Abstract
Free full text

Complementary Genomic Screens Identify SERCA as a Therapeutic Target in NOTCH1 Mutated Cancer
Associated Data
SUMMARY
Notch1 is a rational therapeutic target in several human cancers, but as a transcriptional regulator, it poses a drug discovery challenge. To identify Notch1 modulators, we performed two cell-based, high-throughput screens for small-molecule inhibitors and cDNA enhancers of a NOTCH1 allele bearing a leukemia-associated mutation. SERCA calcium channels emerged at the intersection of these complementary screens. SERCA inhibition preferentially impairs the maturation and activity of mutated Notch1 receptors and induces a G0/G1 arrest in NOTCH1-mutated human leukemia cells. A small-molecule SERCA inhibitor has on-target activity in two mouse models of human leukemia and interferes with Notch signaling in Drosophila. These studies “credential” SERCA as a therapeutic target in cancers associated with NOTCH1 mutations.
INTRODUCTION
Selective expression of transcription factors directs the hierarchical specification of the hematopoietic lineage, and acquired mutations that perturb the function of these factors have a central role in leukemia pathogenesis. A prime example involves Notch1, a surface receptor that is essential for T cell progenitor specification and maturation. Acquired mutations that activate Notch1 are found in 40% to 70% of childhood and adult T-ALL (Lee et al., 2005; Mansour et al., 2006; Weng et al., 2004). Moreover, recent reports identified NOTCH1 activating mutations in 10–15% of chronic lymphocytic leukemia (CLL) (Di Ianni et al., 2009; Puente et al., 2011) and mantle cell lymphoma (Kridel et al., 2012).
Notch receptors regulate many aspects of normal development and tissue homeostasis (reviewed in (Kopan and Ilagan, 2009)). Mammalian Notch receptors are processed during maturation by a furin-like protease, leading to the formation of two, non-covalently associated subunits. Signaling is normally initiated by binding of the Notch ectodomain to a ligand of the DSL family expressed on a neighboring cell. This interaction triggers two additional, successive proteolytic cleavages in the Notch transmembrane subunit. The first, mediated by ADAM-10 or ADAM-17 (Brou et al., 2000), occurs within a juxtamembrane negative regulatory region (NRR) at a site that is protected in the Notch off state (Gordon et al., 2009; Gordon et al., 2007). This cleavage within the Notch transmembrane domain creates a short-lived intermediate that is primed for secondary cleavage by the γ-secretase complex, an event that liberates the intracellular domain of Notch1 (ICN). ICN translocates to the nucleus, associates with the DNA-binding factor RBPJ, and recruits coactivators of the Mastermind-like (MAML) family to activate expression of target genes.
Each of the proteolytic steps involved in the activation of Notch receptors is a potential therapeutic target. Indeed, γ-secretase inhibitors (GSIs) have anti-TALL activity in vitro (Weng et al., 2004) and in vivo (Cullion et al., 2009; Real et al., 2009). The GSI MK-0752 was tested in a phase I clinical trial in patients with relapsed acute leukemia (DeAngelo et al., 2006). This trial was halted, however, due to gastrointestinal toxicity thought to be related to chronic pan-Notch receptor inhibition in gut progenitor cells (Wong et al., 2004). Thus, other approaches to Notch1 inhibition are desirable.
Historically, it has been difficult to develop high-throughput assays for small molecules that disrupt protein-DNA or protein-protein interactions (Darnell, 2002). Recently, there has been renewed interest in cell-based screening to address the problem of “undruggable” targets using various approaches (Carpenter, 2007; Inglese et al., 2007; Stegmaier et al., 2004). Gene Expression-based High-Throughput Screening (GE-HTS) is a chemical genomic approach in which gene expression signatures serve as surrogates for cellular states (Hahn et al., 2008; Stegmaier et al., 2004). One application of GE-HTS is for the identification of small molecules that modulate transcriptional signatures produced by aberrantly activated transcription factors (Corsello et al., 2009). A limitation of cell-based assays is that identifying the biologically relevant target of the small molecule can be a daunting task. One way to overcome this challenge is to design multiple, integrated cell-based screens and then to focus on common emerging hits. With the availability of genome-scale cDNA collections, over-expression screens have also proven to be powerful tools to probe biological pathways and to identify the protein targets of small molecules. Here, we used complementary GE-HTS and cDNA overexpression screens to search for small- molecule modulators of Notch1 signaling in T-ALL.
RESULTS
Development of a GE-HTS Assay for Notch1 Inhibitors
Figure 1A outlines our approach. We first defined a robust Notch1 transcriptional signature for the GE-HTS assay. We selected a set of genes that defined the Notch1 activation state from genome-wide expression profiling of 7 NOTCH1-mutated T-ALL cell lines treated with the GSI compound E (Cpd E) (Palomero et al., 2006b). From a set of approximately 500 genes with differences of p<0.01 by two-sided Student’s t-test, 28 genes with mean fold changes >1.5 between the Notch1 on versus off states (Figure 1B) and 4 invariant control genes were selected and validated (Figure S1A). To confirm that the signature reports on Notch1 inhibition and not GSI-treatment per se, we transduced DND41 cells with Notch1-specific shRNA and demonstrated induction of the Notch1 off signature (Figures S1B–D). This is consistent with prior work in which we showed that GE-HTS identified a Notch1 off signature in T-ALL cells treated with a Notch1-specific inhibitory antibody (Aste-Amezaga et al., 2010). To ensure that the signature does not identify generic growth inhibitors or cellular toxins, we treated DND41 cells with drugs known to be active against T-ALL cells. These drugs inhibited growth but did not induce the Notch1 off signature (Figure S1E–F).
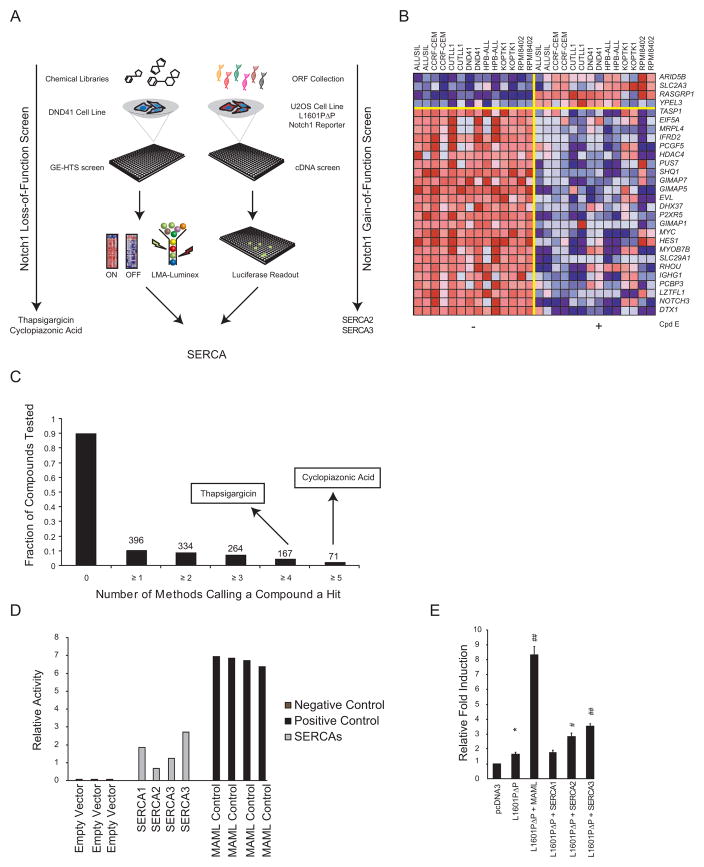
(A) Notch1 inhibitory modulators were identified using GE-HTS in DND41 cells, and these results were integrated with results from a cDNA library screen for factors enhancing the signaling activity of the leukemogenic NOTCH1 allele, L1601PΔP.
(B) Notch1 off signature genes derived from the expression profiling of T-ALL cell lines treated with Cpd E. The columns represent individual cell lines treated in duplicate with Cpd E (0.5 μM) or vehicle for 24 hr. The rows represent the 28 genes selected for the Notch off signature. Dark red indicates high gene expression and dark blue indicates low gene expression by Affymetrix microarray profiling.
(C) The x-axis indicates the number of methods (summed score, weighted summed score, naïve Bayes, KNN and SVM) and the y-axis the fraction of chemicals that scored for the number of methods. SERCA inhibitors are indicated. The number of compounds scoring by the indicated number of methods is noted above each column.
(D) Notch1 gain-of-function primary screen data for the negative (empty vector) and positive (MAML) controls versus SERCA-encoding cDNAs hits. Depicted is the fold-induction of luciferase signal for each replicate normalized to the population median of reporter readout values of pcDNA3-L1601PΔP (n=184).
(E) Retesting of cDNAs scoring in the Notch-sensitive luciferase reporter. Luciferase activity is expressed as relative activity compared to the pcDNA3 control. Errors bars denote the mean ± SD of 10 replicates. Statistical significance relative to pcDNA3 (*p<0.05) and to pcDNA3-L1601PΔP (#p<0.05; ##p<0.01) was determined by Student’s t-test.
Identification of Small Molecules that Modulate Notch1
We screened 3801 drugs or drug-like compounds in the human T-ALL cell line DND41. GE-HTS data was collected after 72 hr of treatment as previously described (Peck et al., 2006). Because true positives are more likely to score by multiple scoring metrics, we applied a consensus classification system requiring hits to score in multiple algorithms: summed score, weighted summed score, K-nearest neighbor, naïve Bayes classification and support vector machine (Figure 1C, Figure S1G) (Banerji et al., 2012). A total of 16 compounds (Table S1) selected for validation based on these criteria were retested in a two-fold dose-response series in DND41, MOLT4 and PF382 cell lines. Notably, multiple compounds reported to modulate calcium ion flux scored as dose-dependent Notch pathway inhibitors in all of the cell lines tested (Figure S1H–J).
A cDNA Library Screen Identifies SERCA as a Notch Signaling Enhancer
A complementary cDNA library screen for factors that enhance the signaling activity of the Notch1 mutant L1601PΔP was simultaneously conducted in the osteosarcoma cell line U2OS. L1601PΔP contains the same heterodimerization mutation that is present in the MOLT4 and KOPTK1 cell lines in cis with a PEST domain deletion (Chiang et al., 2008), a combination that is found in approximately 10–15% of human T-ALL (Weng et al., 2004). U2OS cells were selected for the screen because they are readily transfected and have very low basal Notch signaling tone, a feature that produces favorable signal-to-noise ratios. A total of 18,000 open-reading frames were scored for their ability to enhance L1601PΔP-dependent activation of a Notch luciferase reporter. Among the top hits were ATP2A1, ATP2A2 and ATP2A3 (Figure 1D), which encode SERCA1, SERCA2, and SERCA3, respectively. SERCAs are closely related, inwardly directed, ATP-dependent calcium pumps that localize to the endoplasmic reticulum. Retesting confirmed that ATP2A2 and ATP2A3 potentiate L1601PΔP-dependent signaling (Figure 1E). Of note, loss-of-function mutations in a Drosophila SERCA homolog, Ca-P60A, have been reported to produce Notch loss-of-function phenotypes in this model organism by altering Notch trafficking (Periz and Fortini, 1999). Thus, calcium modulators emerged at the nexus of two complementary screens.
Thapsigargin Targets the Notch Pathway
One of the small molecules that scored in our GE-HTS screen across four scoring metrics was thapsigargicin, an analog of thapsigargin, a highly potent natural product inhibitor of SERCA. Low nanomolar concentrations of thapsigargin induced the Notch1 off signature in a dose-dependent fashion in NOTCH1 mutant T-ALL cells (Figure 2A) and reduced the expression of the direct Notch1 target genes MYC, HES1 and DTX1 (Figure 2B). Subnanomolar concentrations of thapsigargin also inhibited the expression of a Notch reporter by L1601PΔP in U2OS cells (Figure 2C).
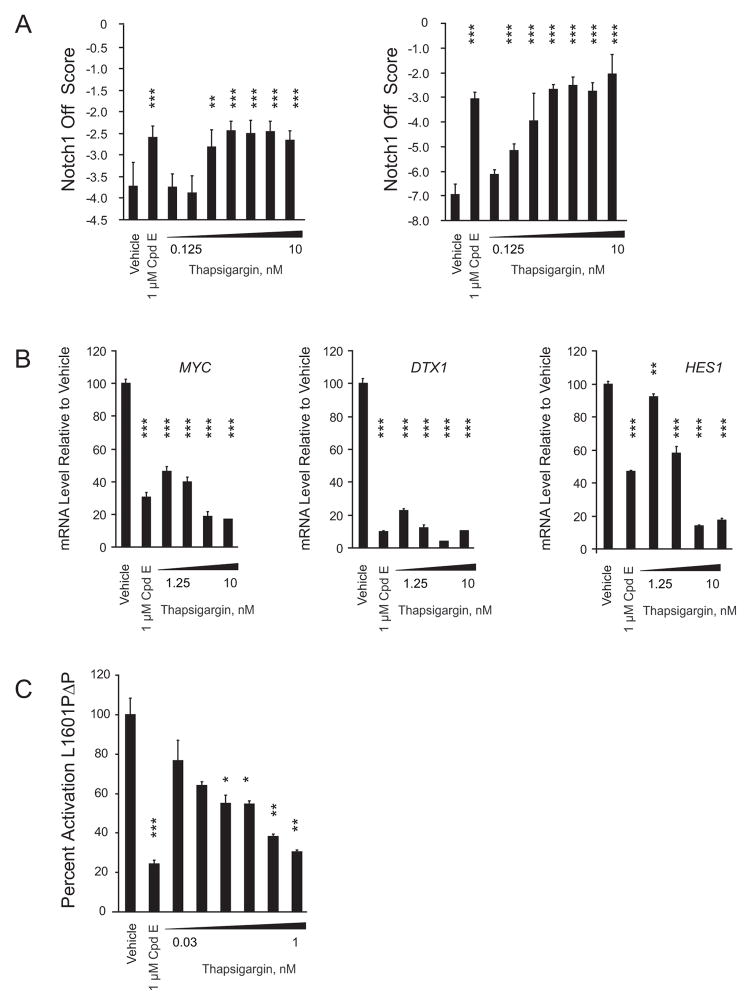
(A) Induction of the Notch1 off Score (weighted summed score) measured by GE-HTS in DND41 (left) and MOLT4 (right) cells treated with thapsigargin in 2-fold dilution for 72 hr. Error bars denote the mean ± SD of 12 replicates for vehicle-treated (DMSO 0.06%) cells and 6 replicates for thapsigargin-treated cells.
(B) Expression of indicated Notch1 target genes in DND41 treated in 2-fold dliution for 24 hr was determined by qRT-PCR. Error bars indicate the mean ± SD of 3 replicates. Data were analyzed using the ΔΔCT method and plotted as a percentage relative to the control gene RPL13A.
(C) Effects of thapsigargin on the activation of a Notch1 luciferase reporter by L1601PΔP in U2OS cells. Normalized luciferase activity relative to a Renilla control was expressed as a percentage of vehicle treatment. Error bars denote mean ± SD of 4 replicates. Statistical significance (*p<0.05; **p<0.01; ***p<0.001) in all panels was determined by one-way ANOVA using Bonferroni’s correction for multiple comparison testing.
Notch1 inhibition results in G0/G1 arrest in human T-ALL cells (Weng et al., 2004) and decreased T-ALL cell size (Palomero et al., 2006b; Weng et al., 2006). As expected, thapsigargin also induced a G0/G1 arrest (Figure 3A) and a decrease in cell size (Figure 3B) in NOTCH1-mutated T-ALL cell lines. We next studied the effect of thapsigargin in a panel of T-ALL cell lines that contain activating mutations in the heterodimerization domain (HD) of Notch1 and/or deletions in the degradation domain (PEST). Three T-ALL cell lines reported to be highly sensitive to GSI (ALL/SIL, DND41, and KOPTK1) were more sensitive to thapsigargin as measured by inhibition of cell growth and induction of apoptosis compared to two cell lines with intermediate sensitivity to GSI (MOLT4 and PF382) (Figure 3C). Furthermore, 24 hr of thapsigargin treatment decreased ICN1 levels in T-ALL cells (Figure 4A). As a further test of the idea that thapsigargin acts by preventing Notch1 activation, the Notch1-dependent T-ALL cell lines MOLT4 and DND41 were transduced with an empty MigR1 vector or with MigR1-ICN1 (Figure 4B). Transduction of ICN1, which lies downstream of the γ-secretase cleavage step in Notch activation, prevented induction of the Notch1 off signature (Figure 4C), growth inhibition (Figure 4D), G0/G1 cell cycle arrest (Figure 4E) and induction of apoptosis (Figure 4F) by thapsigargin. In contrast, empty MigR1 had no effect on these readouts of Notch inhibition. These results are consistent with a Notch pathway inhibitory effect of thapsigargin at low nanomolar concentrations.
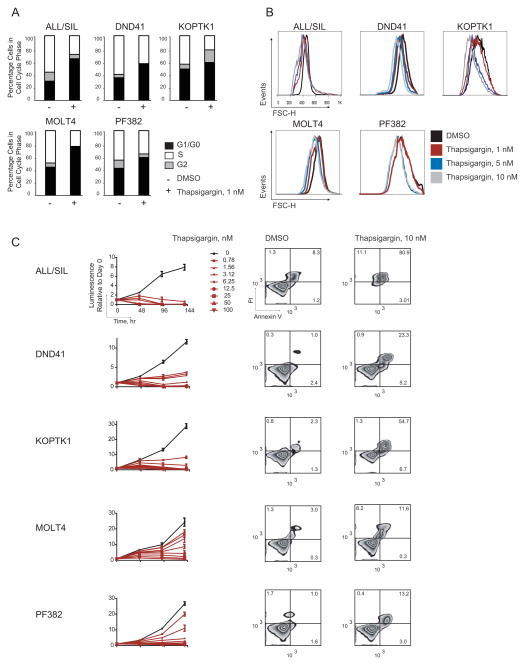
(A) Effect of thapsigargin treatment (6 days) on cell cycle of T-ALL cell lines, as assessed by measurement of DNA content on the viable fraction of cells.
(B) Effect of thapsigargin treatment for 24 hr on cell size as measured by forward scatter flow cytometry.
(C) Effect of thapsigargin treatment on cell growth (left) and induction of apoptosis (right). Error bars denote mean ± SD of 4 replicates. Annexin V/propidium iodide staining of T-ALL cells following 72 hr of treatment with 10 nM thapsigargin.
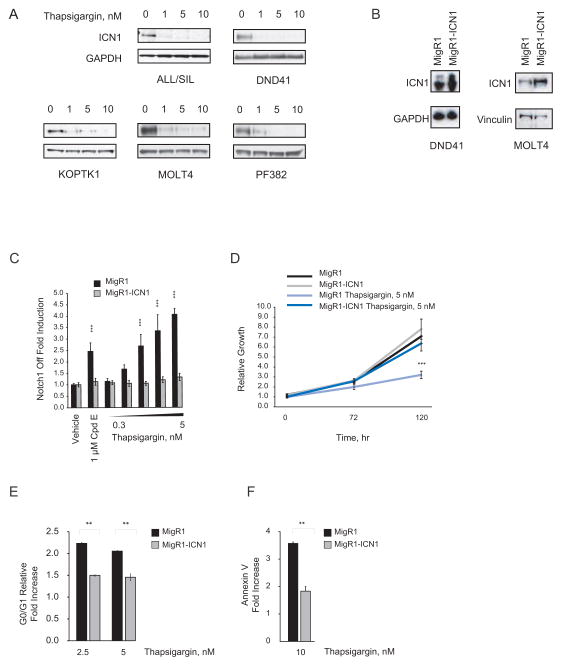
(A) Effect of 24 hr of thapsigargin treatment on the ICN1 level in T-ALL cells. The immunoblot was stained with anti-ICN1 antibody.
(B) ICN1 level in MigR1- or MigR1-ICN1-transduced cells. ICN1 is detected using an anti-ICN1 antibody.
(C) The weighted summed score fold induction is shown for the Notch1 off signature after treatment of cells with thapsigargin or Cpd E for 48 hr. Error bars indicate the mean ± SD of 12 replicates for vehicle-treated and 6 replicates for GSI- or thapsigargin-treated cells. Statistical significance (***p<0.001) was determined by one-way ANOVA using Bonferroni’s correction for multiple comparison testing.
(D) Effect of thapsigargin on the growth of MigR1- or MigR1-ICN1-transduced DND41 cells. Normalized data are plotted relative to time 0. Error bars indicate mean ± SD of 4 replicates. Statistical significance (***p<0.001) was determined by two-way ANOVA with Bonferroni’s correction for multiple comparison testing.
(E) Effect of 3 days of thapsigargin treatment on DNA content of MigR1- or MigR1-ICN1-transduced MOLT4 cells. Error bars indicate mean ± SD of 2 replicates with results expressed relative to vehicle treatment. Statistical significance was determined by Student’s t-test (**p<0.01).
(F) Effect of 3 days of thapsigargin treatment on apoptosis of MigR1- or MigR1-ICN1-transduced DND41 cells. Error bars indicate mean ± SD of 2 replicates with results expressed relative to vehicle. Statistical significance was determined by Student’s t-test (**p<0.01).
Thapsigargin Interferes with Notch1 Maturation
Multiple compounds scoring in our screen, including thapsigargin, are predicted to alter intracellular calcium. For example, thapsigargin and cyclopiazonic acid are known Ca2+ATPase inhibitors, impairing calcium entry into the endoplasmic reticulum. Of note, multiple EGF repeats and all three Lin12/Notch repeats in the extracellular domains of Notch receptors require calcium ions for proper folding (Aster et al., 1999; Gordon et al., 2007; Hambleton et al., 2004; Rand et al., 1997). We thus hypothesized that thapsigargin, by altering ER Ca2+ concentrations, would inhibit Notch1 maturation in T-ALL cells.
To test this hypothesis, we first determined if thapsigargin affected furin processing of Notch1, an event that occurs in the late Golgi compartment. Lysates from T-ALL cell lines treated with thapsigargin were immunoblotted with an antibody specific for the cytoplasmic portion of Notch1 that recognizes both unprocessed Notch1 (~270 kDa) and the furin-processed transmembrane subunit (~110 kDa). Thapsigargin reduced the levels of the furin-processed transmembrane Notch1 subunit, but not the unprocessed full-length Notch1 precursor, in multiple T-ALL cell lines (Figures 5A). Similar results were observed with the less potent SERCA inhibitor cyclopiazonic acid (Figure S2). Misfolded Notch1 receptors are expected to be retained in the ER/Golgi compartment. Immunostaining revealed that thapsigargin treatment reduced the levels of Notch1 on the cell surface (Figure 5B and C) and resulted in the co-localization of Notch1 and giantin, a Golgi membrane protein (Figure 5D). Thus, thapsigargin leads to defective Notch1 maturation in cultured T-ALL cells.
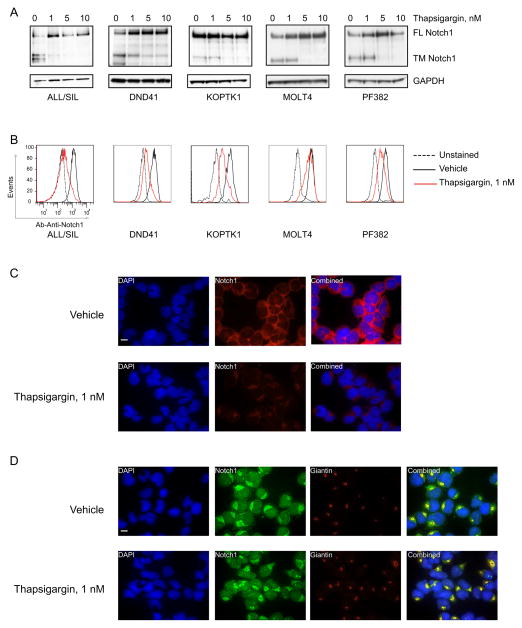
(A) Effect of thapsigargin treatment (24 hr) on Notch1 processing in T-ALL cell lines all with HD mutations (DND41 and ALL/SIL (L1594PΔPEST), KOPTK1 and MOLT4 (L1601PΔPEST), and PF382 (L1575PΔPEST). The blot was stained with an antibody against the C-terminus of Notch1 that recognizes both the furin-processed Notch1 transmembrane subunit (TM) and the unprocessed Notch1 precursor (FL).
(B, C) Effect of thapsigargin treatment (24 hr) on Notch1 cell surface staining, as assessed by flow cytometry (B) and staining of non-permeabilized cells (C). Scale bar represents 10 μm.
(D) Effect of thapsigargin treatment (24 hr) on the subcellular localization of Notch1. Double-immunofluorescence staining of permeabilized DND41 cells stained with anti-Notch1 (green) and Giantin (red) is shown. Co-localization is indicated by yellow signal. Scale bar represents 10 μm.
See also Figure S2.
SERCA Antagonism Inhibits Notch Function and T-ALL Growth In Vivo
To confirm that chemical and genetic inhibition of SERCA lead to Notch inactivation in vivo, we evaluated a Drosophila intestinal stem cell model in which Notch inhibition perturbs differentiation. The adult midgut is maintained by pluripotent stem cells that give rise to two populations of terminally differentiated daughter cells: a large class of polyploid enterocyes (EC) and a smaller class of diploid enteroendocrine (ee) cells (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2007). The stem cells express escargot (esg), a transcription factor, and Delta, a membrane bound ligand of the Notch receptor. High levels of Notch activation are required for daughter cells to adopt the EC cell fate, whereas lower levels of Notch activation specify daughters to adopt the ee fate. Thus, when Notch is completely inhibited, daughter cells fail to differentiate and remain as stem cells. By contrast, when Notch signaling is partially inhibited, daughter cells fail to differentiate into ECs and instead remain as stem cells or differentiate into ee cells.
To enhance the sensitivity of fly-based drug assays, we used transgenic flies that express a human RAF gain-of-function cDNA (RAF(gof)) in their intestinal stem cells. Expression of the RAF(gof) cDNA results in rapid expansion of the esg+ population, which is composed of not only diploid stem cells but also polyploid EC daughter cells. When Notch is inhibited by feeding flies the GSI DAPT or Cpd E, stem cell daughters fail to differentiate into EC cells and instead give rise mostly to additional stem cells, as well as some ee daughters (Figure 6A). Cyclopiazonic acid and thapsigargin treatment also expanded the stem cell and ee cell populations, thus phenocopying the effects of GSI (Figure 6B).
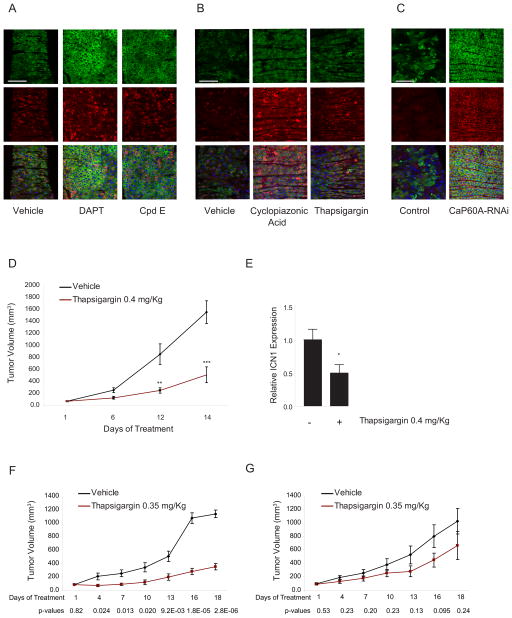
(A–C) Immunofluorescence staining of Drosophila midguts expressing GFP (green) and stained with anti-Delta (membrane red), anti-prospero (nuclear red) and DAPI (blue) is depicted. Treatment was with DAPT (400 μM) or Cpd E (100 μM) for 7 days in (A) and with cyclopiazonic acid (1 mM) or thapsigargin (100 μM) for 7 days in (B). In (C) effects of knockdown of Ca-P60A are shown. Scale bars represent 75 μm.
(D) Effect of thapsigargin on the growth of xenografted MOLT4 tumors. Error bars indicate mean ± SD of 6 replicates for the thapsigargin-treated and 9 replicates for the vehicle-treated mice. Statistical significance (**p<0.01; ***p<0.001) was determined by two-way ANOVA using Bonferroni’s correction for multiple comparison testing.
(E) Effect of thapsigargin treatment on ICN1 levels in xenografted MOLT4 tumors was measured by western blotting, and statistical significance was determined by Student’s t-test (*p<0.05). Error bars represent the mean ± SD of 6 replicates for each group.
(F, G) Effect of thapsigargin on the growth of DND41 cells transduced with MigR1 (F) or MigR1-ICN1 (G) xenografted in NSG mice. Error bars indicate mean ± SD of replicates for each cohort. Statistical significance was determined by Student’s t-test as indicated
See also Figure S3.
If thapsigargin inhibits Notch signaling through effects on calcium channels, then genetic modulation of SERCA should produce similar phenotypes. Indeed, knockdown of Ca-P60A, the only SERCA expressed in Drosophila, produced effects on stem cell and ee cell pools similar to those induced by GSI, thapsigargin, or cyclopiazonic acid treatment (Figure 6C). Thus, the results of chemical and genetic studies are consistent with a model in which SERCA inhibition by thapsigargin impairs Notch signaling in vivo.
We next tested thapsigargin in a human T-ALL xenograft model. Systemic administration of thapsigargin to SCID-beige mice bearing MOLT-4 tumors inhibited tumor growth compared to vehicle-treated controls (Figure 6D). In addition, ICN1 protein levels were diminished in the thapsigargin-treated tumors compared to vehicle-treated tumors (Figure 6E and S3A), linking growth inhibition to Notch inhibition.
To further demonstrate that thapsigargin impairs leukemic progression via Notch signaling inhibition we established a second T-ALL xenograft model in which DND41 cells were transduced with MigR1 or MigR1-ICN1 and subsequently propagated in NSG mice. Thapsigargin treatment markedly decreased the growth of control tumors but had little effect on tumors expressing ICN1 (Figure 6F and G), indicating that tumor growth suppression by thapsigargin is mediated by inhibition of Notch1 signaling in the leukemic cells.
Prior studies demonstrated that gastrointestinal toxicity and lack of sustained response were the major limitations of first generation GSIs (DeAngelo et al., 2006). It was hypothesized that gastrointestinal toxicity was due to blockade of wild-type Notch1 and Notch2 in the gut leading to intestinal secretory metaplasia, increased number of goblet cells and arrested proliferation in the crypts of the small intestine (Milano et al., 2004; Real et al., 2009). Mice treated with thapsigargin did not develop gastrointestinal toxicity (Figure S3B and C), suggesting that HD-mutated Notch1 receptors were more sensitive to the effects of thapsigargin than wild-type Notch1/Notch2 receptors expressed in normal cells. These pre-clinical studies support SERCA as a possible therapeutic target in T-ALL.
NOTCH1 Mutational Status Influences Thapsigargin Sensitivity
The results above suggest that thapsigargin inhibits signaling through wild-type and mutated Notch receptors but may have stronger effect for mutated Notch1. Prior work has shown that many activating HD mutations found in T-ALL result in destabilization of the Notch negative regulatory region and have deleterious effects on Notch1 folding and maturation (Malecki et al., 2006). Because the LNR repeats of the Notch negative regulatory region rely on calcium for folding and function (Aster et al., 1999), mutated Notch1 might be more sensitive to reduced calcium availability than the wild-type Notch1, providing a therapeutic window for SERCA inhibitors.
One simple prediction of the above model is that constitutively active forms of Notch1 lacking Ca2+ binding modules should be insensitive to SERCA inhibitors. To test this prediction, we performed Notch1 reporter assays in U2OS cells transfected with a plasmid encoding ΔEGFΔLNR, a membrane-tethered form of Notch1 lacking the extracellular EGF repeats and Lin12/Notch repeats, or ICN1. As anticipated, co-expression of SERCA did not enhance reporter gene activation by ΔEGFΔLNR (Figure 7A), nor was reporter gene activation by ΔEGFΔLNR (Figure 7B) or ICN1 (Figure 7C) affected by thapsigargin. In contrast, ΔEGFΔLNR was highly sensitive to GSI (Figure 7B). Taken together, these results indicate that at low nanomolar concentrations thapsigargin inhibits Notch1 through a mechanism that requires the Ca2+-binding modules of the Notch1 extracellular domain.
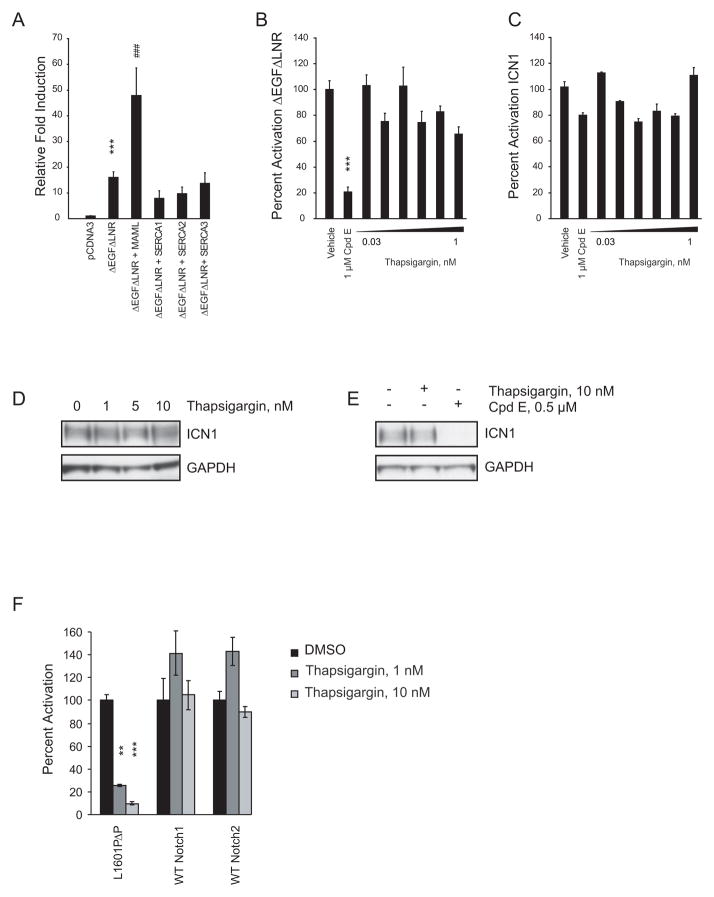
(A) Effect of SERCA co-expression on the activity of ΔEGFΔLNR in a Notch reporter assay. Normalized firefly luciferase activity was expressed as fold induction relative to the empty plasmid. Error bars denote the mean ± SD of 10 replicates. Statistical significance relative to pcDNA3 (***p<0.001) and to pcDNA3-ΔEGFΔLNR (###p<0.001) was determined by one-way ANOVA using Bonferroni’s correction for multiple comparison testing.
(B, C) Effects of thapsigargin or Cpd E on the activity of ΔEGFΔLNR (B) and ICN1 (C) in a Notch reporter assay. Assay conditions and interpretations were as in (A). Errors bars denote mean ± SD of 4 replicates. Statistical significance (***p<0.001) was determined by one-way ANOVA using Bonferroni’s correction for multiple comparison testing.
(D, E) The ICN1 protein level in SUPT1 cells treated for 24 hr as indicated was determined using immunoblots stained with anti-ICN1 antibody.
(F) Ligand-mediated activation of Notch activity was determined using U2OS cells stably (Notch1 and Notch2) or transiently (L1601PΔP) expressing Notch-Gal4 fusion receptors and treated as indicated. Normalized Gal4 firefly luciferase activity was expressed relative to vehicle treated control. Errors bars denote mean ± SD of 4 replicates. Statistical significance (**p<0.01, *** p<0.001) was determined by Student’s t-test.
To determine if the Notch1 extracellular domain is important for the ability of thapsigargin to inhibit leukemia cell growth, we studied the human T-ALL cell line SUPT1, which has two copies of a t(7;9)(q34, q34) fusing the 3′ end of NOTCH1 with enhancer/promoter elements of the T-cell receptor β locus (T) CRB and has no normal NOTCH1 allele (Ellisen et al., 1991). The rearranged NOTCH1 alleles in SUPT1 cells drive the expression of a series of truncated mRNAs encoding N-terminally deleted polypeptides lacking the Notch1 extracellular domain, some of which are inserted into membranes and require γ-ecretase cleavage for activation (Das et al., 2004). As anticipated, thapsigargin had no effect on ICN1 levels in SUPT1 cells (Figure 7D), whereas Cpd E eliminates the generation of ICN1 (Figure 7E). This result prompted us to speculate that protein structure might affect drug response. Thapsigargin failed to inhibit wild-type Notch1 or Notch2 at concentrations that impaired signaling of Notch1 bearing leukemogenic HD mutations (Figure 7F).
To further test whether thapsigargin inhibits wild-type Notch1 maturation we tested two Notch1 wild-type T-ALL and one chronic myelogenous leukemia (CML) cell lines in which high expression of Notch1 was previously reported (Palomero et al., 2006a). Compared to HD mutant, wild-type proteins appear to be less affected by thapsigargin treatment (Figures 8A, B, and C). The effects of thapsigargin on cell viability were then determined in a larger panel of human T-ALL cell lines of known NOTCH1 mutational status (Weng et al., 2004). Cell lines carrying NOTCH1 alleles with HD domain mutations were more sensitive to thapsigargin than cells with wild-type NOTCH1 alleles or lacking the Notch1 extracellular domain (Figure 8D–F).
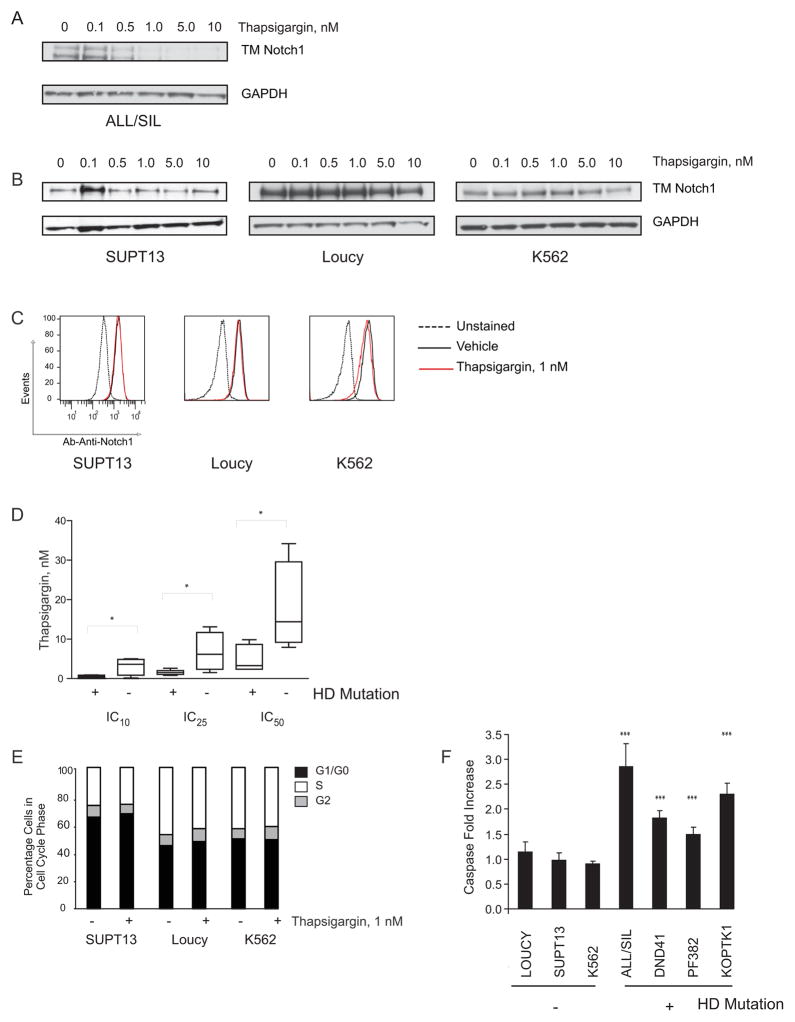
(A, B) Effect of thapsigargin treatment (6 hr) on processing of HD mutant (A) or wild-type (B) Notch1. Notch1 was detected with an antibody against the C-terminus of Notch1 that recognizes the furin-processed Notch1 transmembrane subunit (TM).
(C) Effect of thapsigargin treatment (24 hr) on Notch1 cell surface staining, as assessed by flow cytometry.
(D) The relative growth of thapsigargin-treated T-ALL cell lines with wild-type (LOUCY, MOLT16, SUPT13) or rearranged alleles (SUPT1) of NOTCH1 versus those with HD mutations (ALL/SIL, DND41, KOPTK1, MOLT4, PF382). The line in the box plots represents the median. The upper edge (hinge) of the box indicates the 75th percentile of the data set, and the lower hinge the 25th percentile. The ends of the vertical line indicate the minimum and the maximum data values. Statistical significance was determined by Student’s t-test (*p<0.05).
(E) Effect of thapsigargin treatment on cell cycle progression in Notch1 wild-type cell lines, as assessed by measurement of DNA content on the viable fraction of cells.
(F) Effect of thapsigargin treatment (1 nM) on apoptosis induction as assessed by the lumininescence Caspase 3/7 assay. Errors bars denote mean ± SD of 4 replicates. Statistical significance (***p<0.001) was determined by one-way ANOVA using Bonferroni’s correction for multiple comparison testing.
In summary, these data suggest that Notch1 receptors bearing leukemogenic HD domain mutations are more sensitive to SERCA inhibitors, such as thapsigargin, than normal receptors.
DISCUSSION
Integrating Cell-based Screens for Small Molecule and Protein Target Discovery
While there is a strong rationale for target-based therapies for cancer, with the exception of the nuclear hormone receptors, transcription factors have largely been refractory to conventional small-molecule screening approaches due to the challenges in developing high-throughput, robust screening assays. By definition, any therapeutic agent that modulates a transcription factor must alter the expression of its target genes. While there have been significant advances in our ability to assess global gene expression changes, almost all existing approaches cannot yet be applied to large-scale screening efforts due to cost and throughput limitations. Recognizing these shortcomings, we developed an approach that allows measuring the expression of hundreds of endogenous genes in 384-well format and applied it to identify antagonists of leukemogenic increases in Notch signaling in T-ALL.
A limitation of both phenotypic and expression-based screening, however, is that identification of the relevant target of lead compounds can be difficult. The development of alternative genomic and chemical proteomic approaches for identifying protein targets holds the promise of accelerating the elucidation of underlying mechanism. Integrating results of a cDNA screen with GE-HTS data allowed us to identify SERCAs as Notch1 modulators and potential therapeutic targets in Notch1-associated leukemias.
Altering Maturation of Mutant Notch1 by SERCA Inhibition
We show here that SERCA inhibitors such as thapsigargin cause a Notch1 maturation defect marked by the accumulation of unprocessed Notch1 in the ER/Golgi compartment. The resulting effects on Notch1 signaling and leukemia cell growth depend on the nature of the NOTCH1 mutations. The most common activating NOTCH1 mutations in human T-ALL consist of point substitutions or small in-frame deletions or insertions in the extracellular heterodimerization domain, which disrupt heterodimerization domain structure and permit ligand-independent ADAM-type metalloprotease cleavages (Gordon et al., 2009; Malecki et al., 2006), the so-called Class I NOTCH1 mutations. Folding and maturation of Notch1 are partially impaired by these mutations (Malecki et al., 2006), and it appears that Notch1 receptors bearing such mutations are more sensitive to the inhibitory effects of thapsigargin than wild-type Notch1 receptors. Another possible contributing factor to the greater sensitivity to thapsigargin in cells with mutated Notch1 receptors is that the presence of these mutated polypeptides may itself engender ER stress and thus render these cells more susceptible to the ER stress induced by thapsigargin. Indeed, this may account for the inability to fully rescue the effects of thapsigargin with ICN1 in some of our experiments. Taken together, these data suggest the potential for a therapeutic window for thapsigargin in T-ALLs bearing this type of mutation.
Other types of activating NOTCH1 mutations also exist. Rarely, juxtamembrane in-frame tandem duplications create new “deprotected” ADAM-metalloprotease cleavage sites (Class II mutations) (Malecki et al., 2006), or translocations create NOTCH1 alleles encoding polypeptides that lack the Notch1 extracellular domain (Ellisen et al., 1991; Palomero et al., 2006a). As anticipated, we observed that a T-ALL cell line with two translocated NOTCH1 alleles is relatively resistant to thapsigargin. Our proposed mechanism of action also predicts that murine T-ALLs, which often have Notch1 deletions that remove the Notch1 ectodomain coding sequence (Ashworth et al., 2010), as well as uncommon human breast cancers with NOTCH1 rearrangements (Robinson et al., 2011), will be more resistant to thapsigargin and other SERCA inhibitors. It will be of importance to determine if CLLs, which have recently been reported to have frequent Notch1 PEST domain deletions (Di Ianni et al., 2009; Puente et al., 2011) but lack heterodimerization domain mutations, are sensitive to SERCA inhibitors.
Connecting NOTCH1 and SERCA Mutations in Human Disease
Germline mutations in ATP2A1 are reported in the congenital disorder Brody syndrome, characterized by impaired muscle relaxation and myopathy (Odermatt et al., 1996). ATP2A2 mutations are reported in Darier’s disease (Sakuntabhai et al., 1999), an autosomal dominant skin disorder characterized by loss of adhesion between keratinocytes, scaling due to hyperkeratosis, and thickening of the epidermis due to keratinocyte hyperproliferation. Skin cancers have been reported in patients with Darier’s disease (Robertson and Sauder, 2012). Moreover, in aged Atp2a2+/− mice, tumors develop from the keratinized squamous epithelia (Liu et al., 2001) while the wild-type Atp2a2 allele is retained and expressed, supporting a role for SERCA2 haploinsufficiency in tumor development (Prasad et al., 2005). In addition, thapsigargin acts as a tumor promoter in a skin carcinogenesis mouse model (Hakii et al., 1986).
There is also mounting evidence that Notch signaling suppresses the transformation of squamous epithelial cells. Genetically engineered mice with decrements in Notch signaling in the skin have a high incidence of skin cancers (Nicolas et al., 2003; Proweller et al., 2006). Recent human clinical trial data revealed that semagacestat, a GSI, is associated with an increased risk of skin cancer (discussed in (Crump et al., 2011)), possibly due to inhibition of Notch in the skin by chronic GSI administration. Furthermore, Notch pathway loss-of-function mutations have been reported in squamous cell carcinomas of the head and neck (Agrawal et al., 2011; Stransky et al., 2011), skin and lung (Wang et al., 2011). Of note, at least one NOTCH1 point substitution in human cutaneous squamous cell carcinoma impairs Ca2+-binding and folding of EGF repeat 12 (Hambleton et al., 2004; Wang et al., 2011) and a second (R1549Q) impacts folding of the LNRs (Wang et al., 2011), thus recapitulating the proposed pharmacologic effect of thapsigargin. Additional murine studies indicate that Notch signaling may have a tumor suppressive function in other cell lineages as well, including myeloid progenitors (Klinakis et al., 2011) and endothelial cells (Liu et al., 2011; Yan et al., 2010). Intriguingly, several recent studies report SERCA mutations in head and neck squamous cell carcinoma (Korosec et al., 2008; Stransky et al., 2011), in AML (Yan et al., 2011), and in other malignancies (Korosec et al., 2009), implicating loss-of-function mutations of SERCA as an additional possible mechanism for Notch inactivation in these diseases. Indeed, while NOTCH1 mutations enhance sensitivity to SERCA inhibitors, providing a potential therapeutic window for application of this compound class, wild-type Notch1 is also sensitive to SERCA inhibition but at higher concentrations of the compound. One hypothesis to explain a Notch1 and SERCA functional dependency is by a physical interaction. It has been previously shown that presenilin and SERCA2b co-localize in the endoplasmic reticulum (Green et al., 2008). Since presenilin immunoprecipitation is also reported to preferentially recover the full length Notch1 precursor prior to Notch1 cleavage in the Golgi (Ray et al., 1999), it is possible that Notch1, SERCA and presenilin are part of a macromolecular complex.
Toward Translating SERCA Inhibitors to the Clinic
Our studies and other recent work bring to light a challenge in targeting Notch1 in cancer: its pleiotropic roles. NOTCH1 is an oncogene in some human cancers, such as T-ALL and CLL, whereas it is a tumor suppressor in others, most notably squamous cell carcinomas. Several strategies have been explored to inhibit Notch1 including the use of GSIs, Notch1-directed antibodies, and direct inhibition of the Notch complex with a hydrocarbon stapling approach (reviewed in (Roti and Stegmaier, 2011)). Each of these, however, is anticipated to also inhibit wild-type Notch1. One strategy to mitigate potential cancer-promoting effects in non-diseased cells is intermittent dosing of Notch inhibitors. A second approach is the selective targeting of the oncogenic protein. Our results suggest that common heterodimerization domain mutations in Notch1 render the protein more susceptible to the thapsigargin-induced maturation defect, allowing for a therapeutic window in targeting mutant versus wild-type Notch1 (we observed an anti-leukemia effect with no measureable gut toxicity). The selective targeting of BRAF kinase bearing an activating V600E mutation by vemurafenib in melanoma is an example of successful application of this strategy (Bollag et al., 2010), although acquired resistance with RAS pathway lesions is common (Nazarian et al., 2010; Su et al., 2012). Similarly, reactivation of Notch1 signaling, for example, by altered EGF/LNR repeats, may pose a resistance mechanism in SERCA-targeted therapy in T-ALL.
Given the pervasive role of calcium signaling in normal physiology, it is unlikely that pan-SERCA inhibitors, such as thapsigargin, will have an immediate clinical application unless additional development is pursued. One strategy might be the development of isoform-specific small-molecule inhibitors of SERCA. A second is to derivatize thapsigargin for specific delivery to T-ALL cells. This tactic has already been employed for a derivative of thapsigargin that is designed to treat prostate cancer, which is currently in clinical trials (NCT01056029 and NCT01734681) (Denmeade et al., 2003). Successful “targeted” delivery of SERCA inhibitors to T-ALL cells would further improve the therapeutic window with this class of drugs and enhance the likelihood of clinical translation.
In summary, this study demonstrates the power of an integrative screening strategy to identify alternative ways to target aberrant transcription factors, identify the modulation of SERCA as a tractable approach for inhibiting Notch1 in Notch-driven cancers, and implicate SERCA mutation as another potential pathogenic mechanism for Notch downregulation in human cancers in which the Notch pathway has a tumor suppressive role.
EXPERIMENTAL PROCEDURES
Full experimental details are in the Supplemental Experimental Procedures.
Cell Culture, Compounds, and Antibodies
Cells were cultured in RPMI 1640 (Cellgro) with 10% fetal bovine serum (FBS) (Sigma-Aldrich) and 1% penicillin-streptomycin. Cpd E, thapsigargin, and cyclopiazonic acid were obtained from ENZO Life Sciences and bepridil hydrochloride, ionomycin, salinomycin, methotrexate, dexamethasone, vincristine and DAPT from Sigma-Aldrich. Western blot antibodies against Notch1 were obtained from Cell Signaling and Santa Cruz Biotechnology, Actin (Thermo Scientific), Vinculin (AbCAM) and GAPDH (Santa Cruz Biotechnology). Antibodies for IF include Notch1 [A6] and giantin (AbCAM). Cell surface Notch1 was evaluated by staining non-permeabilized cells with monoclonal anti-human Notch1 antibody (R&D).
Notch1 Off Signature Detection
Marker and control genes for the Notch1 on vs. off signature were chosen using publicly available Affymetrix microarray expression profiling data sets (GEO: GSE5716) (Palomero et al., 2006b). Probes are shown in Supplemental Experimental Procedures. The signature was adapted to GE-HTS as described (Peck et al., 2006). Signature performance was evaluated by calculating the summed score and weighted summed score (Hahn et al., 2008).
Small-molecule Library Screening
DND41 cells were incubated with compounds at approximately 20 μM in DMSO for 72 hr. We screened 3801 compounds in triplicate, including the BSPBio collection (Prestwick, Biomol, and Spectrum libraries) and the HSCI1 collection (Broad Institute). The GE-HTS assay was performed as described (Peck et al., 2006; Stegmaier et al., 2004). Compounds that induce the Notch1 off signature were identified using 5 discrete analytic metrics: summed score, weighted summed score, K-nearest neighbor analysis, naïve Bayes classification, and support vector machine as described (Hahn et al., 2009).
cDNA Library Screen and Validation
The cDNA screening strategy involved the use of three key components: 1) a pcDNA3 plasmid encoding a modestly strong NOTCH1 gain-of-function mutant, L1601PΔP, driven from a CMV promoter, 2) a Notch firefly luciferase reporter, and 3) a pre-plated cDNA library cloned into the Sport6 plasmid. Luminescence was measured 48 hr post-plating.
Viral Transduction
Sequences targeted by each shRNA are listed in Supplemental Experimental Procedures. Viral supernatant production and MigR1 retroviral infections were performed as described (Aster et al., 2000).
Cell Growth, Apoptosis and DNA Content Assays
Cell growth was assessed using the Promega Cell-Titer Glo ATP-based assay (Promega), apoptosis using a Caspase-Glo 3/7 assay (Promega) or Annexin V and propidium iodide (PI) staining by flow-cytometry (eBioscience), and cellular DNA content by staining with propidium iodide. Values for IC10, IC25, IC50 were calculated using Prism 5 Software (version 5.03).
Reporter Gene Assays
Expression plasmids for L1601PΔP (Weng et al., 2004), L1601PΔP-GAL4 (Malecki et al., 2006), ΔEGFΔLNR (Chiang et al., 2008), and ICN1 (Aster et al., 2000) have been described. Co-transfection of U2OS cells with expression plasmids, a Notch firefly luciferase reporter gene, and an internal Renilla luciferase control gene, was performed as described (Aster et al., 2000). A second approach used a Notch1-Gal4 receptor ligand stimulation assay (Malecki et al., 2006).
Real-time RT-PCR
Primers and probes for real-time RT-PCR were obtained from Applied Biosystems. The data were analyzed using the ΔΔCT method and plotted as percentage of transcript compared to vehicle.
Drosophila Experiments
To generate RAF(gof) tumors in the adult Drosophila midgut, we created a stock containing the X-linked UAS-RAF(gof) transgene (Brand and Perrimon, 1994) and the second chromosome-linked esg-Gal4, UAS-GFP, Tubulin, Gal80(ts) transgenes (Micchelli and Perrimon, 2006). Drugs were prepared in DMSO and mixed with fly food 1:100 and fed to flies for 7 days. Flies were given freshly prepared drug every 2–3 days. Drug effects were evaluated by immunofluorescence microscopy.
T-ALL Xenograft Studies
MOLT4 xenografts were established by injecting 1.7×106 cells subcutaneously into 6-week-old female SCID-beige mice (Charles River Labs). Tumor volume was determined by caliper measurements using Vol=0.5 × L × W2. When tumors reached a mean volume of 75 mm3, mice were divided into two groups: thapsigargin 0.4 mg/Kg or vehicle by intraperitoneal injection daily. Three mice that died prematurely due to drug toxicity were excluded from the study, leaving 6 evaluable mice in the thapsigargin-treated arm and 9 in the vehicle arm. DND41 MigR1 and DND41 MigR1-ICN1 xenografts were established by injecting 10×106 cells subcutaneously into NSG mice (n=20 per line). When tumor volume reached over 50 mm3 mice were divided into two groups: thapsigargin 0.35 mg/Kg or vehicle 10 mL/Kg by intraperitoneal injection daily. Mice that were not ready at start of treatment were subsequently added to treatment groups when their tumors reached appropriate sizes. Mice were treated daily through the course of the study and tumors measured every three days. Five thapsigargin-treated mice were found dead during the course of the study with no prior weight loss or clinical signs of illness. All animal studies were performed under a Dana-Farber Cancer Institute IACUC-approved protocol.
Acknowledgments
This work was supported by the Leukemia and Lymphoma Society (LLS) and William Lawrence and Blanche Hughes Foundation (K.S, J.C.A, S.C.B); SynCure Cancer Research Foundation and the NCI P01 CA 98484-11A1 (K.S.); EHA-ASH International Fellowship Award, American-Italian Cancer Foundation PostDoctoral Research Fellowship, Lady Tata International Award for Research in Leukaemia, LLS Special Fellow award and Fondazione Umberto Veronesi (G.R.); LLS Fellow (K.P.); NIH (N.P and M.M.); NIH R01 CA092433 (S.C.B) and Harvard Catalyst (N.P. and M.M.). We thank Zi Peng Fan and Amanda L. Christie for their technical support, Nicola Tolliday for stewardship of the chemical screen, the TRiP at Harvard Medical School for providing transgenic RNAi fly stocks used in this study, and Maria Pia Briziarelli, AULL (Associazione Umbra per lo studio e la terapia delle Leucemie e Linfomi), for grant management (G.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. [Europe PMC free article] [Abstract] [Google Scholar]
- Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, Kelliher M, Kastner P, Chan S, Aster JC. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. [Europe PMC free article] [Abstract] [Google Scholar]
- Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, Huang L, Vitelli S, Vo KT, Haytko P, et al. Characterization of notch1 antibodies that inhibit signaling of both normal and mutated notch1 receptors. PLoS ONE. 2010;5:e9094. [Europe PMC free article] [Abstract] [Google Scholar]
- Aster JC, Simms WB, Zavala-Ruiz Z, Patriub V, North CL, Blacklow SC. The folding and structural integrity of the first LIN-12 module of human Notch1 are calcium-dependent. Biochemistry. 1999;38:4736–4742. [Abstract] [Google Scholar]
- Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20:7505–7515. [Europe PMC free article] [Abstract] [Google Scholar]
- Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, Kakoza RM, Chow KT, Ross L, Alexe G, et al. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. J Clin Invest. 2012;122:935–947. [Europe PMC free article] [Abstract] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. [Europe PMC free article] [Abstract] [Google Scholar]
- Brand AH, Perrimon N. Raf acts downstream of the EGF receptor to determine dorsoventral polarity during Drosophila oogenesis. Genes & development. 1994;8:629–639. [Abstract] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. [Abstract] [Google Scholar]
- Carpenter AE. Image-based chemical screening. Nat Chem Biol. 2007;3:461–465. [Abstract] [Google Scholar]
- Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, Childs ME, Gimotty PA, Aster JC, Pear WS. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. [Abstract] [Google Scholar]
- Corsello SM, Roti G, Ross KN, Chow KT, Galinsky I, DeAngelo DJ, Stone RM, Kung AL, Golub TR, Stegmaier K. Identification of AML1-ETO modulators by chemical genomics. Blood. 2009;113:6193–6205. [Europe PMC free article] [Abstract] [Google Scholar]
- Crump CJ, Johnson DS, Li YM. Target of gamma-secretase modulators, presenilin marks the spot. EMBO J. 2011;30:4696–4698. [Europe PMC free article] [Abstract] [Google Scholar]
- Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, Nikov G, Krishnamoorthy V, Majumder PK, Kelliher MA. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–6181. [Europe PMC free article] [Abstract] [Google Scholar]
- Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. [Abstract] [Google Scholar]
- Das I, Craig C, Funahashi Y, Jung KM, Kim TW, Byers R, Weng AP, Kutok JL, Aster JC, Kitajewski J. Notch oncoproteins depend on gamma-secretase/presenilin activity for processing and function. J Biol Chem. 2004;279:30771–30780. [Abstract] [Google Scholar]
- DeAngelo DJ, Stone RM, Silverman LB, Aster JC. A phase I clinical trial of the Notch inhibitor MK-0752 in patients with T-cell acute lympho- blastic leukemia/lymphoma (T-ALL) and other leukemias. J Clin Oncol. 2006;24:6585. [Google Scholar]
- Denmeade SR, Jakobsen CM, Janssen S, Khan SR, Garrett ES, Lilja H, Christensen SB, Isaacs JT. Prostate-specific antigen-activated thapsigargin prodrug as targeted therapy for prostate cancer. Journal of the National Cancer Institute. 2003;95:990–1000. [Abstract] [Google Scholar]
- Di Ianni M, Baldoni S, Rosati E, Ciurnelli R, Cavalli L, Martelli MF, Marconi P, Screpanti I, Falzetti F. A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br J Haematol. 2009;146:689–691. [Abstract] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. [Abstract] [Google Scholar]
- Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. [Europe PMC free article] [Abstract] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. [Abstract] [Google Scholar]
- Green KN, Demuro A, Akbari Y, Hitt BD, Smith IF, Parker I, LaFerla FM. SERCA pump activity is physiologically regulated by presenilin and regulates amyloid beta production. The Journal of cell biology. 2008;181:1107–1116. [Europe PMC free article] [Abstract] [Google Scholar]
- Hahn CK, Berchuck JE, Ross KN, Kakoza RM, Clauser K, Schinzel AC, Ross L, Galinsky I, Davis TN, Silver SJ, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–294. [Europe PMC free article] [Abstract] [Google Scholar]
- Hahn CK, Ross KN, Warrington IM, Mazitschek R, Kanegai CM, Wright RD, Kung AL, Golub TR, Stegmaier K. Expression-based screening identifies the combination of histone deacetylase inhibitors and retinoids for neuroblastoma differentiation. Proc Natl Acad Sci U S A. 2008;105:9751–9756. [Europe PMC free article] [Abstract] [Google Scholar]
- Hakii H, Fujiki H, Suganuma M, Nakayasu M, Tahira T, Sugimura T, Scheuer PJ, Christensen SB. Thapsigargin, a histamine secretagogue, is a non-12-O-tetradecanoylphorbol-13-acetate (TPA) type tumor promoter in two-stage mouse skin carcinogenesis. J Cancer Res Clin Oncol. 1986;111:177–181. [Abstract] [Google Scholar]
- Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, Handford PA, Downing AK. Structural and functional properties of the human notch-1 ligand binding region. Structure. 2004;12:2173–2183. [Abstract] [Google Scholar]
- Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. [Abstract] [Google Scholar]
- Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Korosec B, Glavac D, Volavsek M, Ravnik-Glavac M. Alterations in genes encoding sarcoplasmic-endoplasmic reticulum Ca(2+) pumps in association with head and neck squamous cell carcinoma. Cancer Genet Cytogenet. 2008;181:112–118. [Abstract] [Google Scholar]
- Korosec B, Glavac D, Volavsek M, Ravnik-Glavac M. ATP2A3 gene is involved in cancer susceptibility. Cancer Genet Cytogenet. 2009;188:88–94. [Abstract] [Google Scholar]
- Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, Gunawardana J, Jenkins C, Cochrane C, Ben-Neriah S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963–1971. [Abstract] [Google Scholar]
- Lee SY, Kumano K, Masuda S, Hangaishi A, Takita J, Nakazaki K, Kurokawa M, Hayashi Y, Ogawa S, Chiba S. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia. 2005;19:1841–1843. [Abstract] [Google Scholar]
- Liu LH, Boivin GP, Prasad V, Periasamy M, Shull GE. Squamous cell tumors in mice heterozygous for a null allele of Atp2a2, encoding the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 Ca2+ pump. J Biol Chem. 2001;276:26737–26740. [Abstract] [Google Scholar]
- Liu Z, Turkoz A, Jackson EN, Corbo JC, Engelbach JA, Garbow JR, Piwnica-Worms DR, Kopan R. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 2011;121:800–808. [Europe PMC free article] [Abstract] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. [Europe PMC free article] [Abstract] [Google Scholar]
- Mansour MR, Linch DC, Foroni L, Goldstone AH, Gale RE. High incidence of Notch-1 mutations in adult patients with T-cell acute lymphoblastic leukemia. Leukemia. 2006;20:537–539. [Abstract] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. [Abstract] [Google Scholar]
- Milano J, McKay J, Dagenais C, Foster-Brown L, Pognan F, Gadient R, Jacobs RT, Zacco A, Greenberg B, Ciaccio PJ. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicological sciences: an official journal of the Society of Toxicology. 2004;82:341–358. [Abstract] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. [Europe PMC free article] [Abstract] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. [Abstract] [Google Scholar]
- Odermatt A, Taschner PE, Khanna VK, Busch HF, Karpati G, Jablecki CK, Breuning MH, MacLennan DH. Mutations in the gene-encoding SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+ ATPase, are associated with Brody disease. Nat Genet. 1996;14:191–194. [Abstract] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. [Abstract] [Google Scholar]
- Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M, Ferrando AA. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006a;20:1279–1287. [Abstract] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006b;103:18261–18266. [Europe PMC free article] [Abstract] [Google Scholar]
- Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. A method for high-throughput gene expression signature analysis. Genome Biol. 2006;7:R61. [Europe PMC free article] [Abstract] [Google Scholar]
- Periz G, Fortini ME. Ca(2+)-ATPase function is required for intracellular trafficking of the Notch receptor in Drosophila. EMBO J. 1999;18:5983–5993. [Europe PMC free article] [Abstract] [Google Scholar]
- Prasad V, Boivin GP, Miller ML, Liu LH, Erwin CR, Warner BW, Shull GE. Haploinsufficiency of Atp2a2, encoding the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 Ca2+ pump, predisposes mice to squamous cell tumors via a novel mode of cancer susceptibility. Cancer Res. 2005;65:8655–8661. [Abstract] [Google Scholar]
- Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, Millar SE, Pear WS, Parmacek MS. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–7444. [Abstract] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. [Europe PMC free article] [Abstract] [Google Scholar]
- Rand MD, Lindblom A, Carlson J, Villoutreix BO, Stenflo J. Calcium binding to tandem repeats of EGF-like modules. Expression and characterization of the EGF-like modules of human Notch-1 implicated in receptor-ligand interactions. Protein Sci. 1997;6:2059–2071. [Europe PMC free article] [Abstract] [Google Scholar]
- Ray WJ, Yao M, Nowotny P, Mumm J, Zhang W, Wu JY, Kopan R, Goate AM. Evidence for a physical interaction between presenilin and Notch. Proc Natl Acad Sci U S A. 1999;96:3263–3268. [Europe PMC free article] [Abstract] [Google Scholar]
- Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. [Europe PMC free article] [Abstract] [Google Scholar]
- Robertson L, Sauder MB. Basal Cell Carcinoma in Type 2 Segmental Darier’s Disease. J Skin Cancer. 2012;2012:839561. [Europe PMC free article] [Abstract] [Google Scholar]
- Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. [Europe PMC free article] [Abstract] [Google Scholar]
- Roti G, Stegmaier K. Targeting NOTCH1 in hematopoietic malignancy. Critical reviews in oncogenesis. 2011;16:103–115. [Abstract] [Google Scholar]
- Sakuntabhai A, Ruiz-Perez V, Carter S, Jacobsen N, Burge S, Monk S, Smith M, Munro CS, O’Donovan M, Craddock N, et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet. 1999;21:271–277. [Abstract] [Google Scholar]
- Stegmaier K, Ross KN, Colavito SA, O’Malley S, Stockwell BR, Golub TR. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36:257–263. [Abstract] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. [Europe PMC free article] [Abstract] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. The New England journal of medicine. 2012;366:207–215. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108:17761–17766. [Europe PMC free article] [Abstract] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. [Abstract] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & development. 2006;20:2096–2109. [Europe PMC free article] [Abstract] [Google Scholar]
- Wong GT, Manfra D, Poulet FM, Zhang Q, Josien H, Bara T, Engstrom L, Pinzon-Ortiz M, Fine JS, Lee HJ, et al. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004;279:12876–12882. [Abstract] [Google Scholar]
- Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB, Niessen K, Plowman GD. Chronic DLL4 blockade induces vascular neoplasms. Nature. 2010;463:E6–7. [Abstract] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ccr.2013.01.015
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1535610813000378/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.ccr.2013.01.015
Article citations
Depolarization induces calcium-dependent BMP4 release from mouse embryonic palate mesenchymal cells.
Nat Commun, 15(1):9806, 12 Nov 2024
Cited by: 0 articles | PMID: 39532850 | PMCID: PMC11558011
Targeting Acute Myeloid Leukemia Stem Cells through Perturbation of Mitochondrial Calcium.
Cancer Discov, 14(10):1922-1939, 01 Oct 2024
Cited by: 0 articles | PMID: 38787341
CAD204520 Targets <i>NOTCH1</i> PEST Domain Mutations in Lymphoproliferative Disorders.
Int J Mol Sci, 25(2):766, 07 Jan 2024
Cited by: 2 articles | PMID: 38255842 | PMCID: PMC10815907
Comprehensive analysis of the proximity-dependent nuclear interactome for the oncoprotein NOTCH1 in live cells.
J Biol Chem, 300(1):105522, 01 Dec 2023
Cited by: 0 articles | PMID: 38043798 | PMCID: PMC10788534
The reciprocal regulation between mitochondrial-associated membranes and Notch signaling in skeletal muscle atrophy.
Elife, 12:RP89381, 15 Dec 2023
Cited by: 0 articles | PMID: 38099641 | PMCID: PMC10723794
Go to all (99) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Clinical Trials (2)
- (1 citation) ClinicalTrials.gov - NCT01734681
- (1 citation) ClinicalTrials.gov - NCT01056029
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE5716
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Cell Chem Biol, 27(6):678-697.e13, 07 May 2020
Cited by: 23 articles | PMID: 32386594 | PMCID: PMC7305996
Leukemia-specific delivery of mutant NOTCH1 targeted therapy.
J Exp Med, 215(1):197-216, 20 Nov 2017
Cited by: 23 articles | PMID: 29158376 | PMCID: PMC5748843
Targeting oncogenic Notch signaling with SERCA inhibitors.
J Hematol Oncol, 14(1):8, 06 Jan 2021
Cited by: 30 articles | PMID: 33407740 | PMCID: PMC7789735
Review Free full text in Europe PMC
Discovery of novel SERCA inhibitors by virtual screening of a large compound library.
Eur J Med Chem, 46(5):1512-1523, 25 Feb 2011
Cited by: 12 articles | PMID: 21353727 | PMCID: PMC3065555
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: R01 CA 092433
Grant ID: R21 CA098484
Grant ID: P01 CA 068484-11A1
Grant ID: P30 CA016520
Grant ID: R01 CA092433
NCRR NIH HHS (2)
Grant ID: UL1 RR025758
Grant ID: 5 UL1 RR025758





