Abstract
Free full text

Plenary paper
Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1
Associated Data
Abstract
Point mutations that trigger ligand-independent proteolysis of the Notch1 ectodomain occur frequently in human T-cell acute lymphoblastic leukemia (T-ALL) but are rare in murine T-ALL, suggesting that other mechanisms account for Notch1 activation in murine tumors. Here we show that most murine T-ALLs harbor Notch1 deletions that fall into 2 types, both leading to ligand-independent Notch1 activation. Type 1 deletions remove exon 1 and the proximal promoter, appear to be RAG-mediated, and are associated with mRNA transcripts that initiate from 3′ regions of Notch1. In line with the RAG dependency of these rearrangements, RAG2 binds to the 5′ end of Notch1 in normal thymocytes near the deletion breakpoints. Type 2 deletions remove sequences between exon 1 and exons 26 to 28 of Notch1, appear to be RAG-independent, and are associated with transcripts in which exon 1 is spliced out of frame to 3′ Notch1 exons. Translation of both types of transcripts initiates at a conserved methionine residue, M1727, which lies within the Notch1 transmembrane domain. Polypeptides initiating at M1727 insert into membranes and are subject to constitutive cleavage by γ-secretase. Thus, like human T-ALL, murine T-ALL is often associated with acquired mutations that cause ligand-independent Notch1 activation.
Introduction
Notch receptors participate in a signaling pathway of broad importance in development, immunity, and disease, including cancer. The clearest association of Notch and cancer is in T cell acute lymphoblastic leukemia/lymphoma (T-ALL).1 Somatic gain-of-function mutations in Notch1 occur in the majority of human and murine T-ALLs, but the most common mutations reported to date differ in kind between the 2 species. In human T-ALL, the most frequent Notch1 mutations are point substitutions or small in-frame insertions or deletions in the Notch1 negative regulatory region (NRR),2 an extracellular domain composed of 3 Lin12/Notch repeats and a heterodimerization domain that holds Notch receptors in the “off-state” in the absence of ligand.3 NRR mutations abrogate NRR function4,5 and lead to successive ligand-independent cleavages. The first cleavage is carried out by metalloproteases of the ADAM family6 at a site just external to the transmembrane domain, which primes the protein for cleavage within its transmembrane domain by γ-secretase.7 γ-Secretase cleavage releases intracellular Notch1 (ICN1), allowing it to translocate to the nucleus and form a transcription activation complex with the DNA-binding factor CSL and coactivators of the Mastermind-like family.
In contrast, the most common Notch1 mutations in murine T-ALL described to date are stop codon or frameshift mutations that result in deletion of a C-terminal PEST degron domain. PEST deletions occur at frequencies of 30% to 80% in murine T-ALL, depending on the genetic background.8–12 PEST deletions also occur in 10% to 20% of human T-ALLs, often in cis with NRR mutations in the same allele.2 When combined with NRR mutations, PEST deletions cause synergistic increases in Notch1 signal dose by stabilizing ICN1, but PEST deletions alone have little or no intrinsic signaling activity and are not oncogenic.12 Of note, most cell lines derived from murine T-ALLs have detectable ICN1 and are sensitive to γ-secretase inhibitors (GSIs),8–12 indicating a dependency on Notch signaling for growth and survival. Given the absence of NRR mutations, the basis for Notch1 activation in murine T-ALL has been unclear. A clue comes from studies of murine T-ALLs arising after irradiation or in the context of RAG or ATM deficiency.13–15 Such tumors often have deletions involving 5′ portions of Notch1, but the relevance of these events to Notch1 activation in other genetic contexts has not been explored.
In this report, we describe 2 types of somatic deletions in the 5′ end of Notch1 in murine T-ALLs that cause ligand-independent Notch1 activation. Both types of deletions create Notch1 alleles that express truncated mRNAs encoding Notch1 polypeptides lacking the NRR. These findings highlight 2 common mechanisms of Notch1 activation in murine T-ALL and support the existence of strong selection for ligand-independent activation of Notch1 in both human and murine disease.
Methods
Cell culture
Mouse T-ALL cell lines were cultured in Opti-MEM medium supplemented with 8% fetal bovine serum, 1% penicillin/streptomycin, 1mM glutamine, and 0.1% β-mercaptoethanol. Human CUTLL1 T-ALL cells were grown in RPMI medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. U2OS cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All cell lines were maintained at 37°C less than 5% CO2.
Cell growth assays
Approximately 1 × 104 cells/well in 96-well plates were cultured in the presence of human IgG (10 μg/mL), anti-Notch1 inhibitory antibodies (10 μg/mL), or 1μM compound E (Tocris). Cell growth was assessed 24, 48, and 72 hours after treatment using the Cell Titer Glo viability assay (Promega). Treatments were performed in triplicate.
ICN1 reconstitution assays
MigRI retroviruses were used to transduce T-ALL cells as described.2 Transduced cells were treated with vehicle or the GSI compound E (1μM; Tocris) for 72 hours. Cells were stained with propidium iodide, and sub-G0/G1 fractions were determined by flow cytometry as described.2
Northern blot analysis
Total RNA (50 μg) was isolated with Trizol (Invitrogen) and subjected to polyA+ selection on magnetic oligo-dT beads (Invitrogen). PolyA-RNA was denatured, electrophoresed in formaldehyde–0.8% agarose gels, and transferred onto nylon membranes (GE Healthcare, Hybond-XL) in 5× saline sodium citrate–10mM NaOH. Membranes were UV cross-linked and prehybridized (Stratagene, QuikHyb) at 68°C for 10 minutes. DNA probes (10 μg) were labeled with α[32P]-dCTP using Ready-To-Go beads (GE Healthcare). Hybridization was performed at 68°C for 1 hour followed by 2 washes with 2 times saline sodium citrate for 15 minutes at 68°C and one wash with 1 time saline sodium citrate for 30 minutes at 68°C.
Southern blot analysis
Genomic DNA (10 μg) was digested with EcoRI (8 U/μg DNA) at 37°C overnight. After electrophoresis in 0.8% agarose gels, the DNA was depurinated for 20 minutes in 0.25M HCl and denatured in 0.4M NaOH for 1 hour at 25°C. DNA was transferred onto nylon membranes overnight in 0.4M NaOH. Hybridization conditions, DNA probe labeling, and subsequent washing were carried out as described for Northern blot analysis.
Western blot analysis
Whole-cell detergent lysates or T-ALL cells or transiently transfected U2OS cells were analyzed on Western blots stained with antibodies against activated ICN1-specific antibody (V-1744, Cell Signaling Technologies), β-actin (Cell Signaling Technologies), or the N-terminus of Ikaros (H-100, Santa Cruz Biotechnology).
Notch1 and Ikaros mutation analysis
Exons 26, 27, and 34 of murine Notch1 were amplified from genomic DNA according to Lin et al10 and sequenced. The Notch1 mutational status of the Tal1 transgenic cell lines has been described.16 The region of Ikaros encoding the zinc finger domains (nucleotides 373-1001) was polymerase chain reaction (PCR) amplified from T-ALL cell line cDNAs and sequenced.
5′-RACE
5′-Rapid amplification of cDNA ends (RACE) with total RNA (5 μg) was done using the FirstChoice RLM-RACE kit (Ambion). Products were amplified using primer specific for the 5′-adapter and Notch1 exon 27 with the Expand Long Template kit (Roche Diagnostics) and the following conditions: 94°C for 2 minutes; 94°C for 15 seconds, 60°C for 30 seconds, 68°C for 5 minutes, 32 cycles.
Notch1 cDNA construction
cDNAs from cell lines expressing truncated Notch1 mRNAs were PCR amplified, sequenced, and subcloned into the vector pcDNA3. Mutagenesis was performed using the Quikchange Site-Directed Mutagenesis kit (Stratagene).
PCR-based detection of type 1 Notch1 deletions
Genomic DNA was amplified using the Expand Long Template PCR System (Roche Diagnostics), the sense primer ATGGTGGAATGCCTACTTTGTA, and the antisense primer CGTTTGGGTAGAAGAGATGCTTTAC. PCR cycling conditions were: 94°C for 1 minute; 35 cycles of 94°C for 15 seconds, 58°C for 30 seconds, and 68°C for 45 seconds; and a final extension at 68°C for 5 minutes.
Notch1 reporter gene assays
Dual luciferase assays were performed using a kit (Promega) on lysates prepared from U2OS cells as described.17 The GSI N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) was the gift of Michael Wolfe, Brigham and Women's Hospital.
Ratiometric quantitative RT-PCR
A total of 1 μg of total RNA was reversed transcribed using the I-Script cDNA synthesis kit (Bio-Rad). Reverse-transcribed (RT)–PCR was performed using oligonucleotide primer pairs specific for exons 23 and 24 or exons 30 and 31 of Notch1. The PCR conditions were: 95°C for 3 minutes, followed by 40 cycles of 95°C for 10 seconds, and 60°C for 30 seconds. Notch1 and control primers used were: exon 23 sense, 5′ TGTGCAGCGTGTTAATGACT; exon 24 antisense, 5′ CAGGGCACCTACAGATGAAT; exon 30 sense, 5′ GGATGTCAATGTTCGAGGAC; exon 31 antisense, 5′ CAGCAGGTGCATCTTCTTCT; hypoxanthine phosphoribosyl transferase sense, 5′ GTTGGATACAGGCCAGACTTTGTTG; hypoxanthine phosphoribosyl transferase antisense, 5′ GATTCAACTTGCGCTCATCTTAGGC.
Relative threshold cycle values for 5′ and 3′ transcripts were measured and normalized to hypoxanthine phosphoribosyl transferase to obtain relative ΔΔ threshold cycle values.
Primers and probes
The sequences of primers and probes used for Southern and Northern blot analyses are available on request.
Results
GSI-sensitive murine T-ALLs are resistant to Notch1-selective inhibitory antibodies
Given the absence of known abnormalities involving the Notch1 ectodomain in murine T-ALL, we anticipated that Notch1-selective inhibitory antibodies specific for the ligand-binding domain (EGF repeats 11-13) or the NRR of Notch118 would suppress the growth of GSI-sensitive murine T-ALL lines. However, all 3 GSI-sensitive T-ALL cell lines12,19,20 tested were completely resistant to both types of inhibitory Notch1 antibodies (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To confirm that GSI was acting through Notch1 inhibition, we tested whether ICN1, which is downstream of γ-secretase, could rescue the lines from γ-secretase inhibition. Supplemental Figure 1 shows a representative result, in which transduction of ICN1 into the cell line 330 abrogated GSI-induced cell death.
Murine T-ALLs express truncated Notch1 transcripts
The insensitivity of murine T-ALLs to inhibitory antibodies suggested that these lines expressed constitutively active Notch1 polypeptides without intact ectodomains. To investigate this possibility, we collected 14 murine T-ALL cell lines derived from tumors arising in diverse genetic backgrounds (Table 1), and initially selected 10 of these cell lines for detailed study. These lines all have detectable ICN1 (supplemental Figure 2).
Table 1
Murine T-ALL cell lines
| Line | Genetic background | GSI-sensitive | PEST mutation |
|---|---|---|---|
| 140 | Kras-G12D | + | − |
| 144 | Kras-G12D | + | + |
| 171 | Kras-G12D | + | + |
| 330 | Kras-G12D | + | + |
| 385 | Kras-G12D | + | − |
| 430 | Kras-G12D | + | − |
| 435 | Kras-G12D | + | − |
| SCID-adh | SCID | + | − |
| G4A2 | Bcr-Abl retrovirus | + | − |
| 130 | Tal1 transgenic/Heb+/− | + | + |
| 135.1 | Tal1 transgenic/Heb+/− | + | + |
| 135.2 | Tal1 transgenic/Heb+/− | + | + |
| 720 | Tal1 transgenic/Heb+/− | + | + |
| 5406 | Tal1 transgenic/p16+/− | + | + |
Northern blot analysis performed with a probe from exon 34, the most 3′ Notch1 exon, revealed that in addition to full-length Notch1 transcripts of approximately 10 kb in size, all 10 cell lines expressed major Notch1 transcripts of approximately 5.5 kb and approximately 4.5 kb in size (Figure 1A and data not shown). Additional Northern blots revealed that the short Notch1 mRNAs were of 2 types. In 7 cell lines, the short transcripts hybridized to an exon 26-specific probe; whereas in 3 cell lines (SCID-adh and the lines 135.1 and 135.2, subclones derived from a single primary tumor), no hybridization was observed (Figure 1B). The exon 26 probe also detected a Notch1 transcript of approximately 9 kb in the cell line G4A2, whereas no long Notch1 transcripts were detected in the cell line 135.2 (Figure 1B). By contrast, an exon 1-specific probe hybridized to the short transcripts in SCID-adh, 135.1, and 135.2 cells and not to the short transcripts in the other 7 cell lines (Figure 1C; and data not shown). We designated the short transcripts that contain exon 26 and lack exon 1 “type 1” transcripts and the short transcripts that contain exon 1 and lack exon 26 “type 2” transcripts. Tsuji et al used RT-PCR to detect Notch1 transcripts in murine T-ALLs that initiated in alternative 5′ exons they designated 1a and 1b.13 However, probes specific for exons 1a and 1b failed to detect Notch1 transcripts on Northern blots in any of our lines (data not shown), suggesting that such transcripts are of low abundance and unlikely to be of functional significance.
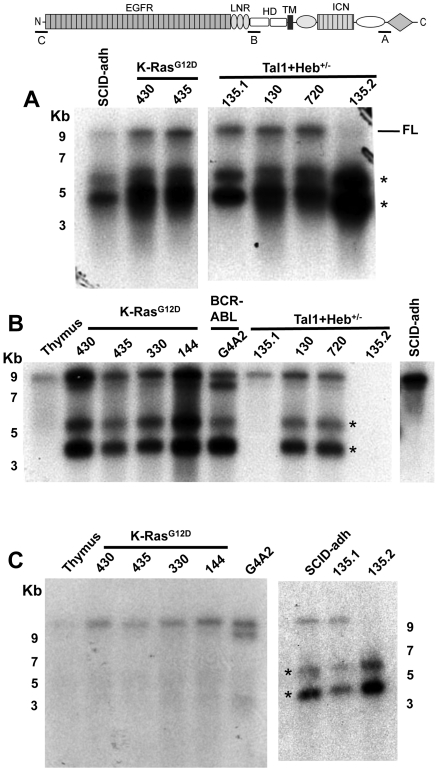
Northern blot analysis of murine T-ALL cell lines reveals 2 types of aberrant Notch1 transcripts. Northern blot analyses used polyA RNAs from normal murine thymus or the indicated cells lines. Panels A, B, and C correspond to Northern blots hybridized to probes A (exon 34), B (exon 26), or C (exon 1), which are homologous to the regions shown in the cartoon of the Notch1 locus above the blots. Numbers correspond to the position of RNA size markers in kilobases. FL indicates full-length Notch1 transcript. *Short Notch1 transcripts.
Two polyadenylation sites have been identified in the 3′-untranslated region of Notch1.13 Only the longest type 1 and type 2 transcripts hybridized to a probe lying 3′ of the first polyadenylation site (supplemental Figure 3; and data not shown). The same probe detected the approximately 10-kb Notch1 transcripts in normal thymus and the T-ALL cell lines (data not shown) but did not hybridize to the approximately 9-kb Notch1 transcript in G4A2 cells (supplemental Figure 3). These results are consistent with 2 alternative 3′ polyadenylation sites in the type1 and type 2 Notch1 transcripts, as well as in the long Notch1 transcripts expressed in G4A2 cells.
Structure of truncated Notch1 transcripts
The absence of exon 1 sequences from type 1 transcripts suggested an origin from an internal transcriptional start site. 5′-RACE using cDNA prepared from 144 cells produced several products, all with 5′ ends within exon 25 at nucleotides 35184, 35219, and 35243, respectively (Figure 2A); here and elsewhere, the nucleotide numbering used for the Notch1 is relative to the A residue in the ATG start codon in exon 1, which is designated position 1. The longest RACE product began at an A residue within the sequence TCAACTT, a sequence conserved between mouse and humans that matches the consensus sequence for an initiator (Inr) element (YYANWYY). Additional Northern blots showed that the major type 1 transcripts hybridized to a 3′ exon 25 probe and failed to hybridize to a 5′ exon 25 probe (Figure 2B). Thus, type 1 transcripts initiate mainly within or immediately 3′ of exon 25, which encodes the second Lin12/Notch repeats of Notch1.

Structure of type 1 and type 2 Notch1 transcripts. (A) Products amplified from 144 cells, a line expressing type 1 Notch1 transcripts, using a 3′ exon 27 primer and 5′ linker primer, are shown after agarose gel electrophoresis. Sequences of 5′RACE products are shown below; the basepair numbering is in relationship to the A residue in the ATG start site in exon 1 of the Notch1 gene, which is designated position 1. (B) Northern blot analysis of lines expressing type 1 Notch1 transcripts with probes from the 3′ end of exon 25 and the 5′ end of exon 25. The diagram shows the regions of the Notch1 that are encoded by exons 25 and 27, respectively. (C) Sequences of RT-PCR products obtained from murine T-ALL cell lines expressing type II transcripts. Sequencing of products obtained from 130.1 and 130.2 cells revealed a transcript in which exon 1 is spliced out of frame to exon 27. In SCID-adh cells, a product was obtained in which exon 1 is spliced to an intron 1 sequence that contains an in-frame stop codon, which is in turned joined to an internal sequence within exon 28.
Guided by the Northern blot data, we were able to determine the structure of type 2 transcripts by performing RT-PCR with primers specific for exon 1 and exon 28. All lines expressing type 2 transcripts yielded RT-PCR products (Figure 2C), whereas cell lines expressing type 1 transcripts did not. Sequencing of the RT-PCR products from 135.1 and 135.2 cells revealed a transcript in which exon 1 was spliced out of frame to the exon 27 via the normal splice donor and acceptor sites. In contrast, SCID-adh cells yielded a transcript consisting of exon 1 and 81 bp of noncontiguous intron 1 sequence joined to exon 28 at a site 12 bp 3′ of the exon 28 splice acceptor site. The point of joining of exon1 to intron 1 lay immediately 3′ of a consensus splice acceptor sequence (CAG) in intron 1 germline sequences, consistent with splicing of exon1 to a cryptic intron 1 acceptor site.
Association of aberrant transcripts with 5′ Notch1 deletions
To determine the origin of type 1 and type 2 transcripts, we assessed the integrity of Notch1 by Southern blot analysis (Figure 3). Probe A, specific for a region lying approximately 12 kb 5′ of exon 1, detected genomic rearrangements in all 7 cell lines expressing type 1 transcripts, whereas cell lines expressing type 2 transcripts gave germline bands (Figure 3A). The rearranged fragment detected with probe A in the cell line 130 was slightly smaller than the rearranged fragments in other “type 1” cell lines, whereas the rearranged fragment in the G4A2 cell line was amplified at least several-fold relative to the germline fragment.
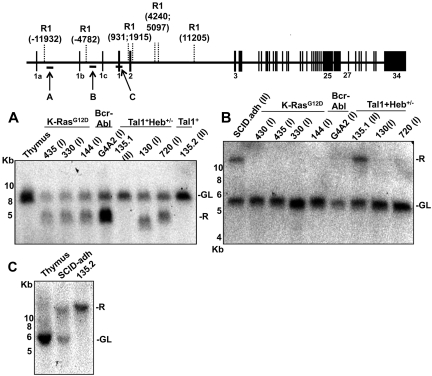
Southern blot analysis reveals 2 types of 5′ Notch1 genomic rearrangements in murine cell lines expressing type 1 (I) or type 2 (II) aberrant transcripts. Genomic DNA (10 μg) from the indicated cell lines was digested with Eco R1 and analyzed on Southern blots. Panels A, B, and C correspond to Southern blots hybridized to probes A, B, and C, which are homologous to the regions shown in the diagram of the Notch1 locus above the blots. The positions of genomic Eco R1 sites (R1) around the Notch1 locus are expressed relative to the position of the ATG start codon in exon 1, which is designated position 1. The positions of murine-specific alternative 5′ Notch1 exons 1a, 1b, and 1c are also shown. R indicates rearranged; and GL, normal genomic locus.
Southern blots hybridized to probes specific for sequences lying approximately 4.8 kb 5′ of exon 1 (B) or within exon 1 (C) detected genomic rearrangements in cell lines expressing type 2 transcripts, but not in cell lines expressing type 1 transcripts (Figure 3B-C). The cell line 135.2 showed a rearranged band and no germline band with probe C, which is consistent with the absence of normal Notch1 transcripts in this line (Figure 1). Additional Southern blots (data not shown) hybridized to probes specific for the region from intron 2 to intron 8 revealed that 135.1 and SCID-adh DNA yielded germline-sized restriction fragments of reduced signal intensity and that 135.2 cells produced no hybridization signals, suggesting that these lines all have intragenic Notch1 deletions involving regions 3′ of exon 1.
In the course of our studies, we become aware of the work of Jeannet et al showing that the expression of truncated Notch1 transcripts could be induced by Cre-mediated deletion of exon 1 and proximal Notch1 promoter sequences.21 Based on this insight, the Southern blot data, and prior work by Tsuji et al,13–15 we designed PCRs that allowed us to deduce a common deleted region in cell lines expressing type 1 transcripts (Figure 4A). The 5′ DNA break in all of the lines was located 8191 bp 5′ of the ATG start site in exon 1 of Notch1. In 6 of 7 cell lines, the 3′ DNA break was located 3575 bp 3′ of the ATG start codon, whereas in the line 130 the 3′ break occurred 1681 bp 3′ of the ATG start codon. Notably, all of the cloned rearranged sequences had features pointing to RAG involvement, including occurrence immediately adjacent to sequences resembling RAG signal sequences (RSSs); various degrees of exonucleolytic “nibbling”; addition of random bases consistent with N-nucleotides; and the presence of P nucleotides. We subsequently screened 4 additional cell lines (140, 171, 385, and 5406, described in Table 1) for the presence of type 1 deletions by PCR, all of which were all positive (Figure 4A). In addition, using this PCR method, Jeannet et al detected deletions involving the same RSS sites in approximately 75% of T-ALLs arising in an Ikaros hypomorphic background.21

Evidence of RAG involvement in Notch1 rearrangements in murine T-ALL. (A) Rearrangements in Notch1 deduced from sequencing of PCR products generated from 11 cell lines with type 1 deletions (top) and 3 cell lines with type 2 deletions (bottom). GL is the sequence of the germline DNA flanking the breakpoints. Nucleotide positions are expressed relative to the ATG start codon in exon 1 of Notch1. Flanking sequences resembling RAG recognition sequences are boxed. Residues matching the consensus RAG signal sequence (a CACATGT heptamer followed by a 12- or 23-bp spacer and the nonameric sequence ACAAAAAAC) are denoted with an asterisk. N nucleotides and P nucleotides (underlined) are also shown. The point of joining in SCID-adh contains a single cytosine residue (underlined) of unknown origin. (B) Distribution of RAG2 binding and H3K4 trimethylation across the murine Notch1 locus. ChIP-Seq was performed with antibodies specific for RAG2 and H3K4-me3 on DNA immunoprecipitated from normal thymocytes (αRAG2 WT), thymocytes expressing a RAG1 D708A mutant (αRAG2 Mut) that binds chromatin but is catalytically inactive, and homozygous RAG2 knockout thymoctyes (αRAG2−/−).22 Histograms showing sequence reads that aligned to the murine genome are superimposed on a diagram of the Notch1 locus. The y-axis of each histogram corresponds to the number of aligned reads per 106 total reads.
To further explore the possible role of RAG in type 1 rearrangements, we mined the ChIP-Seq data of Ji et al22 to determine whether RAG2 associates with the 5′ end of Notch1 in normal thymocytes. Binding of RAG2 near the transcriptional start site in exon 1 of Notch1 was readily detectable in wild-type thymocytes and in thymocytes expressing RAG1 with a D708A mutation that is permissive for binding to chromatin but abolishes RAG catalytic function (Figure 4B). The major RAG2 binding peak overlapped with the ChIP-Seq peak for histone H3K4 trimethylation (H3K4-me3), an activation mark that may be recognized by the plant homology domain of RAG2.22 Thus, RAG2 associates with Notch1 in thymocytes near the ectopic RSSs that give rise to type 1 rearrangements.
Sequencing of RT-PCR products and Southern blot results made it probable that the deletions associated with the type 2 transcripts extended close to exon 27 in 135.1 cells and 135.2 cells and exon 28 in SCID.adh cells. A PCR strategy based on this reasoning amplified the DNA sequences flanking the deleted regions in these cell lines (Figure 4A). In SCID-adh cells, the 5′ DNA break was located within intron 1 at a position 80 bp 3′ of the cryptic intron 1 splice site, and the 3′ break occurred 12 bp 3′ of the start of exon 28. Joining of the sequences is predicted to result in an approximately 38-kb deletion. A single cytosine nucleotide of unknown origin was inserted at the site of nonhomologous DNA joining. The 135.1 and 135.2 cells harbored the same complex genomic rearrangement (Figure 4A). In both lines, breakpoints lying within intron 1 and exon 26 were joined via a 184-bp insertion derived from a region approximately 21 kb 5′ of exon 1. The 3′ end of the inserted region contains a 7-bp sequence homologous to DNA at the site of breakage in exon 26, suggesting the involvement of microhomology-mediated end-joining, a mechanism implicated in many chromosomal aberrations found in lymphoid malignancies.23 None of the genomic breakpoints in lines expressing type 2 transcripts has recognizable flanking RSS-like sequences.
Ikaros mutations are common, but not universal, in cell lines expressing aberrant Notch1 transcripts
Jeannet et al note that aberrant Notch1 transcripts are expressed by tumors arising in Ik L/L mice bearing a germline hypomorphic Ikaros mutation and a conditional deletion of exon 1 of Notch1 catalyzed by CD4-Cre.21 The same floxed Notch1 allele produced loss-of-function phenotypes in prior work, suggesting that loss of Ikaros function might be a key factor in the activation of transcription from the 3′ end of Notch1. Ikaros function can be lost because of mutations that create splice variants encoding smaller, dominant negative protein isoforms, or by point mutations involving the zinc-finger region that abrogate binding to DNA. Western blots (supplemental Figure 4) revealed Ikaros isoforms consistent with dominant negative polypeptides in 2 of the type 2 lines (135.1 and 135.2) and one of the type 1 lines (130). In addition, SCID.adh cells (a type 2 line) contained decreased amounts of an Ikaros isoform of approximately normal size. Five of the remaining cell lines were analyzed by sequencing of Ikaros cDNAs. A heterozygous T to C mutation that results in a L188P substitution in the third zinc finger was found in the 435 line, and a heterozygous G to A mutation that results in a R162Q in the second zinc finger was found in the 720 line (data not shown). Both mutations are close to the position of other Ikaros mutations described in murine T-ALL in previous studies by other workers in the field24 and probably have deleterious effects. The remaining lines, 130, 330, and 430, had wild-type Ikaros sequences.
These data are consistent with past work showing that Ikaros loss-of-function mutations are found in a substantial fraction of murine T-ALL,24 and are strongly selected in the context of Notch1 activation.25,26 However, because 3 of the 6 lines with type 1 deletions we analyzed expressed apparently normal forms of Ikaros, loss of Ikaros function does not appear to be essential for activation of transcription from the 3′ end of Notch1.
An intramembranous methionine residue serves as the translational start site in aberrant Notch1 transcripts
Murine T-ALLs are generally Notch-dependent and sensitive to inhibition by γ-secretase (supplemental Figure 1 and data not shown),8–10,12,16 which cleaves intramembranously.7 Virtual translation of type 2 transcripts revealed that the start codon in exon 1 was either spliced out of frame (135.1 and 135.2 cells) or was followed by an intronic stop codon (SCID-adh cells; Figures 2C, C,5A).5A). As a result, the first in-frame ATG in both kinds of type 2 transcripts encodes M1727, a residue lying within the N-terminal portion of the Notch1 transmembrane domain. Virtual translation of the longest type 1 transcript showed the presence of 3 in-frame ATG codons, M1616, M1659, and M1727 (Figure 5A). M1616 and M1659 are located in the C-terminal portion of the HD domain, making it is uncertain whether the resulting polypeptides would insert into membranes. In contrast, a polypeptide initiating with M1727 would have a hydrophobic N-terminus that could promote membrane insertion.
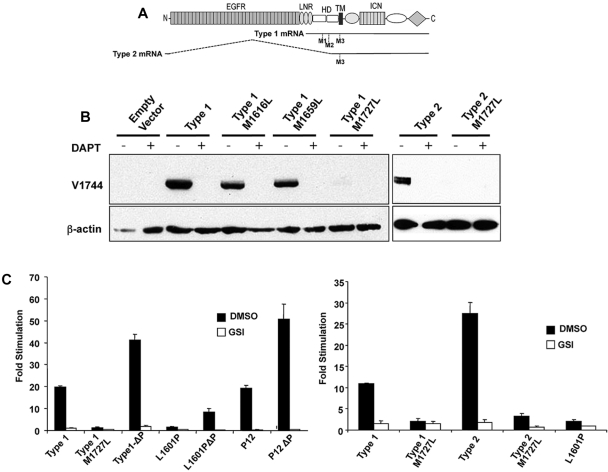
M1727 serves as the translational start site in aberrant type 1 and type 2 Notch1 transcripts. (A) Schematic showing the positions of codons M1616 (M1), M1659 (M2), and M1727 (M3) within type 1 and type 2 Notch1 transcripts relative to the full-length Notch1 polypeptide. (B) Western blot analysis of 293 cells transiently transfected with empty pcDNA3, type 1 transcript cDNA (corresponding to nucleotides 4438-7665 of murine Notch1), type 1 transcript cDNAs bearing M to L point mutations in codons 1616, 1659, or 1727, type 2 transcript cDNA from SCID.adh cells, or type 2 transcript cDNA with an M to L point mutation in codon 1727. Blots were stained with antibodies specific for activated Notch1 (V1744 antibody, Cell Signaling Technology) or β-actin. (C) Notch reporter gene studies conducted in U2OS cells transiently transfected in triplicate with equivalent amounts of a CSLx4 firefly luciferase reporter gene, an internal Renilla luciferase control gene, and empty pcDNA3 vector or pcDNA3 vectors containing the indicated cDNAs. The left-hand panel compares the activities of a murine type 1 Notch1 transcript; a murine type 1 Notch1 transcript containing a M1727L mutation; a murine type 1 Notch1 transcript containing a stop mutation introduced at codon 2415 (ΔP) that deletes the C-terminal 116 amino acids of murine Notch1; a full-length human NOTCH1 cDNA containing a weakly activating NRR mutation, L1601P, with and without a ΔP frameshift mutation at codon 2473 that deletes the C-terminal 83 amino acids of human Notch1; and a full-length human NOTCH1 cDNA containing a strongly activating in-frame insertion in the NRR designated P12, with and without a ΔP frameshift mutation at codon 2473. The right-hand panel compares the activities of murine type 1 and type 2 transcripts with a human NOTCH1 cDNA bearing a L1601P deletion. Each cDNA was tested in the presence and absence vehicle (dimethyl sulfoxide [DMSO]) or vehicle plus the GSI DAPT (10μM). After normalization to an internal Renilla luciferase control, firefly luciferase activities were expressed relative to the empty vector control, which was arbitrarily set to 1. Error bars represent SD. The results shown are representative of 2 independent experiments.
To test the role of M1727, type 1 and type 2 transcripts were transiently expressed in U2OS cells. Western blot analysis showed that the type 1 and type 2 transcripts drive ICN1 expression in a GSI-sensitive fashion. Furthermore, a M1727L mutation markedly decreased ICN1 production, whereas M1616L and M1659L mutations in type 1 transcripts had little or no effect on ICN1 production (Figure 5B). The M1727L mutation also markedly decreased activation of a Notch reporter gene by type 1 and type 2 transcripts (Figure 5C). In direct comparisons, the degree of Notch reporter gene activation produced by the type 1 transcript was comparable with that of a strong gain-of-function Notch1 NRR mutation first identified in the human T-ALL cell line P12 Ichikawa (P12), and substantially greater than that of a relatively weak Notch1 NRR mutation, L1601P (Figure 5C).2,4,12 Like the human T-ALL-associated NRR mutations,2,4,12 reporter gene activation by a type 1 transcript was increased several-fold by inclusion of a PEST deletion in cis. Finally, GSIs such as DAPT (Figure 5C) and compound E (not shown) sharply reduced Notch reporter gene activation by both type 1 and type 2 transcripts.
M1727 is conserved in human Notch1 (where it is encoded by codon 1738), suggesting that it might serve as an internal translational start site in human T-ALLs in which Notch1 is activated through mechanisms other than NRR mutations. The cell line CUTLL1 is derived from a human T-ALL with a (7,9) chromosomal translocation in which DNA breakpoints lying 3′ of the TCRB Jβ2.4 joining segment and within intron 27 of NOTCH1 are joined to create a TCRB-NOTCH1 fusion gene.27 CUTLL1 cells express high amounts of ICN1 and are GSI sensitive,28 indicating that the fusion gene must encode a membrane-tethered form of Notch1. Prior work identified a chimeric transcript in which Jβ2.4 is spliced in-frame to exon 28 of NOTCH127 but did not describe the structure of the 5′ end of the fusion transcript. 5′-RACE using CUTLL1 cell mRNA identified a chimeric transcript consisting of 45 bp of uncertain origin linked to Jβ2.4 sequences followed by exon 28 of NOTCH1 (supplemental Figure 5). M1738 is the only possible translational start site in the fusion transcript, suggesting that it initiates translation of γ-secretase-dependent Notch1 polypeptides in CUTLL1 cells.
Notch1 deletions and truncated transcripts appear before extrathymic spread of T-ALL
Intrathymic T-cell development depends on activation of Notch1 by ligands, such as DLL4,29 expressed on thymic stromal cells. If ligand is limiting within the thymic niche, cells acquiring Notch1 mutations that lead to ligand-independent activation would gain a selective advantage during intrathymic stages of T-ALL development. To explore this issue, we analyzed primary thymic T-ALLs arising in Tal1/Lmo2 transgenic mice. PCR amplification of genomic DNAs from 2 of 3 primary thymic T-ALLs yielded products that were proven by sequencing to contain “type 1” Notch1 deletions (Figure 6A). A ratiometric quantitative RT-PCR assay that compares the levels of Notch1 5′ and 3′ transcripts revealed a significant excess of 3′ transcripts in the 2 tumors with type 1 deletions as well as in positive control T-ALL cell lines expressing type 1 (144) or type 2 (135.2) transcripts (Figure 6B). By contrast, normal thymocytes and a primary tumor lacking Notch1 deletions expressed equal numbers of 5′ and 3′ transcripts. These results indicate that deletions that produce ligand-independent Notch1 activation are selected during intrathymic stages of T-ALL development.
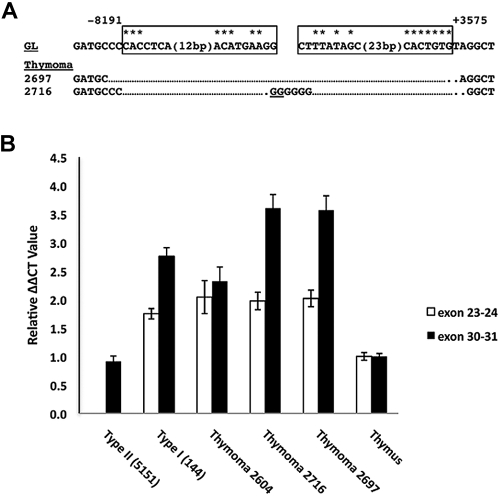
Detection of 5′ deletions and aberrant Notch1 transcripts in primary murine “thymomas.” (A) Sequences of PCR products obtained by amplification of genomic DNA isolated from 2 thymic lymphomas with primers flanking the most common breakpoints associated with type 1 aberrant transcripts. Sites of DNA breakage and joining, as deduced from sequencing of PCR products, are shown. Residues matching the consensus RAG recognition sequence (CACAGTG followed by a 12- or 23-bp spacer and the sequence ACAAAAAAC) are denoted with an asterisk. N nucleotides and P nucleotides (underlined) are also shown. GL indicates germline DNA flanking the breakpoints. Boxes represent sequences resembling RAG signal sequences. (B) Ratiometric Notch1 quantitative RT-PCR analysis. The relative amounts of transcripts containing 5′ (exons 23 and 24) and 3′ (exons 30 and 31) Notch1 sequences were determined for the tumors in panel A and normal murine thymus, a cell line with a homozygous type 2 deletion (135.2), and a cell line with a heterozygous type 1 deletion (144). Each determination was made in triplicate. The results shown are representative of 2 independent experiments.
Discussion
We have described 2 deletion-based mechanisms of Notch1 activation in detail (summarized in Figure 7). “Type 1” deletions appear to be RAG-mediated, remove the 5′ proximal promoter and exon 1 of Notch1, and activate transcription from internal sites in or adjacent to exon 25. RAG activity appears early in T-cell development in CD4− CD8−ckit+CD25− “double-negative” cells30 and is maintained through the early CD4+CD8+ double-positive stage of T-cell development, defining a developmental window within which these deletions occur. Of note, “ChIP-Seq” analyses show RAG2 binding to the 5′ end of Notch1 in normal thymocytes in a distribution that is largely coincident with H3K4-me3 marks,22 which are enriched in the proximal promoters of transcribed genes. Presumably, once RAG2 binds the Notch1 promoter, it can recruit RAG1, forming a functional recombinase that may catalyze deletions involving nearby ectopic RSSs. “Type 2” deletions remove DNA between exon 2 to exons 26 to 28, leading to expression of aberrant Notch1 mRNA splice variants from the 5′ proximal promoter. These deletions are similar to those seen in radiation-induced murine T-ALLs13–15 and presumably arise through random DNA breaks and either nonhomologous end joining reactions or microhomology-based DNA repair.
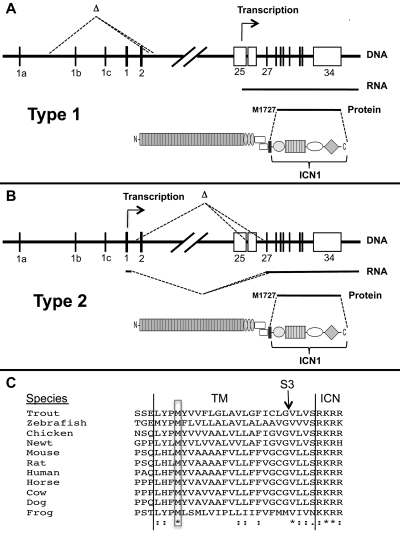
Mechanisms of ligand-independent ICN1 production in T-ALLs bearing Notch1 deletions. (A-B) Structure and functional consequences of type 1 and type 2 Notch1 deletions. (C) Conservation of M1727 in vertebrate Notch1 receptors. *Identical residues. “:” indicates conserved residues. TM indicates transmembrane domain; S3, site of intramembranous γ-secretase cleavage; and ICN, intracellular Notch.
Prior work has pointed to the importance of truncated Notch1 transcripts similar to those described here. In the original report,7,9 chromosomal translocations involving NOTCH1 in human T-ALL, the DNA breaks in NOTCH1 clustered in intron 24, which was joined to TCRβ enhancer/promoter sequences.31 The NOTCH1 transcriptional start sites in these T-ALLs mapped to exon 25, close to the conserved initiator (Inr) site identified in murine tumors with type 1 deletions. Other studies have identified the 5′ portion of Notch1 spanning the region from exon 25 through intron 27 as a common site of retrovirus or transposon insertion in murine T-ALL.32–34 Finally, Tsuji et al described T-ALLs arising in irradiated ATM−/− and SCID mice that were associated with 5′ deletions in Notch1 and abnormally short Notch1 transcripts.13–15 Our work extends these studies to show that 5′ deletions in Notch1 are common in tumors arising independent of irradiation in a variety of T-ALL-prone genetic backgrounds.
In tumors with type 2 deletions, the expression of Notch1 transcripts is presumably governed by the 5′ promoter elements and associated factors that regulate normal Notch1 expression. Of greater interest is the transcriptional regulation of Notch1 in T-ALLs with type 1 deletions. The deleted region contains several binding sites for positive regulators of Notch1 expression,35 including CSL/ICN1 sites at approximately −5 kb, approximately −1 kb, and approximately 1.4 kb, and 2 E2A-binding sites located at approximately −3.0 kb and approximately −5.6 kb. Not all these sites need to be removed for type 1 transcripts to be expressed, as Jeannet et al show that Cre-mediated deletion of the region from approximately −2.7 kb to approximately 0.8 kb is sufficient to drive the expression of type 1 transcripts.21 How type 1 deletions activate transcription from the cryptic promoter in exon 25 is uncertain. Chromatin marks that are laid down during the RNA elongation are thought to suppress internal transcription initiation.36 Type 1 deletions remove this inhibitory influence and might also relieve promoter competition and/or alter chromatin structure so as to permit loading of positive regulators onto chromatin near exon 25.
The aberrant transcripts resulting from type 1 and type 2 deletions appear to initiate translation at M1727, a conserved residue in the N-terminal portion of the transmembrane domain of Notch1 (Figure 7C). Of note, M1727 was previously shown to serve as an internal translational start site in an engineered murine Notch1 cDNA,37 and proviral insertions into 5′ portions of Notch1 in murine T-ALL drive the expression of transcripts in which the first in-frame ATG encodes M1727.32,34,38 The structure of the TCRB-NOTCH1 fusion transcript in the human T-ALL cell line CUTLL1 suggests that the same methionine serves as a translational start site in some human T-ALLs as well. Interestingly, a leukemogenic form of Notch2 transduced by feline leukemia virus also initiates translation from a methionine located within the transmembrane domain of Notch2.39 Thus, a latent ability to express truncated polypeptides that generate ligand-independent signals is a feature of several vertebrate Notch receptors and warrants further study.
One model for Notch activation proposes that nicastrin binds the extracellular “stub” produced by metalloprotease cleavage of Notch and thereby delivers Notch to the active site of γ-secretase.40 The activity and γ-secretase dependence of the Notch1 polypeptide created by translational initiation at M1727 argues against this model and is consistent with other recent genetic studies indicating that nicastrin stabilizes γ-secretase, but is nonessential for γ-secretase recognition and cleavage of Notch.41,42
Although murine T-ALL is commonly associated with 5′ deletions in Notch1, analysis of close to 200 human T-ALLs (on Affymetrix arrays, with either 615 000 features (250K + 50K arrays) or SNP 6 arrays (1.8 million features) has not revealed focal copy number variants in NOTCH1 (Dr Charles Mullighan, St Jude Children's Research Hospital, written communication, August 4, 2010). It thus appears that the relatively large deletions that are observed frequently in murine Notch1 are rare, if they occur at all, in NOTCH1 in human T-ALL, with the important caveat that additional studies are needed to exclude the presence of smaller deletions or point mutations involving the 5′ end of the gene. The paucity of deletions involving NOTCH1 in human T-ALL may be related to divergence in the ectopic RSSs, particularly the heptamer sequences of the 2 RSSs that are most commonly involved in murine Notch1 deletions (supplemental Figure 6).
A proposed strategy for selective targeting of Notch receptors in cancer and other diseases is the use of inhibitory antibodies that recognize either the ligand-binding domain or NRR.18,43 The expression of truncated polypeptides lacking these regions, which is common in murine T-ALL and also occurs in human T-ALLs bearing the t(7;9), reveals limitations in this strategy and suggests a possible mechanism for resistance to therapeutics directed against the ectodomain of Notch1. Based on these insights, a multipronged approach to Notch targeting, including protease inhibitors and novel agents that target Notch in the nucleus,44 seems prudent.
Acknowledgments
The authors thank Yiping He for technical support, Grace Teng and David Schatz (Yale University) for providing RAG2 and H3K4-me3 ChIP-Seq data across the Notch1 locus, and Dr Hans Huber and Dr Jose Aste-Amezaga (Merck Inc) for providing Notch1 inhibitory antibodies.
This work was supported by the National Institutes of Health (J.C.A., W.S.P., and S.C.B.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.D.A., L.X., J.M., M.Y.C., and P.K. performed the research and assisted with the collection and analysis of data; W.S.P. and M.K. provided critical reagents; and W.S.P., S.C.B., P.K., S.C., and J.C.A. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.Y.C. is Division of Hematology and Oncology, University of Michigan Cancer Center, Ann Arbor, MI.
Correspondence: Jon C. Aster, Department of Pathology, Brigham and Women's Hospital, Room 630E, New Research Bldg, 77 Ave Louis Pasteur, Boston, MA 02115; e-mail: ude.dravrah.hwb.scir@retsaj.
References
Articles from Blood are provided here courtesy of The American Society of Hematology
Full text links
Read article at publisher's site: https://doi.org/10.1182/blood-2010-05-286328
Read article for free, from open access legal sources, via Unpaywall:
https://ashpublications.org/blood/article-pdf/116/25/5455/1462103/zh805110005455.pdf
Free to read at www.bloodjournal.org
http://www.bloodjournal.org/cgi/content/abstract/116/25/5455
Free after 12 months at www.bloodjournal.org
http://www.bloodjournal.org/cgi/content/full/116/25/5455
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1182/blood-2010-05-286328
Article citations
NOTCH1 fusions in pediatric T-cell lymphoblastic lymphoma: A high-risk subgroup with CCL17 (TARC) levels as diagnostic biomarker.
Hemasphere, 8(7):e117, 27 Jun 2024
Cited by: 1 article | PMID: 38948925
IKAROS Family Transcription Factors in Lymphocyte Differentiation and Function.
Adv Exp Med Biol, 1459:33-52, 01 Jan 2024
Cited by: 0 articles | PMID: 39017838
Review
Inhibition of the NOTCH and mTOR pathways by nelfinavir as a novel treatment for T cell acute lymphoblastic leukemia.
Int J Oncol, 63(5):128, 06 Oct 2023
Cited by: 0 articles | PMID: 37800623 | PMCID: PMC10609462
Outlier Expression of Isoforms by Targeted or Total RNA Sequencing Identifies Clinically Significant Genomic Variants in Hematolymphoid Tumors.
J Mol Diagn, 25(9):665-681, 05 Jul 2023
Cited by: 2 articles | PMID: 37419244 | PMCID: PMC10488324
IL-15 Prevents the Development of T-ALL from Aberrant Thymocytes with Impaired DNA Repair Functions and Increased NOTCH1 Activation.
Cancers (Basel), 15(3):671, 21 Jan 2023
Cited by: 2 articles | PMID: 36765626 | PMCID: PMC9913776
Go to all (73) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL.
Blood, 116(25):5443-5454, 09 Sep 2010
Cited by: 55 articles | PMID: 20829372 | PMCID: PMC3100247
Cooperation between somatic Ikaros and Notch1 mutations at the inception of T-ALL.
Leuk Res, 35(11):1512-1519, 15 Aug 2011
Cited by: 2 articles | PMID: 21840596
Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors.
PLoS One, 5(2):e9094, 08 Feb 2010
Cited by: 114 articles | PMID: 20161710 | PMCID: PMC2817004
The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia.
Blood, 129(9):1124-1133, 23 Jan 2017
Cited by: 123 articles | PMID: 28115368
Review
 1
1




