Abstract
Free full text

ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death
Associated Data
Abstract
Protein misfolding in the endoplasmic reticulum (ER) leads to cell death through PERK-mediated phosphorylation of eIF2α, although the mechanism is not understood. ChIP-seq and mRNA-seq of activating transcription factor 4 (ATF4) and C/EBP homologous protein (CHOP), key transcription factors downstream of p-eIF2α, demonstrated that they interact to directly induce genes encoding protein synthesis and the unfolded protein response, but not apoptosis. Forced expression of ATF4 and CHOP increased protein synthesis and caused ATP depletion, oxidative stress and cell death. The increased protein synthesis and oxidative stress were necessary signals for cell death. We show that eIF2α-phosphorylation-attenuated protein synthesis, and not Atf4 mRNA translation, promotes cell survival. These results show that transcriptional induction through ATF4 and CHOP increases protein synthesis leading to oxidative stress and cell death. The findings suggest that limiting protein synthesis will be therapeutic for diseases caused by protein misfolding in the ER.
The ER is the organelle essential for calcium storage, lipid synthesis and protein folding and secretion in metazoan cells. The ER has a dynamic capacity to accommodate increases in the demand for protein folding. However, extracellular stimuli and changes in intracellular homeostasis cause protein misfolding in the ER. The ER uses its protein folding status as a signal to orchestrate downstream adaptive or apoptotic responses. The unfolded protein response (UPR) is a cellular adaptive response that evolved to restore protein-folding homeostasis by reducing protein synthesis through phosphorylation of eIF2α and by increasing the ER protein-folding and degradative capacities through transcriptional activation by XBP1 and ATF6α (refs 1–3). If the UPR cannot resolve the protein-folding defect, cells undergo apoptosis. One of the mechanisms of ER stress-induced cell death involves sequential steps of PERK-mediated eIF2α phosphorylation4,5, preferential translation of ATF4/CREB-2 messenger RNA (refs 6–8) and induction of CHOP/GADD153 (refs 9–11). Studies of Chop deletion in mice show that CHOP is required for ER stress-mediated cell death in response to a variety of pathological conditions12–18. However, forced expression of CHOP alone does not induce cell death, but sensitizes to ER stress-induced cell death19–21, suggesting that another yet unidentified signal is required for the apoptotic response, as previously suggested22. Analysis of Atf4–/– cells has generated conflicting results as to having a pro-survival23–25 or a pro-apoptotic role26–30. Here, we identified the roles of ATF4 and CHOP for initiating ER stress-induced cell death.
RESULTS
ATF4 and CHOP mediate ER stress-mediated cell death
To analyse events downstream of eIF2α phosphorylation, we examined the expression of several key proteins in wild-type (WT) mouse embryo fibroblasts (MEFs) on tunicamycin (Tm) treatment to disrupt ER protein folding (Fig. 1a). Phosphorylation of eIF2α transiently increased between 1 and 2 h after Tm treatment, followed by sequential induction of ATF4 and CHOP. Cleavage of caspase 3 (CASP3) and PARP was not detected until 24 h after Tm, indicating that increased ATF4 and CHOP expression precedes cell death, as suggested previously21. To determine whether ATF4 and CHOP can initiate cell death, we examined cell survival after adenovirus-mediated delivery of ATF4 and/or CHOP (Supplementary Fig. S1a). To our surprise, forced expression of CHOP alone did not reduce cell viability, whereas forced expression of ATF4 alone decreased survival, and this was accentuated by co-expression with CHOP (Fig. 1b), suggesting that ATF4 might be the primary signal and CHOP a secondary signal required for ER stress-induced cell death.
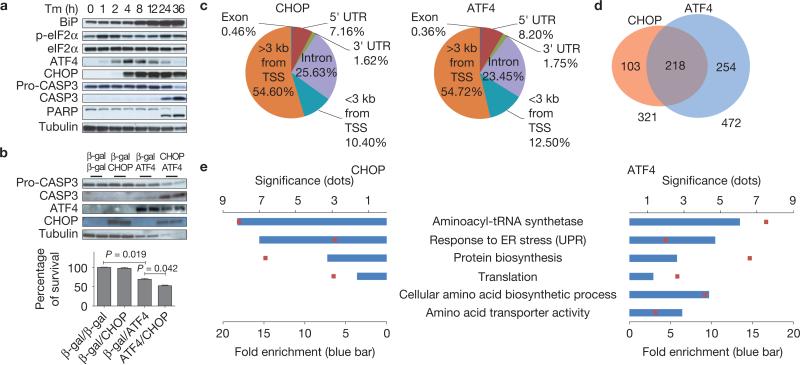
ATF4 and CHOP bind to promoter regions of genes encoding protein synthesis and UPR functions. (a) Protein expression during ER stress-mediated cell death. Cell lysates were collected at the indicated times after Tm (2 μg ml–1) treatment for western blot analysis. (b) Effect of ATF4 and CHOP expression in WT MEFs. MEFs were infected with adenoviruses expressing CHOP and/or ATF4 at an MOI (mode of infection) of 100. At 24 h after infection, cell lysates were analysed by western blotting (upper panel). At 48 h after infection, cell viability was measured by a WST-8 assay (n = 3 independent experiments; lower panel). (c) Distribution of CHOP and ATF4 ChIP-seq peaks across the genome. The peaks were classified as: within introns (Intron), within 3′ untranslated regions (UTRs), within 5′ UTRs, or within coding sequences (Exon), <3 kb from TSSs or >3 kb from TSSs in intergenic regions. The numbers below the annotations represent the percentage of peaks across the genome. (d) Venn diagram showing overlapping and unique sets of ATF4- and CHOP-occupied genes that have peaks <3 kb from the TSS. (e) Functional enrichment analysis of ATF4 and CHOP target genes that have peaks <3 kb from the TSS. All error bars represent means±s.e.m. Uncropped images of blots are shown in Supplementary Fig. S7.
ATF4 and CHOP share target genes
As ATF4 (ref. 6) and CHOP (ref. 31) are basic leucine zipper-containing transcription factors, we reasoned that ATF4 and CHOP target genes that establish the apoptotic program. Therefore, we applied genome-wide chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) and mRNA expression analysis (mRNA-seq) to identify genes directly regulated by ATF4 and CHOP in response to Tm (Supplementary Table S1 and Methods). ChIP-seq found 2,598 peaks for CHOP and 3,023 peaks for ATF4, which are assigned to the nearest transcription start sites (TSSs) of unique genes based on gene annotations of USCS known genes (mm9). Analysis of the ATF4-and CHOP-binding site distribution demonstrated that 10.4% of CHOP- and 12.5% of ATF4-binding sites were located <3 kilobases (kb) from the TSSs, representing a ~2.5- and ~3-fold enrichment over random (4.2%), respectively (Fig. 1c and Supplementary Fig. S1b–e). Thus, ATF4 and CHOP preferentially bind to proximal promoter regions of target genes. Conventional ChIP validated the ChIP-seq results (Supplementary Fig. S1f). Strikingly, a total of 218 genes out of 321 CHOP targets and out of 472 ATF4 targets that have binding sites <3 kb from the TSS were bound by both transcription factors (Fig. 1d and Supplementary Table S2). ChIP-seq identified known ATF4 and CHOP target genes, including Atf3 (ref. 32), Gadd34 (refs 33,34) and Trib3 (ref. 35), as well as many uncharacterized genes (Supplementary Fig. S1g and Table S2). To our surprise, the most significantly enriched biological functions of ATF4 and/or CHOP target genes involve protein synthesis including eleven aminoacyl-tRNA synthetases (Aars, Lars, Yars, Sars, Wars, Vars, Nars, Mars, Gars, Iars and Eprs) and four initiation factors (Eif2s2, Eif3c, Eif4g2 and Eif5), and, as expected, the UPR (Fig. 1e and Supplementary Fig. S1h). It is notable that ATF4 alone targets genes encoding functions in amino acid transport and amino acid biosynthesis (Fig. 1e and Supplementary Table S3). Most strikingly, we did not find enrichment in cell death-related functions in either ATF4 and/or CHOP target genes.
ChiP-seq and mRNA-seq of ATF4 and CHOP identifies protein synthesis as a highly enriched functional group of target genes
mRNA-seq (see Methods) identified genes for which expression was significantly increased (770) or decreased (738) on Tm treatment in WT MEFs (Supplementary Fig. S2a and Table S4). Significantly, a large proportion of genes induced (366 out of 770) or repressed (326 out of 738) on Tm treatment required ATF4 and/or CHOP. Interestingly, many of the Tm-induced genes in WT MEFs (red bar in Fig. 2a) were directly bound by ATF4 and/or CHOP (171 out of 770 genes), whereas the gene cluster that was repressed on Tm treatment (green bar in Fig. 2a) had few binding sites for either ATF4 or CHOP (14 out of 738 genes). This suggests that ATF4- and/or CHOP-mediated repression does not require DNA binding, and may involve squelching36. In addition, 92 of the 97 genes that bound both ATF4 and CHOP (Group C) were upregulated by Tm treatment (Fig. 2b and Supplementary Table S5), indicating that ATF4 and CHOP activate their target genes through DNA binding. It is notable that the most enriched functional groups of common regulated target genes (Fig. 2c, Group C) encode amino acid metabolic processes, mRNA translation and the UPR. Finally, among several functional categories, genes in the response to oxidative stress category were repressed in response to Tm in an ATF4- and/or CHOP- dependent-, as well as -independent, manner (Supplementary Fig. S2b).
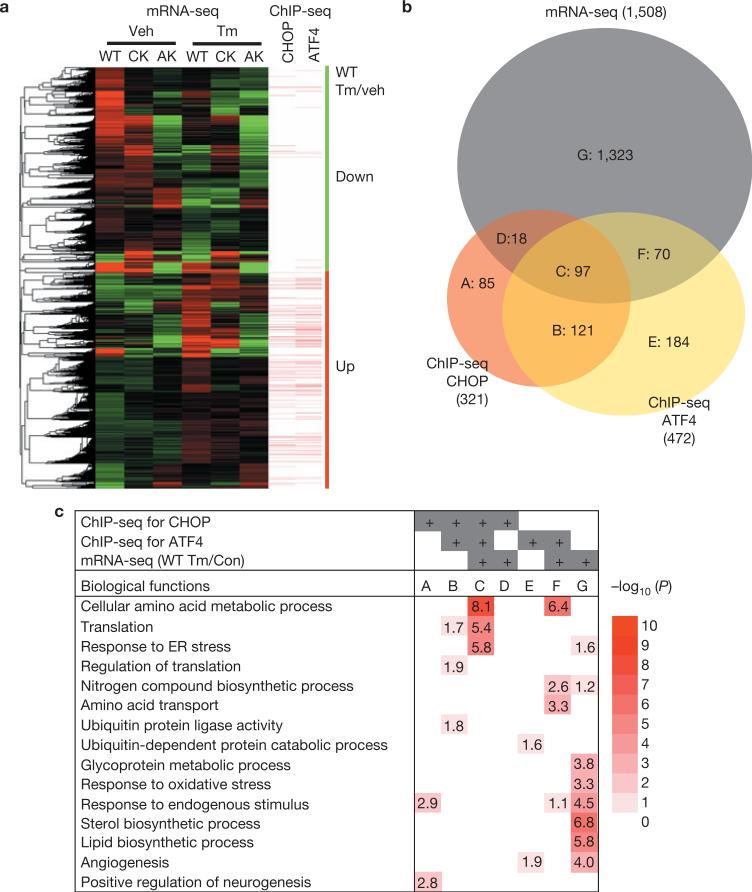
ATF4 and CHOP upregulate expression of genes encoding protein synthesis and UPR functions. (a) Clustered mRNA-seq expression data presented in a heat map with ChIP-seq peaks aligned. Tm-induced and -repressed genes in WT, Chop–/– (CK), and Atf4–/–-(AK) MEFs are shown. Genes containing ATF4- and CHOP- binding sites <3 kb from the TSSs are denoted by bars. Green vertical bar represents downregulated genes, whereas red vertical bar represents upregulated genes in WT MEFs in response to Tm (2 μg ml–1) when compared with vehicle (dimethylsulphoxide). (b) Venn diagram illustrating overlaps of gene sets from mRNA-seq and ChIP-seq for ATF4 and CHOP. A: genes bound by CHOP without expression changes in response to Tm (2 μg ml–1). B: genes bound by both ATF4 and CHOP without expression changes. C: genes bound by both ATF4 and CHOP with differential expression. Genes in C are shown in Supplementary Table S4. D: genes bound by CHOP that exhibit differential expression. E: genes bound by ATF4 that do not change expression. F: genes bound by ATF4 that exhibit differential expression. G: genes with differential expression but do not bind ATF4 or CHOP. (c) Functional enrichment analysis of gene sets in b. The EASE scores of DAVID functional enrichment of selected GO terms are represented in a heat map with –log10 (EASE score) as a colour index and number. White corresponds to an EASE score of 1.0 (no number in the block) with no statistical significance; dark red corresponds to an EASE score of 1.0×10–10 or lower.
ATF4 and CHOP interact to induce their target genes
An unbiased motif search was performed using the sequences of peaks <3 kb from the TSS. The localization of motif sequences was enriched in the middle of the binding peaks (Supplementary Fig. S2c), indicating that these motifs represent binding sites for these transcription factors37. The motifs for ATF4 and CHOP were strikingly similar (Fig. 3a and Supplementary Fig. S2d) and were located at the same position in 75.9% of the overlapped target genes (Supplementary Table S6). Co-immunoprecipitation of overexpressed (Fig. 3b), as well as endogenous (Fig. 3c), ATF4 and CHOP demonstrated that they interact. In addition, sequential ChIP with anti-CHOP and anti-ATF4 and quantitative PCR (qPCR) demonstrated that endogenous ATF4 and CHOP co-occupy the promoter regions of the common target genes Wars, Trib3 and Atf3, consistent with previous reports38,39 (Fig. 3d).
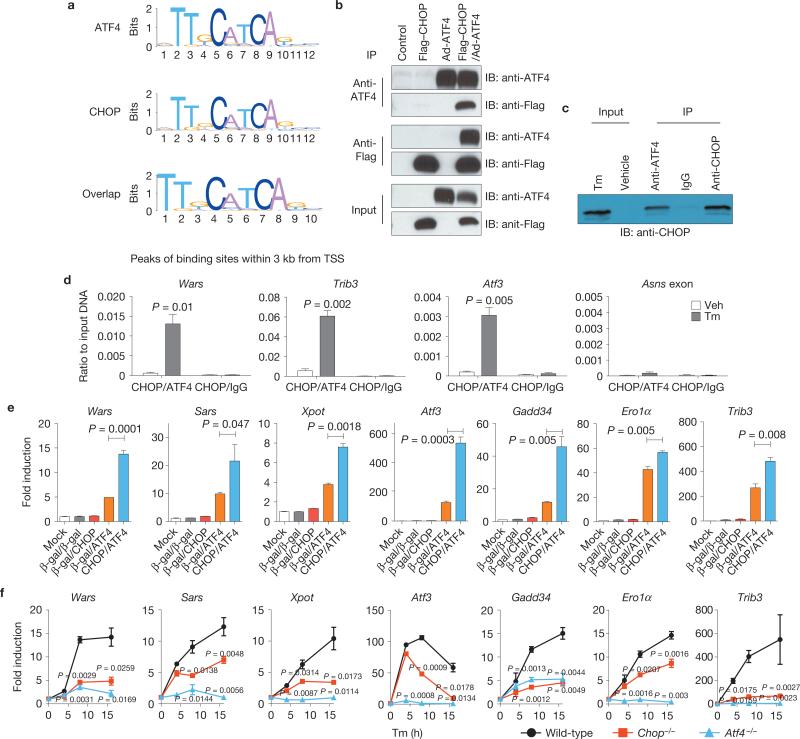
ATF4 and CHOP interact to induce target genes involved in protein synthesis and the UPR. (a) Motif analysis of ATF4- and CHOP-binding sites. Sequences from CHOP- and ATF4-binding regions <3 kb upstream and downstream of a TSS were analysed. Significantly over-represented motifs from each gene set are shown. (b, c) Interaction of ATF4 and CHOP. (b) Flag–CHOP and ATF4 were expressed either alone or together in HEK293 cells and immunoprecipitated (IP) using anti-Flag or anti-ATF4 antibodies. Immunoprecipitated proteins were analysed by western blotting (IB) using ATF4 or Flag antibody. (c) MEFs were treated with Tm (2 mg ml–1) and immunoprecipitated using anti-ATF4, normal rabbit IgG or anti-CHOP. Immunoprecipitated proteins were analysed by western blotting using anti-CHOP antibody. (d) Co-occupancy of ATF4 and CHOP in the promoter regions of commonly targeted genes. Cells were treated with Tm for 10 h, followed by sequential ChIP assay (see Methods). The enrichment was determined for re-ChIPed chromatin by qPCR (n = 3 independent experiments). (e) Effect of ATF4 and/or CHOP overexpression on common target genes. ATF4 and CHOP were overexpressed either alone or together in WT MEFs. Gene expression was measured by qRT–PCR using β-gal as a negative control. Data are presented as means±s.e.m. (n = 3 independent experiments). (f) Expression profile of common target genes during Tm treatment. WT, Chop–/– and Atf4–/– MEFs were treated with Tm (2 μg ml–1) for 16 h and total RNAs were prepared at indicated time points for qRT–PCR (n = 3 independent experiments). All error bars represent means±s.e.m. Uncropped images of blots are shown in Supplementary Fig. S7.
We then examined whether ATF4 and CHOP regulate expression of their target genes. Although Tm induction of target genes required CHOP (Supplementary Fig. S2e), induction was not observed on adenovirus-mediated forced expression of CHOP alone (Fig. 3e), indicating that CHOP is necessary, but not sufficient for transcriptional induction. In contrast to CHOP, adenovirus-mediated expression of ATF4 induced ATF4 and CHOP common target genes, which was further increased by co-expression with CHOP (Fig. 3e). Quantitative real-time PCR (qRT–PCR) during Tm treatment confirmed the cooperation between ATF4 and CHOP where induction of the common target genes was significantly attenuated by either Atf4 or Chop deletion (Fig. 3f and Supplementary Fig. S2f). These results suggest that ATF4 and CHOP interact to induce transcription of a common set of target genes involved in protein synthesis and the UPR.
ATF4 and CHOP increased protein synthesis
As ATF4 and CHOP induce genes encoding functions in protein synthesis (Fig. 2c, Group C), we analysed protein synthesis on forced expression of ATF4 and CHOP. Whereas expression of β-gal or CHOP did not alter protein synthesis, expression of ATF4 increased protein synthesis in Chop+/+ MEFs, and to a lesser extent in Chop–/– MEFs (Fig. 4a). In contrast, forced expression of an ATF4 mutant that lacks DNA binding (ATF4ΔRK; ref. 27) did not increase protein synthesis, indicating that ATF4 exerts its effect on protein synthesis through transcriptional activation (Supplementary Fig. S3a,b). The ATF4-mediated increase in protein synthesis was further enhanced by expression of CHOP (Fig. 4a), suggesting that CHOP is required for maximal ATF4-stimulated protein synthesis. The increased protein synthesis on ATF4 and CHOP overexpression is probably through induction of their common target gene, Gadd34/Ppp1r15a, encoding a regulatory subunit of protein phosphatase 1 (PP1) that directs eIF2α dephosphorylation33,34,40 (Fig. 4b, see lanes 3 and 6). Consistent with this idea, GADD34 induction by ATF4 and CHOP prevented transient Tg-induced translation attenuation (Fig. 4b,c and Supplementary Fig. S3c). However, because ATF4 and CHOP overexpression significantly increased protein synthesis in Gadd34–/– MEFs, but to a lesser extent than in WT MEFs (Fig. 4d and Supplementary Fig. S3d,e), we conclude that ATF4 and CHOP increase protein synthesis in both GADD34-dependent, as well as -independent, manners41.
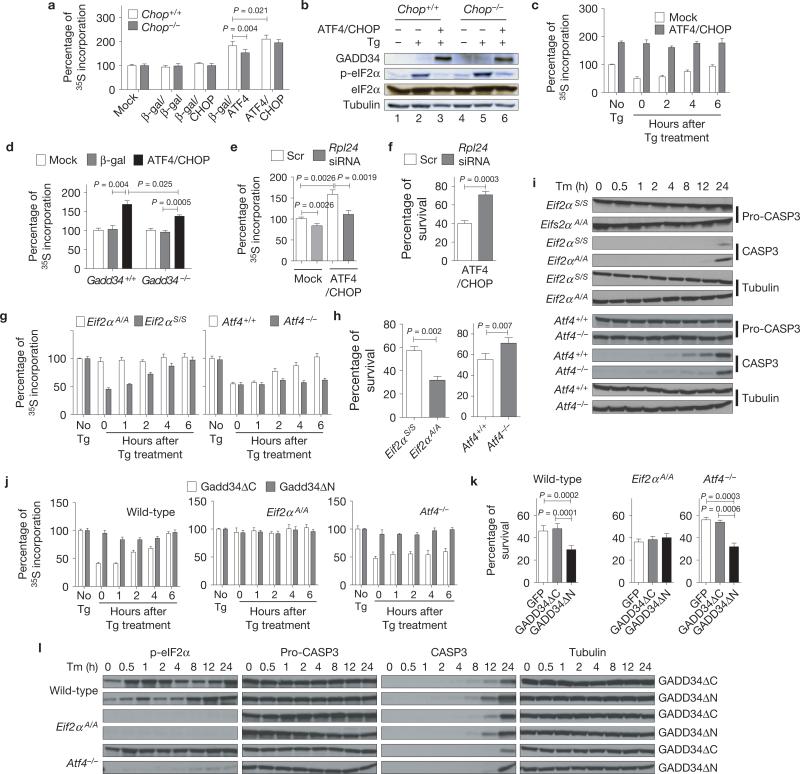
ATF4 and CHOP increase protein synthesis leading to cell death. (a) Increased protein synthesis by ATF4 and CHOP. ATF4 and CHOP were expressed either alone or together in Chop+/+ or Chop–/– MEFs. After 24 h, cells were pulse-labelled with [35S]methionine/cysteine (n = 3 independent experiments; see Methods). (b, c) Ablation of translation attenuation by ATF4- and CHOP-mediated GADD34 induction. After forced expression of ATF4 and CHOP, MEFs were treated with 1 μM thapsigargin (Tg) for an hour and analysed by western blotting (b) and metabolic labelling (n = 3 independent experiments; c). (d) Effect of Gadd34 deletion on ATF4/CHOP-mediated increase in protein synthesis. ATF4 and CHOP were expressed in WT and Gadd34–/– MEFs, followed by metabolic labelling (n = 3 independent experiments). (e, f) Effect of translation attenuation on cell survival. Cells were transfected with scrambled (Scr) or Rpl24 siRNAs, followed by Ad-ATF4 and Ad-CHOP infection. Protein synthesis at 24 h (e) and cell viability at 48 h (f) were measured (n = 3 independent experiments). (g–i) Effect of unrestricted protein synthesis on survival. (g) Translational recovery after one hour Tg (1 μM) treatment in Eif2αA/A and Eif2αS / S (left panel) or Atf4+/+ and Atf4–/– MEFs (right panel; n = 3 independent experiments). (h) MEFs were treated with Tm (2 μg ml–1) for 24 h, and cell viability was measured (n = 3 independent experiments). (i) Cell lysates were collected at indicated times for western blot analysis. (j–l) Effect of GADD34 overexpression on cell survival. (j) Translational recovery after one hour Tg (1 μM) treatment in WT, Eif2αA/A and Atf4–/– MEFs after forced expression of either Gadd34ΔC or Gadd34ΔN (n = 3 independent experiments). (k) Indicated MEFs were treated with Tm (2 μg ml–1) for 24 h and viability was measured (n = 3 independent experiments). (l) Western blot analysis of cell lysates from MEFs treated with Tm (2 μg ml–1) for the indicated times. All error bars represent means ± s.e.m. Uncropped images of blots are shown in Supplementary Fig. S7.
Increased protein synthesis by ATF4 and CHOP induces cell death
Studies suggest that protein synthesis is coupled with cell death42–46. Therefore, we examined whether the ATF4- and CHOP-mediated increase in protein synthesis reduces cell survival. Protein synthesis was attenuated by knockdown of the ribosomal genes Rpl24 (ref. 47) and Rpl7 (Supplementary Fig. S3f,g). Knockdown of these genes prevented the ATF4- and CHOP-mediated increase in protein synthesis and increased cell survival (Fig. 4e,f and Supplementary Fig. S3h,i). In addition, pharmacological inhibition of protein synthesis also increased cell viability in response to ATF4 and CHOP expression (Supplementary Fig. S3j,k).
Our findings indicate that ATF4- and CHOP-mediated cell death results from increased protein synthesis. To elucidate the mechanistic role of protein synthesis, we analysed the effect of cell stress on the viability of Atf4-null cells and cells with homozygous S51A mutation at the phosphorylation site in eIF2α (Eif2αA/A). Where Atf4–/– MEFs cannot activate ATF4-dependent genes and are defective in recovery of protein synthesis after ER stress34, Eif2αA/A MEFs cannot attenuate protein synthesis in response to cell stress5,48 (Fig. 4g). Eif2αA/A and Atf4–/– MEFs were equally sensitive to oxidative stress (Supplementary Fig. S3l), as previously described23. However, when compared with WT MEFs, Eif2αA/A MEFs were more sensitive and Atf4–/– MEFs were significantly more resistant to Tm (Fig. 4h,i). The expression of genes encoding functions in protein synthesis, ER protein folding, the UPR and protein degradation was not significantly different between Eif2αA/A and Atf4–/– MEFs (Supplementary Fig. S3m). Importantly, these findings indicate that the ATF4-mediated increase in protein synthesis contributes to cell death, whereas eIF2α phosphorylation limits protein synthesis to promote cell survival in response to ER stress.
Our findings suggest that reduced protein synthesis, and not increased ATF4 synthesis, is a critical determinant for cell survival. The requirement for translational attenuation to prevent cell death was also tested by forced expression of GADD34 to promote eIF2α dephosphorylation and impair attenuation of translation in response to ER stress (Fig. 4j). Overexpression of the functional GADD34 carboxy-terminal domain (GADD34ΔN), but not the inactive amino-terminal domain (GADD34ΔC), significantly sensitized WT (Eif2αS/S) and Atf4–/– MEFs to ER stress (Fig. 4k,l). In contrast, Eif2αA/A cells that cannot phosphorylate eIF2α were not affected by expression of GADD34 (Fig. 4k,l), indicating that prolonged attenuation of protein synthesis in Atf4–/– MEFs is beneficial in the context of ER stress.
ATF4 and CHOP induce cell death through oxidative stress and ATP depletion
Increased protein synthesis would require increased disulphide bond formation in the ER, where electrons are shuttled through protein disulphide isomerase and ERO1α to O2 to generate H2O2 (ref. 49). Dichlorofluorescein staining detected increased reactive oxygen species (ROS) following Tm treatment, as previously described23. Overexpression of ATF4 and CHOP increased ROS to a similar level as Tm treatment (Fig. 5a,b). Treatment with the antioxidant butylated hydroxyanisole (BHA; Fig. 5a,b) or Rpl24 silencing (Supplementary Fig. S4b) reduced ROS, and significantly increased cell survival (Fig. 5c). In addition, short interfering RNA (siRNA)-mediated knockdown of ERO1α (Supplementary Fig. S4d) increased cell survival on overexpression of ATF4 and CHOP or Tm treatment (Fig. 5d). Consistent with the mRNA-seq results (Supplementary Fig. S2b), Tm treatment significantly reduced expression of several anti-oxidative stress response genes, including Sod3, Sod2 and Ucp2, in an ATF4- and CHOP-dependent, as well as -independent manner (Fig. 5e), which may also contribute to increased oxidative stress and reduced survival19. We also analysed ATP content in response to overexpression of ATF4 and CHOP. At 24 and 48 h after ATF4 and CHOP expression, the ratio of ATP/ADP was significantly reduced (Fig. 5f), which was mitigated by Rpl24 silencing (Supplementary Fig. S5). These results show that increased protein synthesis produces ROS and depletes ATP.
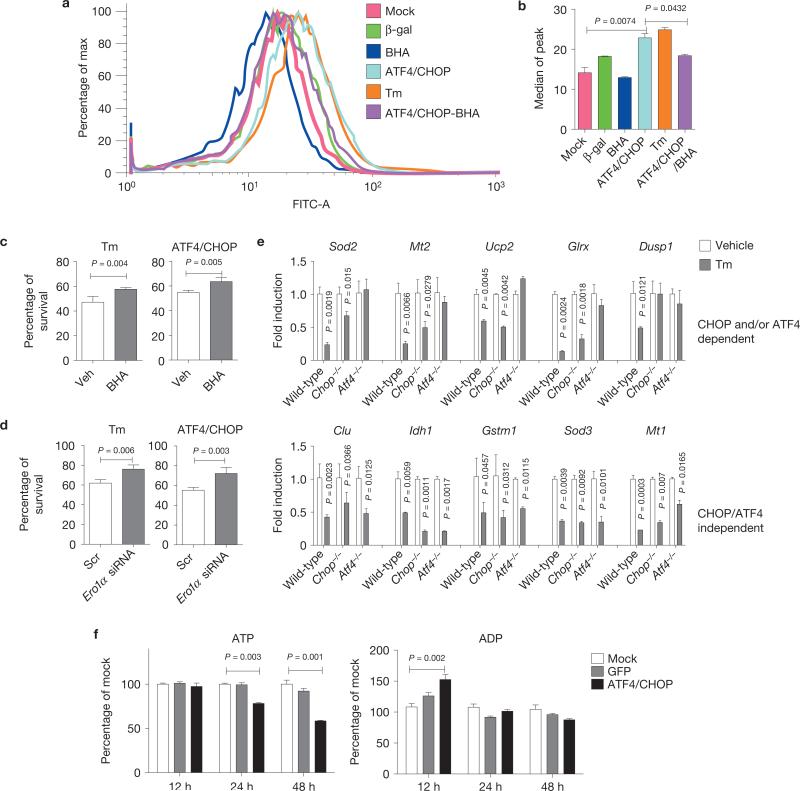
ATF4 and CHOP increase oxidative stress and deplete ATP. (a, b) Oxidative stress induced by ATF4 and CHOP overexpression. (a) WT MEFs were mock-infected or infected with Ad-ATF4 and Ad-CHOP for 24 h and stained with CM-H2DCFDA for analysis by flow cytometry. Where indicated, 100 μM BHA was added to the medium at time of infection. Cells were treated with Tm (2 μg ml–1) at 24 h before analysis. (b) Histogram for median peaks in a (n = 3 independent experiments). (c) Effect of BHA treatment on cell viability. MEFs were treated with vehicle (Veh) or BHA (100 μM) 24 h before infection with Ad-ATF4 and Ad-CHOP for 48 h or Tm treatment (2 μg ml–1) for 24 h and cell survival was measured (n = 3 independent experiments). (d) Effect of Ero1α knockdown on cell viability. Survival was measured at 48 h after forced expression of ATF4 and CHOP or at 24 h after Tm treatment (2 μg ml–1; n = 3 independent experiments). (e) Expression profiles of anti-oxidant genes in response to ER stress. MEFs were treated with Tm (2 μg ml–1) for 10 h and total RNAs were extracted for qRT–PCR (n = 3 independent experiments). (f) ATP and ADP levels measured at the indicated times after infection with Ad-ATF4 and Ad-CHOP or Ad-GFP as control (n = 3 independent experiments). All error bars represent means±s.e.m.
ATF4 and CHOP increase protein synthesis, oxidative stress and apoptosis in vivo
We examined the physiological significance of our findings in vivo by adenovirus-mediated delivery of ATF4 and CHOP to the liver in mice. Stable isotope (2H2O) labelling50 and carbonyl analysis demonstrated that ATF4 and CHOP expression increase protein synthesis and cause protein oxidation in vivo (Fig. 6a,b). In addition, adenovirus-mediated expression of dominant-negative ATF4ΔRK in the liver (Supplementary Fig. S6a) reduced protein synthesis and cell death in response to Tm injection (Fig. 6c–e). These results show that ATF4 and CHOP increase protein synthesis to cause oxidative stress and cell death in vivo. We further investigated the role of protein synthesis on beta cell function in vivo by comparison of pancreatic islets in mice with Atf4 deletion or with beta-cell-specific Eif2αA / A mutation43 (Supplementary Fig. S6b). When compared with the significantly reduced beta cell mass and impaired glucose tolerance in mice with beta-cell-specific Eif2αA / A mutation, beta cell mass and glucose tolerance were not altered in Atf4–/– mice (Fig. 6f–i). These results emphasize the significant requirement for tight regulation of protein synthesis to prevent cell death in response to ER stress in vivo.
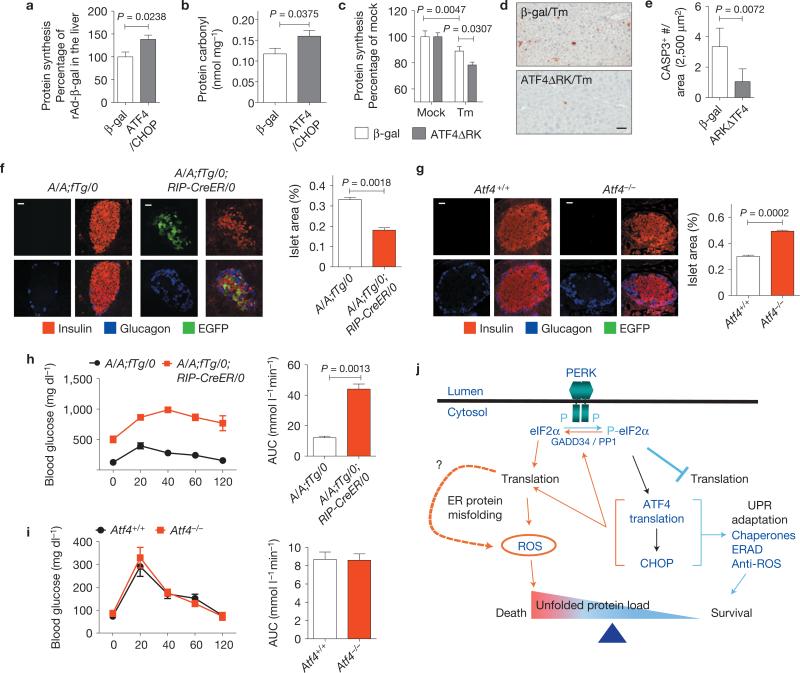
ATF4 and CHOP increase protein synthesis and oxidative stress in vivo. (a) Protein synthesis was measured in the liver at 3 days after injection of Ad-β-gal (n = 6 mice) or Ad-ATF4/Ad-CHOP (n = 5 mice) applying the 2H2O tracer method (see Methods). (b) Carbonyls in the livers isolated from mice at 3 days after adenovirus infection (n = 6 mice per group). (c–e) Effect of inhibition of ATF4 function on protein synthesis and cell death. At 3 days after injection of Ad-β-gal or Ad-ATF4ΔRK, Tm was administered to each group of mice for 24 h followed by collection of liver and plasma for protein synthesis measurement (c) and staining for cleaved CASP3 (d). Scale bar, 50 μm. (e) Quantification of cleaved CASP3-positive cells was performed on cells in d (n = 3 mice per group). (f, g) Insulin and glucagon immunofluorescence staining. Insulin and glucagon staining were performed using pancreatic sections from A/A;fTg/0 (having the intact eIF2α transgene; n = 4 mice) and A/A;fTg/0;RIP-CreER/0 (having a WT Eif2α transgene deleted by tamoxifen injection; n = 4 mice) (f) and 4-month-old Atf4+/+ (n = 3 mice), and Atf4–/– mice (n = 3 mice) (g). Scale bars, 20 μm. Islet area was measured using dotSlide software. The percentage indicates the total islet area over the pancreas area in each section. (h, i) Glucose tolerance tests were performed and the area under curve from the glucose tolerance tests was quantified in A/A;fTg/0 (n = 6 mice) and A/A;fTg/0;RIP-CreER/0 (n = 4 mice) (h) mice at 12 weeks after tamoxifen injection or in Atf4+/+ (n = 5 mice) and Atf4–/– (n = 4 mice) mice at the age of 4 months (i). (j) Mechanism for ATF4- and CHOP-mediated cell death. On ER stress, eIF2α phosphorylation by PERK promotes preferential translation of ATF4 for subsequent induction of CHOP. ATF4 and CHOP act together to upregulate target genes encoding functions in protein synthesis to restore general mRNA translation. If the adaptive UPR effectively reduces the unfolded protein load, restoration of protein synthesis promotes cell survival. However, if protein synthesis increases before restoration of proteostasis, ROS are produced as a signal to promote cell death. Blue colour indicates pro-survival pathways and orange colour indicates pro-death pathways. All error bars represent means±s.e.m.
DISCUSSION
We have identified increased protein synthesis as one mechanism by which ATF4 and CHOP mediate cell death in response to ER stress. Although CHOP is known to be a pivotal player in ER stress-mediated death, CHOP binding to genes known to function in cell death, including Bcl2l11 (Bim; ref. 51), Bcl2 (ref. 19), Bax and Bad (refs 19,51,52) was not observed. However, it is possible that CHOP regulates these genes in other cell types or contexts. For example, CHOP induced the transcription of Ero1α, which then activates calcium-mediated apoptosis in macrophages53. In this study, we identified another function of CHOP as a transcription factor that interacts with ATF4 to bind promoter regions of genes encoding proteins that increase protein synthesis. In addition, although it was previously known that ATF4 regulates amino acid biosynthesis and transport54,55, we show that ATF4 itself increases protein synthesis and causes cell death. Although there were small differences in gene expression between the Eif2αA/A MEFs and Atf4–/– MEFs, only the Eif2αA/A MEFs, which cannot attenuate protein synthesis, are more susceptible in response to ER stress than Atf4–/– MEFs, indicating that the inability to attenuate protein synthesis, and not defective Atf4 mRNA translation, induces cell death. We found that increased protein synthesis generates ROS, which are a necessary signal to induce apoptosis in response to ER stress.
It has been a long-standing enigma how phosphorylation of eIF2α promotes both survival and death in response to ER stress. Our results emphasize the dual role of eIF2α phosphorylation as a molecular switch to induce cell death through induction of ATF4 and CHOP to regulate protein synthesis. In light of recent findings56, our results support the idea that eIF2α phosphorylation needs to be finely tuned to ensure protein-folding homeostasis (proteostasis) in the ER (Fig. 5j). Immediately after an insult, eIF2α is transiently phosphorylated to acutely and transiently attenuate protein synthesis. Subsequently, induction of ATF4 and CHOP and their downstream gene targets functions to restore protein synthesis. It is noteworthy that Atf4 and Chop mRNAs are preferentially translated when eIF2α is phosphorylated7,8,57. If protein synthesis increases before restoration of proteostasis, ROS are produced that are a necessary signal for apoptosis. This ATF4- and CHOP-dependent cell autonomous apoptosis may have evolved to selectively eliminate stressed, damaged or infected cells from the organism. These findings suggest that agents that reduce protein synthesis may be therapeutic for protein-misfolding diseases.
METHODS
ChIP
Chop+/+, Chop–/–, Atf4+/+ and Atf4–/– MEFs were treated with Tm (2 μg ml–1; Sigma) for 10 h, followed by crosslinking with 1% formaldehyde for 10 min and subsequent ChIP using anti-CHOP (s-575, Santa Cruz) or anti-ATF4 antibodies as described previously38. Immunoprecipitated DNAs were analysed by real-time qRT–PCR with primers listed in Supplementary Table S5. Sequential ChIP analysis was performed as described previously38. Purified, immunoprecipitated DNA was analysed by qPCR.
ChIP-seq assay
Immunoprecipitated DNA fragments for ChIP-seq were prepared from MEFs treated with Tm (2 μg ml–1) for 10 h, followed by crosslinking with 1% formaldehyde for 10 min and ChIP using anti-CHOP or anti-ATF4 antibodies. DNA fragments (200 base pairs) were prepared for Solexa (Illumina) sequencing according to the manufacturer's instructions. Cluster generation and sequencing were performed by the DNA Sequencing Core at the University of Michigan using an Illumina Solexa Genome Analyser II sequencing machine. Reads were aligned to the mouse reference genome (mm9) using the Eland aligner (Illumina) with the –multi option. Binding peaks for CHOP and ATF4 were identified using ERANGE (ref. 58) software with default parameters or MACS (ref. 59) software for ChIP-seq (see Supplementary Note) and the reads from the Chop–/– and Atf4–/– experiments, respectively, as controls. Reads from replicate lanes were concatenated before analysis.
mRNA-seq assay
MEFs were treated with vehicle or Tm (2 μg ml–1) for 10 h, followed by RNA isolation. Complementary DNAs were prepared for mRNA-seq according to the manufacturer's instructions (Illumina). cDNA fragments were prepared for Solexa sequencing as described above for ChIP-seq. Two samples were prepared for each condition, and each sample was run on one lane for sequencing, with the number (proportion) of successfully aligned reads from a sample ranging from 10.6 million (86%) to 18.4 million (89%). Bowtie was employed to align reads to the mouse reference genome (version mm9) plus known splice junctions, created by ERANGE scripts and UCSC known genes (http://genome.ucsc.edu). Counts of reads and RPKM (reads per kilobase pair per million reads mapped) values for each gene were determined using ERANGE software and were tested for differential expression in R using the limma package and the IBMT (intensity-based moderated t-statistic) method60. Testing was performed using log2 read counts normalized to the total number of aligned reads for each sample. We tested the following comparisons: WT Tm versus WT untreated, Atf4–/– Tm versus Atf4–/– untreated, and Chop–/– Tm versus Chop–/– untreated. The IBMT method is an empirical Bayesian method that provides improved estimates of variance for experiments with small samples sizes, while taking into account the relationship between variance levels and the total read count. The false discovery rate (FDR) for each comparison was calculated using the Benjamini–Hochberg method. To consider a gene as significantly differentially expressed, we used the following set of criteria: >2-fold change either upregulated or downregulated, FDR < 0.05 and average RPKM of all 12 mRNA-seq experiments ≥1.
Heat-map generation
A heat map was generated to illustrate the gene expression profiles of genes differentially expressed between treated and untreated WT MEFs using the Java TreeView program (http://jtreeview.sourceforge.net/; Fig. 2a). A second heat map was generated to illustrate the significance levels of over-represented biological functions across multiple gene sets (Fig. 2c). The statistical significance values (–log10-transformed P values) were colour-indexed (white for no significance to dark red for the highest significance).
Gene set enrichment testing
Functional enrichment analyses were performed on genes identified by ChIP-seq or those differentially expressed in mRNA-seq experiments to identify significantly over-represented biological functions defined by Gene Ontology (GO) terms and pathways. The Database for Annotation, Visualization and Integrated Discovery61 (DAVID) and ConceptGen62 were employed for each gene set with an FDR < 0.05 as the significance cutoff. Human orthologous genes were used in these enrichment analyses after mapping mouse genes through the National Center for Biotechnology Information (NCBI) HomoloGene (http://www.ncbi.nlm.nih.gov/homologene) and Mouse Genome Informatics (MGI) Mammalian Orthology databases (http://www.informatics.jax.org/orthology.shtml).
Motifs search
ChIP-seq peak sequences were downloaded from the UCSC Genome Database (mm9, July 2007). De novo motif discovery was carried out using the MEME suite63 (see also Supplementary Note). The parameters used were -dna -nmotifs 2 -mod zoops -minw 6 -maxw 12 -maxsize 20000000 -revcomp, which specify the number of motifs for which to search (2), the zoops assumption (zero or one occurrence per peak sequence), a minimum motif length of 6, a maximum motif length of 12 and a maximum data set size of 20,000,000 characters. Sequences were searched in both forward and reverse complement orientations.
Cell culture and MEF generation
MEFs were prepared and were cultured as described previously21. HEK293 cells were obtained from ATCC.
Plasmid and viral vector production
Murine Chop cDNA was amplified by PCR using mouse genomic DNA and the primers shown in Supplementary Table S5. Flag–CHOP plasmid was made by transferring the confirmed Chop cDNA into the pFLAG–CMV-4 expression vector (Sigma). Adenoviral vector expressing CHOP was made using the AdEasyTM (Qbiogene). Adenovirus expressing ATF4 and ATF4ΔRK was kindly provided by R. Ratan (Cornell University, USA)27. Recombinant retroviral vectors that express enhanced green fluorescent protein (EGFP), the 299 C-terminal amino acids (299–590) of hamster GADD34 (ΔN) or the 536 N-terminal amino acids (121 C-terminal amino acid deletion) of hamster GADD34 (ΔC) were kindly provided by D. Ron (University of Cambridge, UK). Adenoviruses expressing β-gal and GFP were purchased from the Viral Vector Core at the University of Michigan.
Western blot analyses
Cells were collected in cell lysis buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate and protease inhibitors (Roche Diagnostics)). Primary antibodies were as follows: anti-caspase 3 (#9664, Cell Signaling), anti-PARP (#9542, Cell Signaling), anti-p-eIF2α (44728G, Invitrogen), anti-eIF2α (AHO1182, Invitrogen), anti-tubulin (T5168, Sigma), anti-ATF4 (M.S.K.), anti-Flag (F1804, Sigma), anti-CHOP (Sc-575, Santa Cruz), anti-GADD34 (Sc-825, Santa Cruz) and anti-KDEL (ab12223, Abcam).
Immunoprecipitation
HEK293 cells that overexpress Flag–CHOP and/or ATF4 were collected in lysis buffer (20 mM Tris–HCl (pH 7.5), 300 mM NaCl, 1% NP-40, 10% glycerol, 0.5 mM EDTA and protease inhibitors). Supernatants were immunoprecipitated with 5 μg of anti-Flag, anti-ATF4 or anti-CHOP overnight and then incubated with protein A beads for another 1 h at 4 °C. Individual proteins were detected as described for western blot analyses.
Cell viability assay
Relative cell viabilities were determined using either Cell counting kit-8 (Dojindo) or CellTiter-Glo Luminescent Cell Viability Assay (Promega). Relative cell viability was determined using a microplate reader (Molecular Devices).
Measurements of protein synthesis by metabolic labelling with [35S] methionine and cysteine
After pre-incubation for 15 min, medium was replaced with labelling medium with 125 μCi [35S]cysteine/methionine ml–1 (TRANS35S-LABEL, MP Biomedical)] for 15 min. Cell lysates were taken for measurements of total protein synthesis by trichloroacetic acid precipitation on Whatman filter paper. Radiolabelled lysates were assayed for total protein using the Bio-Rad DC protein assay kit.
Gene expression analysis by quantitative real-time (qRT)–PCR
The relative amounts of mRNA were calculated from the comparative threshold cycle (Ct) values relative to β-actin. Real-time primer sequences are shown in Supplementary Table S7.
siRNA knockdown experiments
siRNAs were purchased from Invitrogen (Ero1α) and Dharmacon (Rpl24 and Rpl7). siRNAs (200 pmol) were transfected using the MEF nucleofactor kit 1 (AMAXA) with program T-20.
Islet morphology and immunohistochemistry
Insulin and glucagon were identified using guinea-pig anti-insulin antibody (4011-01F, Linco) with goat anti-guinea-pig Texas red as the secondary antibody (106-075-003, Jackson Immuno Research) and rabbit anti-glucagon antibody (4030-01F, Linco) with goat anti-rabbit Alexa Fluor 488 as the secondary antibody (A11008, Invitrogen), respectively. For measurement of islet area, the slides were observed under a digital virtual microscope (dotSlide; Olympus) at the UNIST-Olympus Biomed Imaging Center, and the islet area was measured using dotSlide software.
Quantification of carbonylated proteins
Adenoviruses (1011 viral particles) were administered into mice intravenously. At 3 days after injection, protein carbonyls in the liver were measured according to the manufacturer's instruction (Cell Biolabs).
Flow cytometry analysis
Cells were loaded with CM-H2DCFFDA (Molecular Probe) for 30 min followed by recovery for 30 min before fluorescence measurements. A FACS Caliber (BD) and Flowjo software were used for analysis.
In vivo measurement of protein synthesis using 2H2O labelling of protein-bound alanine
Mice were treated as described in each experiment. Liver, heart and plasma were obtained and processed as described before50. GC–MS analysis provided the 2H-labelling of body water percentage enrichment and 2H-labelled protein percentage enrichment for each mouse plasma and liver tissue, respectively. These data were used to calculate the protein synthesis rate, percentage of newly made per hour, as previously described50.
Animal experiments
Atf4–/– and Atf4–/– mice were previously described64. For adenovirus injection, 2- to 3-month-old male mice were used. A/A;fTg/0 and A/A;fTg/0;RIP-CreER/0 mice were described previously43. Three- to five-month-old male mice were used for deletion of fTg in pancreatic β cells as described previously43. Animals were housed with 12 h light and 12 h dark cycles in the Unit for Laboratory Animal Medicine at the University of Michigan and SBMRI. All animal care and procedures were conducted according to the protocols and guidelines approved by the University of Michigan Committee on the Use and Care of Animals or the SBMRI Institutional Animal Care and Use Committee. Blood glucose was measured using a OneTouch Ultra glucometer (LifeScan). Glucose tolerance tests were performed on mice fasted for 5–6 h, followed by intraperitoneal injection of indicated amounts of glucose (2 mg kg–1 body weight).
Statistical analysis
All data are presented as the means ± s.e.m. Statistical significance of the difference between groups was evaluated using Student's t-test. P < 0.05 was considered significant. * P < 0.05, ** P < 0.01.
All of the next-generation sequencing results were deposited in GEO with the accession number GSE35681 and the raw data are available from SRA as SRP010861.
Supplementary Material
Table 1
Table 2
Table 3
Table 4
Table 5
Table 6
Table 7
08
ACKNOWLEDGEMENTS
We thank the University of Michigan Sequencing Core for next-generation sequencing, D. Ron (University of Cambridge, UK) for recombinant retroviral vectors expressing GADD34 derivatives and GFP and Ratan (Cornell University, USA) for adenovirus expressing ATF4 and ATF4ΔRK. We are grateful to members of the R.J.K. laboratory for assistance, advice and stimulating discussions. This work was supported by a University of Michigan CCMB Pilot Grant (J.H.), NIH grants (HL057346, DK042394, DK088227, HL052173, DK093074 (R.J.K.)), (DK092062, DK094729 (M.S.K.)), (DK60596, DK53307 (M.H.)), and the National Research Foundation of Korea (NRF) grants 2010-0001199, 2011-0011433, 2012M3A9C3048686 (S.H.B.).
Footnotes
AUTHOR CONTRIBUTIONS
J.H., S.H.B. and R.J.K. designed all experiments and performed most of them. J.H., Y-H.L and M.A.S. performed bioinformatic analysis and contributed to preparation of figures and tables. R.G. performed western blot and cell viability assays using Eif2αA / A and Atf4–/– MEFs and GADD34 overexpression experiments. J.S. and M.S.K. generated and characterized the ATF4 antibody and performed co-immunoprecipitation and sequential ChIP experiments. C.L.Y., D.K. and M.H. measured in vivo protein synthesis using 2H2O. S.W. analysed protein synthesis and western blots. J.H., S.H.B. and R.J.K. prepared the manuscript.
Supplementary Information is available in the online version of the paper
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb2738
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3692270?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Concerning influences of micro/nano plastics on female reproductive health: focusing on cellular and molecular pathways from animal models to human studies.
Reprod Biol Endocrinol, 22(1):141, 11 Nov 2024
Cited by: 0 articles | PMID: 39529078 | PMCID: PMC11552210
Review Free full text in Europe PMC
ATF4/NUPR1 axis promotes cancer cell survival and mediates immunosuppression in clear cell renal cell carcinoma.
Discov Oncol, 15(1):607, 31 Oct 2024
Cited by: 0 articles | PMID: 39480570 | PMCID: PMC11528094
Metabolic RNA Labeling and Translating Ribosome Affinity Purification for Measurement of Nascent RNA Translation.
Bio Protoc, 14(20):e5091, 20 Oct 2024
Cited by: 0 articles | PMID: 39512885 | PMCID: PMC11540052
Integrated stress response plasticity governs normal cell adaptation to chronic stress via the PP2A-TFE3-ATF4 pathway.
Cell Death Differ, 30 Sep 2024
Cited by: 0 articles | PMID: 39349971
The cytoplasmic-nuclear transport of DDX3X promotes immune-mediated liver injury in mice regulated by endoplasmic reticulum stress.
Cell Death Dis, 15(9):702, 30 Sep 2024
Cited by: 0 articles | PMID: 39349420 | PMCID: PMC11442484
Go to all (1,014) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE35681
Nucleotide Sequences
- (1 citation) ENA - SRP010861
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Translational and posttranslational regulation of XIAP by eIF2α and ATF4 promotes ER stress-induced cell death during the unfolded protein response.
Mol Biol Cell, 25(9):1411-1420, 12 Mar 2014
Cited by: 74 articles | PMID: 24623724 | PMCID: PMC4004591
The role of de novo protein synthesis and SIRT1 in ER stress-induced Atf4 and Chop mRNA expression in mammalian cells.
Biochimie, 138:156-167, 04 May 2017
Cited by: 7 articles | PMID: 28478109
The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.
Curr Mol Med, 16(6):533-544, 01 Jan 2016
Cited by: 370 articles | PMID: 27211800 | PMCID: PMC5008685
Review Free full text in Europe PMC
Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α.
J Biol Chem, 289(18):12593-12611, 19 Mar 2014
Cited by: 91 articles | PMID: 24648524 | PMCID: PMC4007450
Funding
Funders who supported this work.
NHLBI NIH HHS (4)
Grant ID: HL057346
Grant ID: R01 HL052173
Grant ID: HL052173
Grant ID: P01 HL057346
NIDDK NIH HHS (16)
Grant ID: DK088227
Grant ID: DK093074
Grant ID: DK53307
Grant ID: R01 DK094729
Grant ID: R01 DK060596
Grant ID: DK094729
Grant ID: R37 DK060596
Grant ID: DK042394
Grant ID: R01 DK042394
Grant ID: R01 DK053307
Grant ID: DK60596
Grant ID: R01 DK088227
Grant ID: R24 DK093074
Grant ID: R37 DK042394
Grant ID: DK092062
Grant ID: R01 DK092062





