Abstract
Free full text

Role of Human Immunodeficiency Virus Type 1 Matrix Phosphorylation in an Early Postentry Step of Virus Replication
Abstract
The matrix domain (MA) is important for targeting of human immunodeficiency virus type 1 Gag assembly to the plasma membrane, envelope incorporation into virions, preintegration complex import into the nucleus, and nuclear export of viral RNA. Myristylation and phosphorylation are key regulatory events for MA function. Previous studies have indicated that MA phosphorylation at serine (Ser) residues is important for viral replication. This study defines the molecular mechanisms of virus particle assembly and infectivity through a detailed study of the role of MA serine phosphorylation. We show that the combined mutation of Ser residues at positions 9, 67, 72, and 77 impairs viral infectivity in dividing and nondividing cells, although the assembly of these Ser mutant viruses is comparable to that of wild-type virus. This defect can be rescued by pseudotyping these mutant viruses with vesicular stomatitis virus G protein, suggesting that these serine residues are critical in an early postentry step of viral infection. The phosphorylation level of MA in defective mutant viruses was severely reduced compared to that of the wild type, suggesting that phosphorylation of Ser-9, -67, -72, and -77 is important for an early postentry step during virus infection.
The matrix protein (MA) of human immunodeficiency virus type 1 (HIV-1) is derived by proteolytic cleavage of the N-terminal part of the Gag precursor Pr55Gag (10). Virus assembly requires plasma membrane association of Pr55Gag, Gag oligomerization, raft association, and the release of virus particles mediated by components of the vacuolar protein-sorting pathway (6, 11, 19, 26, 28, 29). The viral envelope glycoprotein and genomic RNA accumulate with Pr55Gag and the Gag-Pol precursor proteins at the site of assembly and bud from infected cells. Particle maturation occurs in newly budded virions when a viral protease cleaves Gag into MA, capsid (CA), nucleocapsid (NC), and p6 proteins as well as two small peptides. HIV-1, like other lentiviruses, infects both dividing and nondividing cells. Upon entry into the host cell, uncoating of the virus core is followed by reverse transcription of the viral RNA into cDNA, active transport of the viral cDNA into the nucleus, and integration of the cDNA into the host genome. The MA protein plays a major role in all of these steps of the virus life cycle.
MA is a 132-amino acid (aa) structural protein that is myristylated at the N terminus. The three-dimensional structure of MA has been determined by nuclear magnetic resonance as well as X-ray crystallography (22, 27), and it consists mainly of five α-helices, of which helices 1 to 4 form a compact globular domain while the C-terminal helix (helix 5) projects away from the membrane. The N-terminal myristyl moiety facilitates binding of MA to the membrane. Membrane binding also depends on an electrostatic interaction between the basic domains of MA, which cluster about an extruded cationic loop that connects β-strands 1 and 2, and acidic phospholipids in the inner phase of the lipid bilayer.
MA is required for the incorporation of the envelope glycoprotein into the virion (5, 7, 36). Mutation at MA residue Leu-12 or Leu-30 blocks envelope incorporation in the virion (15). However, this defect is rescued by truncation of the cytoplasmic tail of the Env transmembrane protein gp41 (14, 15). In addition, the highly basic domain of MA (aa 17 to 31) and residues 84 to 88 contain a major determinant for HIV-1 Gag plasma membrane targeting (30). MA is also important in early postentry events of the virus life cycle. Mutation of a highly conserved Leu at MA aa 20 or a deletion at the C terminus causes a significant defect in an early step in the virus life cycle (24, 35). As a component of the preintegration complex (PIC), some studies have suggested that MA is required for PIC transport to the nucleus. Although two nuclear localization signals have been identified in MA (3, 20), it remains controversial whether they are able to carry out this function alone (9, 13).
In addition to myristylation, phosphorylation has been shown to be a critical regulator of MA functions. It was initially proposed that MA was phosphorylated at Tyr-132 and regulated the nuclear localization of the PIC (17, 18). However, other groups were unable to confirm this finding (2, 12). MA is also phosphorylated at serine residues, as protein kinase C was identified as one of the kinases for MA phosphorylation and Ser-111 was identified as a putative phosphoacceptor residue (4). However, the role of this modification in virus replication has not been defined. Bukrinskaya and colleagues reported that at least five different serines are phosphorylated during HIV-1 entry into susceptible cells (2). By using kinase inhibitors, they observed that MA phosphorylation at serine or tyrosine residues regulates nuclear targeting of virus nucleic acids. In addition, Nef has been shown to enhance MA phosphorylation through a Nef-associated serine-threonine kinase (34). However, these studies failed to identify the phosphoacceptor serines and the functions of the modifications of each residue.
The present study examines the contribution of each serine residue to the regulation of virus replication. We found that the mutation of all serine residues other than Ser-6 has no effect on virus assembly. However, mutations at four different serine residues (Ser-9, Ser-67, Ser-72, and Ser-77) impair virus replication. The phosphorylation status of these crucial serine residues was examined, and a direct role of MA phosphorylation is implicated in the virus life cycle at an early postentry step.
MATERIALS AND METHODS
Cells and virus.
293T cells, used for all proviral transfections, were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum and 100 U of penicillin-streptomycin/ml at 37°C in 5% CO2. MAGI-5 cells, which are HeLa reporter cells expressing CD4 and CCR5 and containing the β-galactosidase gene under the control of the HIV-1 promoter, were cultured in the same medium supplemented with puromycin, hygromycin, and G148 (31). PM1 cells were grown in RPMI 1640 (Invitrogen) medium supplemented with 10% fetal bovine serum. Human monocyte-derived macrophages, isolated from whole human blood, were prepared by adherence to plastic followed by culturing in RPMI 1640 medium with 10% human serum and 500 U of macrophage colony-stimulating factor (R&D Laboratories)/ml.
Wild-type (WT) and mutant virus stocks were generated by transfection of 293T cells with proviral clones by use of TransIT-LTI (Mirus Corp.), harvested after 48 h, and normalized for p24Gag content by use of a commercially available p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (Beckman Coulter).
Plasmid construction.
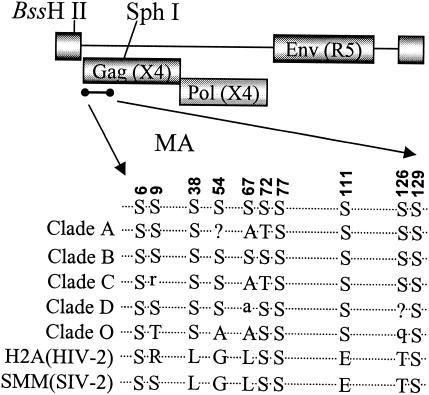
Proviral clones were derived from the HIV-1 NL4-3 strain with a substitution of the YU-2 envelope to generate a macrophage-tropic virus. Serine residues in the BssHII-SphI fragment (nucleotides [nt] 711 to 1446) of the MA region of the proviral clone were mutated to alanine singly or in different combinations by use of a PCR-based site-directed mutagenesis system (23). The mutant fragments were inserted back into the pNL4-3 clone and confirmed by sequence analysis. The primers used for the mutagenesis are available upon request.
Immunoprecipitation.
293T cells transfected with proviral clones were labeled for 16 h with Trans35S-labeling mix (>1,000 Ci/mmol) (ICN Radiochemicals) in Cys- and Met-free DMEM supplemented with 10% dialyzed fetal bovine serum and penicillin-streptomycin. The supernatants were clarified by centrifugation (500 × g, 5 min). Cell lysates and supernatants were prepared in 1× lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, and 1% Triton X-100), and the samples were incubated overnight with AIDS patient sera or rabbit polyclonal sera against the MA protein. The antigen-antibody complexes were precipitated with protein A-agarose beads (Repligen Corp.) for 1 h at 4°C and were washed three times with RIPA buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]). The antigen-antibody complexes were resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer (50 mM Tris-Cl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glucose), heated to 100°C for 3 min, electrophoresed in a 12 to 17.5% gel, and visualized by autoradiography.
Infectivity assay.
Single-cycle infections were carried out with MAGI-5 cells as described previously (31). Briefly, MAGI-5 cells were seeded at 104 cells/well in 96-well plates and infected for 3 h in triplicate with each of three different dilutions of virus. At 24 to 48 h postinfection, the cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 45 min. The substrate was washed off with phosphate-buffered saline and the number of blue cells was counted in each well. Standard errors were calculated for five independent experiments. The virus titers were normalized to the amount of p24. For macrophage infections, the cells were incubated with 20 ng of a p24 equivalent of virus for 24 h, washed, and returned to fresh medium. Supernatants were collected every 3 to 4 days for 6 weeks, and reverse transcriptase (RT) activity was measured as described by Poiesz et al. (32).
Envelope incorporation assay.
After transfection of HeLa cells with proviral clones and labeling of cells with a Trans35S-label as described above, the supernatants were cleared of cell debris by centrifugation (500 × g, 5 min) and the virus particles were pelleted from the supernatant by centrifugation through a 20% sucrose cushion in an SW55Ti rotor (Beckman Coulter) at 45,000 rpm for 45 min at 4°C. Virus particles were resuspended in 1× lysis buffer and immunoprecipitated with AIDS patient sera as described above.
VSV-G and MLV Env pseudotyping assays.
Vesicular stomatitis virus G protein (VSV-G) pseudotyped virions were produced by cotransfection of 293T cells with 500 ng of VSV-G expressed with plasmid pHCMVg along with 2 μg of the proviral clone. Similarly, WT and mutant virions were pseudotyped by expression of the amphotropic murine leukemia virus (A-MLV) Env from pMLVenv (25). The cleared supernatants were used as the source of virus for the MAGI-5 assay described above.
PCR analysis of viral cDNA synthesis.
Newly synthesized viral cDNAs in infected cells were analyzed as described previously (2). Briefly, virus supernatants were treated with DNase I (Worthington Biochemical Corp.) prior to infection to remove residual proviral plasmid DNA. PM1 cells (5 × 106) were infected with 50 ng of p24 from the viral supernatant. Cells (106) were collected at different time points, and the total cellular DNA was extracted by use of a blood DNA mini kit (Qiagen, Valencia, Calif.). Late reverse transcription products were amplified by using the specific primers R (5′ G485GGAGCTCT CTGGCTAACT) and gag (5′ G912GATTAACTGCGAATCGTTC). The PCR conditions were as follows: first denaturation at 94°C for 3 min; 35 cycles of 94°C for 30 s, 66°C for 30 s, and 73°C for 30 s; and a final extension at 73°C for 5 min. PCRs were performed in the presence of [α-32P]dCTP (6,000 Ci/mmol) (ICN Radiochemicals). Samples were electrophoresed in 6% nondenaturing polyacrylamide gels, and autoradiography was performed.
Phosphorylation studies.
For phosphorylation studies, 293T cells were transfected with proviral clones containing WT or mutant MA sequences. At 24 h posttransfection, the cells were labeled for 16 h with 0.5 mCi of [32P]phosphoric acid (ICN Radiochemicals)/ml in phosphate-free DMEM containing Ser-Thr phosphatase inhibitors (20 nM okadaic acid, 50 nM cantharidic acid, and 100 nM calyculin A [Sigma, St. Louis, Mo.]). Cellular supernatants were cleared of debris by centrifugation (500 × g, 5 min) and then were centrifuged through a 20% sucrose cushion to pellet virus particles. The virus samples were lysed in 1× lysis buffer containing phenylmethylsulfonyl fluoride, aprotinin (Sigma), and phosphatase inhibitors and were immunoprecipitated with rabbit polyclonal MA antibodies. After three washes with RIPA buffer, the samples were electrophoresed by SDS-17.5% PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was autoradiographed and subsequently subjected to Western blotting with a rabbit polyclonal MA antibody to determine the amount of MA present in each sample.
RESULTS
Construction of mutants.
There are 10 serine residues present in the MA protein of strain pNL4-3. Most of these serines are conserved among different strains of HIV-1 (Fig. (Fig.1).1). The serines at positions 6, 9, 38, 72, 77, 111, 126, and 129 are more conserved than those at residues 54 and 67. The serines at positions 6, 72, 77, and 129 are also present in HIV-2 and simian immunodeficiency virus MA proteins. The serine at position 6 is important for MA myristylation and therefore for the plasma membrane association of Gag (33). Mutants were generated with the proviral clone of pNL4-3 with a substitution of the YU-2 envelope gene, resulting in a macrophage-tropic strain. Serine residues were changed to alanines as single mutations or in different combinations.
Mutations at serine residues other than Ser-6 have no deleterious effects on viral assembly and maturation.
The production of viruses containing serine mutations was analyzed by transfection of 293T cells with the proviral clones and estimation of the amount of p24Gag in the cell supernatants by p24 antigen ELISA (Fig. (Fig.2).2). The particle production of all the mutants was comparable to that of the WT, except for combinatorial mutants 2S (S126/129A) and 3Sa (S111/126/129A). For assessment of Gag processing, transfected 293T cells were metabolically labeled overnight and cell lysates and supernatants were immunoprecipitated with AIDS patient sera (Fig. (Fig.3).3). No defect in the assembly or processing of Gag was observed for any of the mutants, as was evident by the similar amounts of viral proteins and cleaved Gag polyproteins. It was previously reported that mutation of Ser-6 inhibits the production of virus particles (16); thus, the combinatorial mutant 10S produced no detectable virus (Fig. (Fig.22 and and3).3). The efficiency of virus assembly was assessed by comparing the ratio of immunoprecipitated p24 Gag in the supernatant to Gag protein synthesized in transfected cells. The ratios were equivalent for all the mutants and for WT NL4-3, with the exception of mutant 10S (data not shown). This indicates that the mutation of any serine residue other than residue 6 does not deleteriously affect virus assembly or Gag processing.
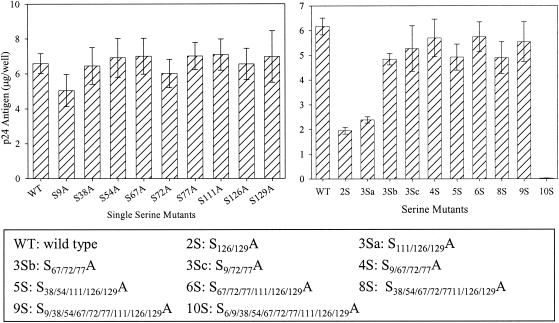
Relative production of virus particles of WT and serine mutants. 293T cells were transfected with the WT or with mutant proviral clones, and the supernatants were harvested at 48 h posttransfection. The amount of virus made from each mutant was measured by determining the p24 antigen level from the culture supernatant by quantitative ELISA. The bars represent standard errors for four different experiments. Locations of Ser-Ala substitutions for each mutant are indicated.
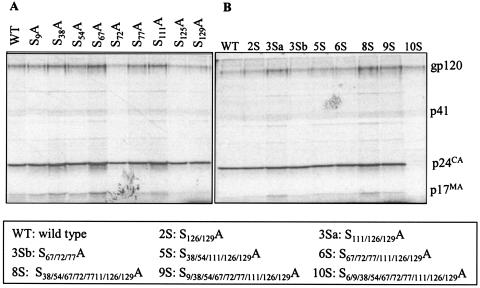
Radioimmunoprecipitation analysis of virion-associated proteins. 293T cells were transfected with the indicated molecular clones and metabolically labeled with a Trans35S-label. Supernatants containing virus particles were immunoprecipitated with AIDS patient sera. The positions of Env glycoprotein (gp120), p41 (partially cleaved Gag), p24 (CA), and p17 (MA) are indicated. (A) Single mutations. (B) Multiple mutations.
Serines 9, 67, 72, and 77 are critical for infection of dividing and nondividing cells.
To test the effects of serine mutations on virus replication in dividing cells, we infected MAGI-5 cells with similar amounts of p24 Gag virus in single-cycle virus replication assays. All mutant viruses with single serine mutations had infectivity titers of at least 70% that of WT virus (Fig. (Fig.4A).4A). Viruses with mutations at multiple Ser residues were also tested (Fig. (Fig.4B).4B). Viruses 2S (S126/129A), 3Sa (S111/126/129A), and 5S (S38/54/111/126/129A) exhibited similar levels of infectivity as the WT virus. However, viruses 3Sb (S67/72/77A), 6S (S67/72/77/111/126/129A), and 8S (S38/54/67/72/77/111/126/129A) exhibited infectivities of approximately 20% that of WT virus. Together, these results indicate that Ser-67, -72, and -77 are critical for virus infection. Moreover, virus 9S (S9/38/54/67/72/77/111/126/129A) exhibited an infectivity of <5% that of WT virus, suggesting that Ser-9 is also critical for infection in vitro. Viruses 4S (S9/67/72/77A) and 3Sc (S9/72/77A) were constructed subsequently, altering only those serine residues which might be deleterious for virus replication. The assembly and Gag processing of both of these viruses were similar to those of WT virus (data not shown). Both of these viruses showed an infectivity of <5% that of WT virus, similar to virus 9S (see Fig. Fig.7A).7A). Virus 10S, with mutations at all 10 Ser residues, produced very low levels of virus particles (Fig. (Fig.22 to to3)3) and no detectable infection of MAGI-5 cells.
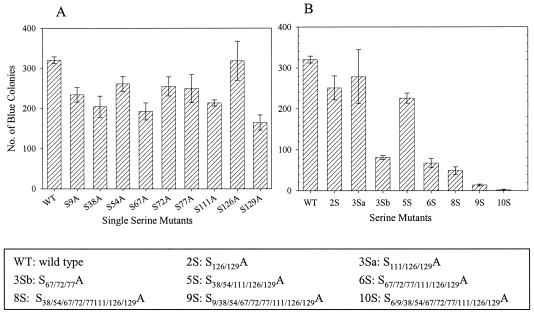
Single-cycle infection assays. MAGI-5 cells were infected with WT or mutant viruses with equivalent amounts of p24 in three different dilutions. At 40 h postinfection, cells were fixed and incubated with β-galactosidase substrate. Blue colonies were counted for each mutant and plotted on the y axis. The error bars represent values for triplicate samples from five independent experiments. (A) Single mutations. (B) Multiple mutations.
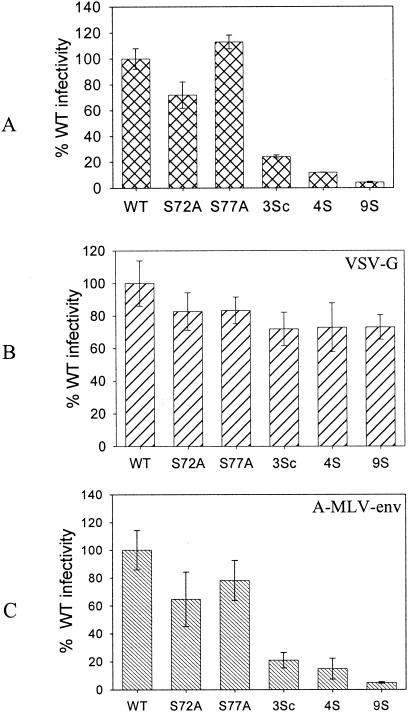
Relative infection of pseudotyped MA mutant viruses. Proviral clones were cotransfected with either a VSV-G or A-MLV Env expression plasmid to make pseudotyped particles. MAGI-5 cells were used to determine the relative levels of infection. (A) Nonpseudotyped viruses. (B) Viruses pseudotyped with VSV-G. (C) Viruses pseudotyped with A-MLV Env. 3Sc virus, S9/72/77A. The other mutants are described in the legend for Fig. Fig.33.
Human macrophages were used to examine the replication of mutant viruses in nondividing primary cells (Fig. (Fig.5).5). The pattern of infection of the mutant viruses in macrophages was similar to that in MAGI-5 cells. Virus 3Sa (S111/126/129A) showed replication in macrophages that was similar to that of WT virus. Viruses 6S (S67/72/77/111/126/129A) and 8S (S38/54/67/72/77/111/126/129A) showed impaired replication in macrophages compared to WT virus. Virus 9S (S9/38/54/67/72/77/111/126/129A) exhibited the most impaired replication in macrophages. No detectable replication in macrophages was observed with virus 10S. Together, these results suggest that serines at positions 9, 67, 72, and 77 are important for virus replication in both dividing and nondividing cells.
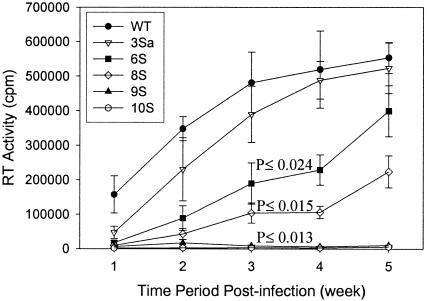
Relative replication of WT and serine mutant viruses in macrophages. Human macrophages were infected with WT or mutant viruses with equivalent amounts of p24. The culture supernatants were collected every week, and the virus replication was determined by measuring the RT activity in those supernatants. P values were determined by a two-sided t test comparing RT values for mutants with those from infection with WT virus.
Envelope incorporation is unaffected in viruses with MA Ser mutations.
Viruses 4S (S9/67/72/77A) and 9S (S9/38/54/67/72/77/111/126/129A) exhibited >90% decreases in infection of MAGI-5 cells and macrophages. This result could be due to a defect in an entry, uncoating, or nuclear import process that depends upon MA functioning. To determine if mutations in MA affected entry, possibly through effects on MA interactions with the envelope, we examined envelope glycoprotein incorporation into mutant 4S and 9S virus particles. Although the results shown in Fig. Fig.3B3B demonstrated that the gp120 envelope was released into the cell supernatant, these results do not differentiate free gp120 from virus-associated gp120. Indeed, the detection of gp120 in the supernatant from cells transfected with mutant clone 10S, despite the lack of detectable released Gag protein, indicates that free gp120 is released from transfected cells. For examination of the level of virus-associated gp120 envelope protein, culture supernatants from transfected and metabolically labeled HeLa cells were pelleted through a 20% sucrose cushion. The virus pellet was resuspended, immunoprecipitated with AIDS patient sera, and electrophoresed, and gp120 was visualized by autoradiography. The Env incorporation in 4S and 9S particles was comparable to that in the WT virus (Fig. (Fig.6).6). A Δenv virus, with the luciferase gene in place of HIV-1 env, was used as a negative control. The absence of detectable gp120 in the 10S lane rules out contamination with free gp120 derived from the supernatant. Therefore, these results suggest that the defect observed does not result from the failure of Env incorporation in virions.
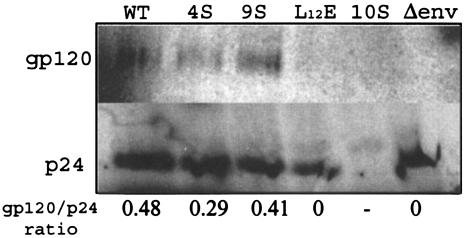
Env incorporation in WT and mutant virions. HeLa cells were transfected with WT and mutant proviral constructs and labeled with a Trans35S-label, and viruses in the culture supernatants were pelleted through a 20% sucrose cushion. The pelleted viruses were lysed, immunoprecipitated with AIDS patient sera, and separated by SDS-PAGE.
VSV-G pseudotypes of defective serine mutant viruses overcome the infection defect.
It has been shown that Nef-defective mutants can be rescued when pseudotyped with VSV-G (1). For testing of similar effects, the defective serine mutant viruses were pseudotyped with VSV-G by cotransfection of proviral clones with a plasmid expressing VSV-G. All of the pseudotyped mutant viruses were as infective as pseudotyped WT virus in a single-round infection assay with MAGI-5 cells (Fig. (Fig.7).7). VSV-G-pseudotyped viruses were several times more infectious than nonpseudotyped viruses, and hence smaller amounts of VSV-G-pseudotyped viruses were used to measure the infection in order to remain within the linear range of the assay. As a control, these viruses were also pseudotyped with the A-MLV envelope. The pseudotyped viruses were two to three times more infectious than the corresponding nonpseudotyped viruses. However, the relative levels of infection of the A-MLV-pseudotyped MA mutants or WT viruses were similar to those of the nonpseudotyped viruses. This was expected, as A-MLV-pseudotyped viruses resulted in pH-independent fusion, similar to that utilized by the HIV envelope glycoprotein. These results indicate that the VSV-G-mediated entry pathway, which employs pH-dependent endocytosis, rescues defective mutant viruses. These observations suggest that the viruses with serine mutations are defective in an early postentry step of the HIV-1 life cycle.
MA serine mutant viruses have a defect in viral cDNA synthesis.
For examination of the early events in HIV entry, the synthesis of viral cDNAs of MA Ser mutant viruses was measured by radiolabeled PCR using long terminal repeat-Gag-specific primers (Fig. (Fig.8)8) (21). Mutant viruses 4S and 9S were found to be defective in viral cDNA production in comparison to WT virus, for which a 427-bp PCR product was evident as early as 6 h after infection (Fig. (Fig.8).8). This demonstrates that the MA Ser mutant viruses 4S and 9S have a defect in an early postentry step of virus replication.
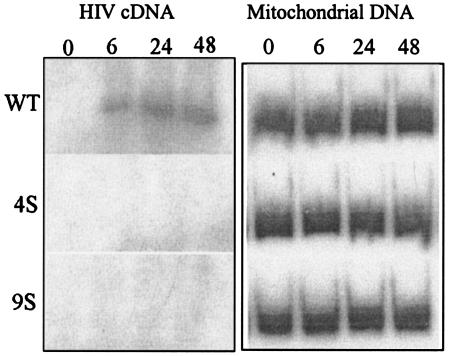
PCR amplification of viral DNA at early times postinfection. PM1 cells were infected with WT or mutant viruses, and equal numbers of cells were harvested at the indicated times. Total DNA was purified from the samples, and PCR was performed with the R and gag specific primers (sequence positions are described in Materials and Methods) in the presence of [α-32P]dCTP. The PCR products were electrophoresed in 6% polyacrylamide gels and autoradiographed.
Mutant viruses exhibit hypophosphorylation of MA.
For determination of the cause of defects associated with mutation of serine residues 9, 67, 72, and 77, the phosphorylation status of the MA mutants was determined. Cells transfected with proviral clones were labeled with [32P]phosphoric acid, and virus particles pelleted from the supernatants were analyzed for MA phosphorylation. The amount of MA protein in each lane was quantitated by Western blotting with a polyclonal MA antiserum. As shown in Fig. Fig.9,9, mutant virus 3Sb (S67/72/77A), 3Sc (S9/72/77A), or 4S (S9/67/72/77A) exhibited markedly reduced levels of MA phosphorylation compared to WT virus or virus 3Sa (S111/126/129A). There was no significant difference in the levels of MA protein (Fig. (Fig.9B).9B). The alteration of single serine residues did not affect the level of phosphorylation (data not shown). The extent of phosphorylation was calculated by comparing band intensities of different samples and normalizing them with the amount of MA in each lane. Virus 4S MA was 5% phosphorylated compared to the WT, whereas the percent phosphorylation for the 3Sb and 3Sc MA proteins was 20 and 10%, respectively. These findings suggest that Ser-9, -67, -72, and -77 are phosphoacceptor residues. The explanation for the presence of two 32P-labeled bands for MA is unclear, but similar results were observed in another study (2).
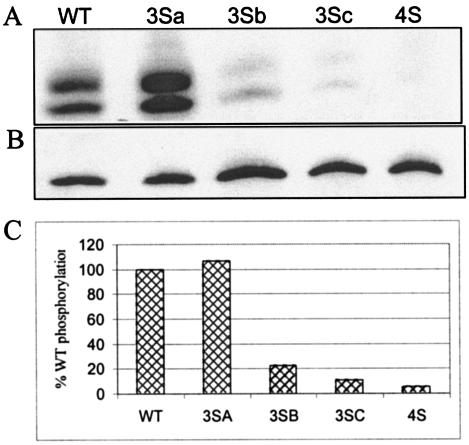
Phosphorylation of HIV-1 MA. 293T cells were transfected with WT or mutant proviral clones. Transfected cells were labeled with [32P]phosphoric acid overnight in the presence of Ser-Thr phosphatase inhibitors. The labeled culture supernatants were spun through a 20% sucrose cushion to pellet virus particles. The samples were lysed in RIPA buffer, immunoprecipitated with anti-MA antibodies, separated by SDS-17.5% PAGE, and electroblotted onto a polyvinylidene difluoride membrane. Phosphorylation was analyzed by autoradiography (A and C), and the amount of MA present in each sample was determined by Western blotting with an anti-MA antibody (B).
DISCUSSION
Our studies further define the role of MA phosphorylation in virus infectivity. Serine and tyrosine residues are phosphoacceptor residues in the MA protein (2). Earlier observations in our laboratory confirmed that serines are the major phosphoacceptor residues in MA (R. Horton and L. Ratner, unpublished observations). To define the serine residues involved in the regulation of HIV infection, we mutated all 10 serine residues to alanines singly or in combination. The serine at position 6 is required for myristylation, and mutation of this residue disrupts virus assembly. In contrast, mutation of the other nine serine residues alone or in combination did not affect virus assembly. Furthermore, the particle density of mutant 9S, with mutations at all serine residues but Ser-6, was similar to that of the WT (data not shown). No alteration in Gag or Env packaging into particles or in processing was detected. However, mutation of the serines at positions 9, 67, 72, and 77 resulted in a profound defect in virus infection of MAGI-5 cells and macrophages. These serine residues are among the most conserved residues in HIV MA (Fig. (Fig.1).1). Nevertheless, single serine-to-alanine mutations at these positions do not significantly affect virus infectivity, suggesting an additive effect of these residues.
Several key steps of the viral life cycle were examined in order to identify the defect in viral infection of the serine mutants. Env incorporation into virus particles is mediated by MA, but the MA serine mutant viruses had comparable amounts of virion-associated envelope glycoprotein compared to WT virus. Although envelope oligomerization, receptor and coreceptor binding, conformational changes, and activation of the fusion process were not examined and cannot be ruled out, there is no precedent for a role of MA in these activities. Pseudotyping of the mutant virions with VSV-G rescued the defect of serine mutants in MAGI-5 cells, suggesting that MA acts at an early step of the HIV-1 life cycle that is either circumvented or facilitated by targeting of virus entry to the endocytic pathway. Mutants pseudotyped with A-MLV Env remained defective, confirming the earlier observations, as the MLV Env-mediated entry pathway is similar to that of HIV-1.
Viral cDNA made by RT after uncoating of the virus core was not observed for the defective MA mutant viruses, suggesting that the defect lies in or prior to the uncoating event. Earlier studies by Bukrinskaya and colleagues suggested that the nuclear transport of viral nucleic acids was defective in the presence of protein kinase inhibitors (2). However, their studies were carried out with a hypophosphorylated virus deficient in phosphorylation of MA and other viral proteins.
MA phosphorylation in the defective serine mutant viruses 3Sb, 3Sc, and 4S was found to be severely reduced compared to that in the WT virus, suggesting a phosphorylation-dependent role of MA during infection. The phosphorylation of single serine mutants was found to be at a level similar to that of the WT virus (data not shown), suggesting that there is an additive effect of multiple serine mutations. A three-dimensional structure was generated (Fig. (Fig.10)10) for monomeric MA with coordinates available from its crystal structure (22). The presence of basic residues, essential for membrane binding, and serines 9, 67, 72, and 77 on the putative membrane-binding surface suggests that phosphorylation at multiple sites might change the overall charge balance on this surface of MA, which may be a prerequisite for its release from the plasma membrane. Our observations confirm and extend the hypothesis that phosphorylation of MA might disrupt the electrostatic interaction between positively charged residues of MA and anionic phospholipid head groups in the membrane that is necessary for successful uncoating of the virus core (2). Although these defects indicate a role of MA phosphorylation in the postentry step, changes in the MA conformation or alterations in protein-protein interactions have not been ruled out. A direct correlation between MA phosphorylation and a replication defect needs to be further confirmed by studying mutants in which the serines at positions 9, 67, 72, and 77 are altered to either threonine or aspartic acid. Changing serine residues to threonine may not alter the phosphorylation pattern, whereas altering these residues to aspartate would add negative charges required for membrane dissociation in the postentry event. Further efforts will characterize the phosphorylation of these residues in infected cells as well as in producer cells. Nevertheless, this is the first study to determine the role of phosphorylation of specific residues in HIV-1 MA and to define the role of these residues in virus replication.
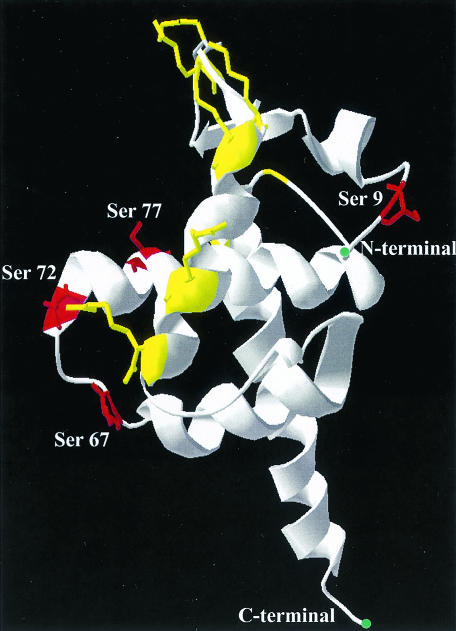
Schematic model for MA showing a view of the putative membrane-binding surface as seen from above. Serine residues are shown in red, with their positions depicted, and basic residues, essential for membrane binding, are shown in yellow. The coordinates for this structure were taken from the work of Hill et al. (22), and the model was generated with DeepView/Swiss-PdbViewer software (http://www.expasy.org/spdbv/).
Similar observations have been made with Nef-defective mutants, suggesting its role in early postentry steps, although these steps are not well understood (1). Nef has been shown to enhance MA phosphorylation by interacting with virion-associated cellular kinases (34), suggesting that MA may be a functional target of Nef, but Nef has also been shown to enhance virus infectivity in the absence of MA (8). However, the functional relationship between MA and Nef can be further examined by using phosphorylation-defective mutants of MA with a Nef deletion. Studies of the effects of different mutations at residues 9, 67, 72, and 77 may provide additional insights into their individual roles in virus infection. Moreover, the effects of MA phosphorylation on cellular protein and lipid interactions may provide a biochemical explanation for the biological effects described in this study. A further understanding of MA phosphorylation, its role in virus replication, and the responsible cellular kinases may uncover new targets for therapeutic interventions.
Acknowledgments
We thank Nancy Vander Heyden, Christopher Brooke, Lingling Jin and Robert Horton for technical help. We thank Michael Belshan for critical review of the manuscript.
This work was supported by PHS grants. R.K. is supported by an American Foundation for AIDS Research (AMFAR) grant.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.78.5.2319-2326.2004
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/78/5/2319.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.78.5.2319-2326.2004
Article citations
Current Status of Biomedical Products for Gene and Cell Therapy of Recessive Dystrophic Epidermolysis Bullosa.
Int J Mol Sci, 25(19):10270, 24 Sep 2024
Cited by: 0 articles | PMID: 39408598 | PMCID: PMC11476579
Review Free full text in Europe PMC
Evolution toward beta common chain receptor usage links the matrix proteins of HIV-1 and its ancestors to human erythropoietin.
Proc Natl Acad Sci U S A, 118(2):e2021366118, 01 Jan 2021
Cited by: 3 articles | PMID: 33372148 | PMCID: PMC7812818
Post-Translational Modifications of Retroviral HIV-1 Gag Precursors: An Overview of Their Biological Role.
Int J Mol Sci, 22(6):2871, 11 Mar 2021
Cited by: 7 articles | PMID: 33799890 | PMCID: PMC8000049
Review Free full text in Europe PMC
Toward Combined Cell and Gene Therapy for Genodermatoses.
Cold Spring Harb Perspect Biol, 12(5):a035667, 01 May 2020
Cited by: 15 articles | PMID: 31653644 | PMCID: PMC7197428
Review Free full text in Europe PMC
Cellular and viral determinants of retroviral nuclear entry.
Can J Microbiol, 62(1):1-15, 16 Oct 2015
Cited by: 24 articles | PMID: 26553381
Review
Go to all (29) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Elucidating the mechanism by which compensatory mutations rescue an HIV-1 matrix mutant defective for gag membrane targeting and envelope glycoprotein incorporation.
J Mol Biol, 427(6 pt b):1413-1427, 07 Feb 2015
Cited by: 20 articles | PMID: 25659909 | PMCID: PMC4844178
HIV-1 Matrix Trimerization-Impaired Mutants Are Rescued by Matrix Substitutions That Enhance Envelope Glycoprotein Incorporation.
J Virol, 94(1):e01526-19, 12 Dec 2019
Cited by: 22 articles | PMID: 31619553 | PMCID: PMC6912099
Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle.
J Virol, 72(5):4116-4126, 01 May 1998
Cited by: 106 articles | PMID: 9557701 | PMCID: PMC109641
Mutation of critical serine residues in HIV-1 matrix result in an envelope incorporation defect which can be rescued by truncation of the gp41 cytoplasmic tail.
Virology, 384(1):233-241, 06 Dec 2008
Cited by: 16 articles | PMID: 19059618 | PMCID: PMC2651518