts a central cationic loop tethered by two flanking β-strands. Several predictions of the model have been fulfilled by experimental data, including circular dichroism and functional activity of crucial deletion and point-mutants of the peptide sequence, as well as covalent crosslinking to specific nicotinic subunits. This 3-dimensional structure may serve as a useful guide in designing further experiments to probe crucial links between this peptide’s structure and function, and may serve as a starting point in the development of nonpeptide mimetics of this endogenous nicotinic cholinergic antagonist. Indeed, synthetic non-competitive nicotinic cholinergic antagonists (or ganglionic blockers) such as trimethaphan or hexamethonium are useful in antihypertensive management [33], though their side effects currently preclude widespread use; hence, ganglionic blockers with enhanced potency or selectivity would be attractive.
Acknowledgements
We appreciate the assistance of the laboratory of Palmer Taylor, Ph.D., University of California, San Diego, in designing these experiments. David Johnson, Ph.D., University of California, Riverside, kindly supplied Torpedo membranes. Circular dichroism experiments were performed with the assistance of Joseph Taulane in the laboratory of Murray Goodman, Ph.D., University of California, San Diego. The authors acknowledge support from the Department of Veterans Affairs, National Institutes of Health and the American Heart Association.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/s0167-0115(98)00040-8
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3676947
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Catestatin: Antimicrobial Functions and Potential Therapeutics.
Pharmaceutics, 15(5):1550, 20 May 2023
Cited by: 4 articles | PMID: 37242791 | PMCID: PMC10220906
Review Free full text in Europe PMC
Chromogranin A and its fragments in cardiovascular, immunometabolic, and cancer regulation.
Ann N Y Acad Sci, 1455(1):34-58, 06 Oct 2019
Cited by: 42 articles | PMID: 31588572 | PMCID: PMC6899468
Review Free full text in Europe PMC
Role of the neuroendocrine antimicrobial peptide catestatin in innate immunity and pain.
Acta Biochim Biophys Sin (Shanghai), 49(11):967-972, 01 Nov 2017
Cited by: 4 articles | PMID: 28981685
Review
Discrepancies between Cyclic and Linear Antimicrobial Peptide Actions on the Spectrochemical and Nanomechanical Fingerprints of a Young Biofilm.
ACS Omega, 2(9):5861-5872, 18 Sep 2017
Cited by: 5 articles | PMID: 30023754 | PMCID: PMC6044769
Granin-derived peptides.
Prog Neurobiol, 154:37-61, 22 Apr 2017
Cited by: 32 articles | PMID: 28442394
Review
Go to all (31) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (3)
-
(5 citations)
PDBe - 8FABView structure
-
(5 citations)
PDBe - 2IG2View structure
-
(5 citations)
PDBe - 1PKMView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Interaction of the catecholamine release-inhibitory peptide catestatin (human chromogranin A(352-372)) with the chromaffin cell surface and Torpedo electroplax: implications for nicotinic cholinergic antagonism.
Regul Pept, 95(1-3):9-17, 01 Nov 2000
Cited by: 22 articles | PMID: 11062327
Primary structure and function of the catecholamine release inhibitory peptide catestatin (chromogranin A(344-364)): identification of amino acid residues crucial for activity.
Mol Endocrinol, 14(10):1525-1535, 01 Oct 2000
Cited by: 67 articles | PMID: 11043569
Conformational preferences and activities of peptides from the catecholamine release-inhibitory (catestatin) region of chromogranin A.
Regul Pept, 118(1-2):75-87, 01 Apr 2004
Cited by: 19 articles | PMID: 14759560
Catestatin is a novel endogenous peptide that regulates cardiac function and blood pressure.
Cardiovasc Res, 80(3):330-338, 09 Jun 2008
Cited by: 44 articles | PMID: 18541522
Review





