Abstract
Free full text

Metformin Inhibits Cell Proliferation, Migration and Invasion by Attenuating CSC Function Mediated by Deregulating miRNAs in Pancreatic Cancer Cells
Abstract
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the United States, which is, in part, due to intrinsic (de novo) and extrinsic (acquired) resistance to conventional therapeutics, suggesting that innovative treatment strategies are required for overcoming therapeutic resistance to improve overall survival of patients. Oral administration of metformin in patients with diabetes mellitus has been reported to be associated with reduced risk of pancreatic cancer and that metformin has been reported to kill cancer stem cells (CSC); however, the exact molecular mechanism(s) has not been fully elucidated. In the current study, we examined the effect of metformin on cell proliferation, cell migration and invasion, and self-renewal capacity of CSCs and further assessed the expression of CSC marker genes and microRNAs (miRNA) in human pancreatic cancer cells. We found that metformin significantly decreased cell survival, clonogenicity, wound-healing capacity, sphere-forming capacity (pancreatospheres), and increased disintegration of pancreatospheres in both gemcitabine-sensitive and gemcitabine-resistant pancreatic cancer cells. Metformin also decreased the expression of CSC markers, CD44, EpCAM, EZH2, Notch-1, Nanog and Oct4, and caused reexpression of miRNAs (let-7a, let-7b, miR-26a, miR-101, miR-200b, and miR-200c) that are typically lost in pancreatic cancer and especially in pancreatospheres. We also found that reexpression of miR-26a by transfection led to decreased expression of EZH2 and EpCAM in pancreatic cancer cells. These results clearly suggest that the biologic effects of metformin are mediated through reexpression of miRNAs and decreased expression of CSC-specific genes, suggesting that metformin could be useful for overcoming therapeutic resistance of pancreatic cancer cells.
Introduction
Pancreatic cancer is one of the most lethal malignancies, and annually 37,000 patients are newly diagnosed with pancreatic cancer, resulting in 34,000 deaths annually in the United States, ranking pancreatic cancer as the fourth leading cause of cancer-related deaths (1). Because of the absence of any specific early symptoms and the lack of early detection methods, pancreatic cancer is usually diagnosed at advanced and incurable stage (2), consequently, the 5-year overall survival rate is less than 5% (1, 2). Among many risk factors, smoking, alcohol consumption, chronic pancreatitis, and family risk factors have been widely recognized as potential risk factors of pancreatic cancer (3, 4). Moreover, epidemiologic studies have shown that diabetes, especially type 2 diabetes mellitus is positively associated with increased risk of pancreatic cancer. Therefore, anti-diabetic drugs have been investigated for their use in the prevention and/or treatment of cancers, such as pancreatic cancer, for more than a decade, although the precise molecular mechanism of action of metformin is still unknown.
The primary systemic effect of metformin for the treatment of diabetes mellitus is to increase insulin sensitivity and to decrease blood glucose levels through reduced hepatic gluconeogenesis and increased glucose uptake in peripheral tissues, including skeletal muscles and adipose tissues (3–5). A large number of epidemiologic data have revealed that the oral use of metformin in patients with diabetes mellitus elicits a protective effect by decreasing incidence of different tumors and improving prognosis of patients with cancers including pancreatic cancer (6, 7). For example, one recent large case–control clinical trial for the treatment of pancreatic cancer with metformin in patients with diabetes mellitus was conducted involving 973 patients with pancreatic cancer (including 259 patients with diabetes mellitus) and 863 controls (including 109 patients with diabetes mellitus; ref. 8) showed that patients with diabetes mellitus who received metformin had a significantly lower risk of pancreatic cancer than those who did not. In contrast, patients with diabetes mellitus who were administered insulin had a significantly higher risk of pancreatic cancer than those without insulin. These recent findings clearly suggest that the oral administration of metformin is positively associated with decreased risk of pancreatic cancer in patients with diabetes mellitus, whereas the use of insulin or other agents is associated with an increased risk. Therefore, metformin appears to exert a protective effect against the development and progression of pancreatic cancer; however, the molecular mechanism by which metformin elicits its biologic effects has not been fully established.
Among several proposed mechanism of action of metformin (9–11), recent in vitro and in vivo studies are compelling showing that metformin can block tumor growth by inactivation of breast cancer stem–like cells (CSC) and epithelial-to-mesenchymal transition (EMT) phenotypic cells that are believed to be the root cause of tumor recurrence and metastasis (12, 13) especially because CSCs or EMT phenotypic cells are highly resistant to conventional therapeutics. Moreover, it is clear that metformin could selectively kill CSCs (12, 14); however, the precise molecular mechanism of action of metformin remains to be elucidated. In the current study, we found that metformin inhibited cell proliferation, migration and invasion, and the CSC self-renewal capacity of both gemcitabine-sensitive and gemcitabine-resistant pancreatic cancer cells. These effects of metformin was, in part, due to decreased expression of CSC marker genes including CD44, EpCAM, EZH2, Notch-1, Nanog, and Oct4, which was associated with reexpression of specific miRNAs (let-7a, let-7b, let-7c, miR-26a, miR-101, miR-200b, and miR-200c) that are typically lost in pancreatic cancer and in pancreatospheres of gemcitabine-resistant cells. We have further shown that reexpression of miR-26a by the transfection of its precursor decreased the mRNA and protein expression of EZH2 and EpCAM in pancreatic cancer cells, suggesting that the reexpression of miR-26a and other specific miRNAs results in the downregulation of CSC-specific genes contributing to the killing of pancreatic cancer cells especially CSCs and EMT-type cells that are highly resistant to conventional therapeutics.
Materials and Methods
Cell culture
Human pancreatic cancer cell lines AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR were used for this study on the basis of their sensitivities to chemotherapeutic drugs, gemcitabine and Tarceva (erlotinib). AsPC-1 and MiaPaCa-2 cells were exposed to 200 nmol/L of gemcitabine and 5 μmol/L of Tarceva for 3 days every other week for about 6 months to establish gemcitabine- and Tarceva-resistant cell lines, named as AsPC-1-GTR and MiaPaCa-2-GTR, respectively. All the cell lines were maintained in Dulbecco's Modified Eagle's Media (DMEM; Invitrogen), supplemented with 5% FBS, 2 mmol/L glutamine, 50 units/mL penicillin, and 50 μg/mL streptomycin. All the cells were maintained in a standard culture condition, as described previously (15). The cell lines have been tested and authenticated in core facility at the Applied Genomics Technology Center of Wayne State University (Detroit, MI) on March 13, 2009. The method used for testing was short tandem repeat (STR) profiling using the PowerPlex 16 System from Promega.
Reagents and antibodies
Metformin was purchased from Sigma Chemicals. Difluorinated curcumin (CDF), a synthetic derivative of curcumin was synthesized as described in our earlier publication (15). Antibodies against CD44 and EpCAM were purchased from Cell Signaling Technology. Antibody against Notch-1 was purchased from Santa Cruz. Antibody against β-actin was acquired from Sigma Chemicals. Antibody against EZH2 was obtained from BD Transduction. Alexa Fluor 488 goat anti-mouse IgG for CD44 and EpCAM were purchased from Invitrogen. The miRNA and mRNA reverse transcription (RT) primers and PCR probes were purchased from Applied Biosystems. Crystal violet was purchased from Sigma.
Cell survival assay
MTT assay was conducted using AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR cells as described previously (15) All the cell lines were exposed to different concentrations of metformin (1–20 mmol/L) for 3 days of treatment. After 3 days, MTT assay was conducted as described previously (15, 16).
Clonogenic assay
Clonogenic assay was conducted to examine the effect of metformin on cell growth and proliferation in all the cells, as described previously (16). A total of 5 × 104 cells were plated in a 6-well plate. After 72 hours of exposure to 20 mmol/L of metformin, the cells were trypsinized, and 1,000 single viable cells were plated in 100-mm Petri dishes. The cells were then incubated for 10 to 12 days at 37° C in a 5% CO2/5% O2/90% N2 incubator. Colonies were stained with 2% crystal violet, washed with water, and counted.
Cell migration and invasion assays
Cell migration (wound-healing) assay was conducted to examine the capacity of cell migration and invasion, as described previously (17). Briefly, the wound was generated when the cells reached 90% to 95% confluency by scratching the surface of the plates with a pipette tip. The cells were then incubated in the absence and presence of metformin (20 mmol/L) for 18 hours and then photographed with a Nikon Eclipse TS100 microscope. Chamber invasion assay was conducted using BD BioCoat Tumor Invasion System, as described previously (16).
Sphere formation/disintegration assay
Sphere formation assay was conducted to assess the capacity of CSC self-renewal as described previously (18). Briefly, single-cell suspensions of pancreatic cancer cells were plated on ultra–low adherent wells of 6-well plates (Corning) at 1,000 cells per well in sphere formation medium (1:1 DMEM/F12 medium supplemented with B-27 and N-2; Invitrogen). After 7 days, the spheres termed as “pancreatospheres” were collected by centrifugation (300 × g, 5 minutes) and counted. The proportion of sphere-generating cells was calculated by dividing the number of cells seeded by the number of pancreatospheres. The sphere formation assay of secondary pancreatospheres was conducted by using primary pancreatospheres. Briefly, primary pancreatospheres were harvested and incubated with Accutase (Sigma) at 37° C for 5 to 10 minutes. Single-cell suspensions of pancreatospheres were plated at 500 cells per well in the sphere formation medium. After 1 or 3 weeks of incubation with metformin, secondary pancreatospheres were harvested for counting as described above.
For sphere disintegration assay, 1,000 cells per well on ultra–low adherent wells of 6-well plate were incubated for 10 days, following 5 days of drug treatment the cells were harvested as described previously (18). The pancreatospheres were collected by centrifugation and counted under a microscope as described above.
CDF, a novel synthetic curcumin-derived compound, has shown a greater bioavailability in mice than curcumin and an antitumor activity of human pancreatic cancer in vivo by inactivation of NF-κB and COX2 and deregulation of miR-21 and miR-200 (15, 16), suggesting that CDF may have an impact on the treatment of human cancers with a better outcome. Thus, our previous publications (15, 16, 19–21) provided the rationale for studying the role of CDF in the current manuscript. To examine the combined effect of CDF and metformin on CSC growth, we also conducted sphere formation and disintegration assays by the combination of metformin and CDF in pancreatic cancer cells.
Immunostaining assay and confocal imaging microscopy
For this experiment, we used enriched CSCs as described below. Single-cell suspensions of secondary sphere cells formed by the cells derived from the tumors of MiaPaCa-2 pancreatospheres developed in a mouse xenograft were plated at 5,000 cells per well in the sphere formation medium as described above. After 7 days of incubation with 20 mmol/L of metformin, the pancreatospheres were collected by centrifugation, washed with 1× PBS, and fixed with 3.7% paraformaldehyde. Monoclonal CD44 and EpCAM antibodies were used for immunostaining following the manufacturer's protocol as described previously (22). The CD44- or EpCAM-labeled pancreatospheres were photographed under a Nikon ESLIPSE E800. Confocal microscopy (Leica TCS SP5) was conducted in the MIRL Core Facility of Wayne State University School of Medicine.
Transfection of miRNA precursor miR-26a
A total of 2 × 105 cells per well of MiaPaCa-2 cells were seeded in 6-well plates and transfected with pre-miR-26a or miRNA negative control (Ambion) at a final concentration of 20 nmol/L using DharmaFECT transfection reagent (Dharmacon), following the manufacturer's protocol as described previously (22, 23). After 3 days of transfection, the transfected cells were harvested for total RNA isolation or protein extraction. The relative levels of miRNA, mRNAs, and proteins were measured using standard methods as detailed below.
Protein extraction and Western blot analysis
Western blot analysis was conducted using whole-cell lysates. Total cell lysates from different experiments were obtained by lysing the cells in the protein lysis buffer containing 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 2 mmol/L sodium fluoride, 2 mmol/L Na3VO42, 1 mmol/L EDTA, 1 mmol/L EGTA, and 1× protease inhibitor cocktail, and Western blot analysis was conducted as previously described elsewhere (24).
TaqMan miRNA real-time reverse transcriptase PCR
To determine the expression of miRNAs (let-7, miR-26a, miR-101, miRNA-200b, and miRNA-200c) in AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR cells, we used TaqMan MicroRNA Assay kit (Applied Biosystems) following manufacturer's protocol. Five nanograms of total RNA was reverse transcribed and real-time PCR reactions were carried out in 10 μL of reaction mixture as described earlier (25), using AB StepOnePlus Real-Time PCR System (Applied Biosystems). Data were analyzed using Ct method and were normalized by RNU6B or RNU48 expression in each sample.
Levels of mRNAs by real time RT-PCR
To determine the mRNA expression of EZH2, Notch-1, EpCAM, Nanog, Sox2, and Oct4 mRNAs in the cells, 2 μg of total RNAs from each sample were used for RT reaction in 20 μL of reaction volume, using a reverse transcription system (Invitrogen) according to the manufacturer's instructions. SYBR Green Assay kit (Applied Biosystems) was used for real-time PCR reaction, following manufacturer's protocol. Sequences of PCR primers were described previously (22,23). Data were analyzed using Ct method and normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each sample.
Animal experiments
The animal protocol was approved by the Animal Investigation Committee, Wayne State University. Female CB17/SCID mice (4 weeks old) were purchased from Taconic Farms and fed Lab Diet 5021 (Purina Mills, Inc.). About 5,000 pancreatospheres of MiaPaCa-2 were implanted in xenograft mouse model (4 weeks) for enriching the CSC population. The tumor was removed, and the cells were used in the formation of secondary pancreatospheres. The secondary tumor pancreatospheres were then treated with metformin for 1 or 3 weeks for assessing the effects of metformin on CSCs.
Statistical analysis
The data with mean and SD as presented here were prepared using GraphPad Prism software (version 4.03). Comparisons of treatment outcome were tested for statistical difference by the paired t test. Statistical significance was assumed at a value of P < 0.05.
Results
Effect of metformin on cell survival and clonogenicity of gemcitabine-resistant pancreatic cancer cells
To investigate the effect of metformin on cell proliferation of pancreatic cancer cells, we conducted MTT and clonogenic assays using gemcitabine-resistant AsPC-1 and MiaPaCa-2 cells (AsPC-1-GTR and MiaPaCa-2-GTR cells). We found that metformin significantly inhibited cell survival by a dose-dependent manner in AsPC-1 and MiaPaCa-2 cells as well as gemcitabine- and Tarceva-resistant cells (AsPC-1-GTR and MiaPaCa-2-GTR cells; Fig. 1A). We also found that metformin significantly decreased the clonogenicity in both drug-sensitive and drug-resistant pancreatic cancer cells compared with those cells without metformin treatment (Fig. 1B). These results suggest that metformin could elicit pronounced cell growth–inhibitory effect on both drug-sensitive and drug-resistant pancreatic cancer cells.
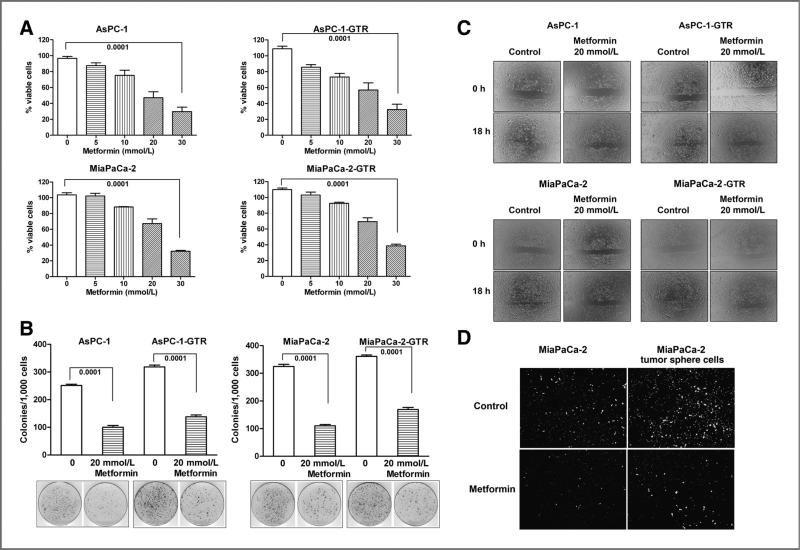
Metformin decreased cell survival (A), clonogenicity (B), wound-healing capacity (C), and invasion (D), in AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR cells. The drug-resistant cell lines AsPC-1-GTR and MiaPaCa-2-GTR were established as described under the Materials and Methods section. Wound-healing assay was conducted to assess the capacity of cell migration and invasion of both drug-sensitive and drug-resistant cells. The cells were incubated with 20 mmol/L of metformin for 18 hours. Chamber invasion assay was conducted in parental MiaPaCa-2 and its tumor sphere cells for 18 hours of incubation.
Metformin inhibited cell migration and invasion of gemcitabine-resistant pancreatic cancer cells
To examine the effect of metformin on cell migration and invasion, we conducted cell migration (wound-healing) assay using gemcitabine-sensitive and gemcitabine-resistant AsPC-1 and MiaPaCa-2 cells. We found that metformin treatment significantly inhibited the capacity of wound healing in both gemcitabine-sensitive and gemcitabine-resistant AsPC-1 and MiaPaCa-2 cells compared with cells without metformin treatment (Fig. 1C), which suggests that metformin could inhibit the cell migration and invasion in drug-resistant pancreatic cancer cells. We also conducted chamber invasion assay in parental MiaPaCa-2 and its tumor sphere cells. The results indicate that MiaPaCa-2 tumor sphere cells had greater capacity of tumor invasion than its parental cells (Fig. 1D). Metformin treatment profoundly inhibited the capacity of tumor invasion in both parental MiaPaCa-2 and its tumor sphere cells compared with the cells without treatment (Fig. 1D).
Effect of metformin on the formation/disintegration of pancreatospheres in drug-sensitive and drug-resistant pancreatic cancer cells
We examined the effect of metformin on self-renewal capacity of CSCs or CSC-like cells by conducting sphere formation assay using both drug-sensitive and drug-resistant pancreatic cancer cells. We found that metformin significantly decreased the formation of pancreatosphere in both drug-sensitive and drug-resistant cells in a dose-dependent manner (Fig. 2A). We also examined the effect of metformin and its combination with CDF, a novel curcumin-derived antitumor agent, on the formation of secondary pancreatospheres using both gemcitabine-sensitive and gemcitabine-resistant AsPC-1 and MiaPaCa-2 cells (Fig. 2B). We found that metformin and its combination with CDF also inhibited the formation of secondary pancreatospheres of gemcitabine-resistant pancreatic cancer cells. To further examine the effect of metformin on disintegration of pancreatospheres, we incubated cells in sphere formation medium for a period of 10 days to establish pancreatospheres, followed by 5 days of metformin treatment and its combination with CDF to assess the disintegration of pancreatospheres that already formed. We found that metformin and its combination with CDF increased the disintegration of pancreatospheres of both gemcitabine-sensitive and gemcitabine-resistant pancreatic cancer cells (Fig. 3A). These results suggest that metformin could exert inhibitory effect on self-renewal capacity of both gemcitabine-sensitive and gemcitabine-resistant pancreatic cancer cells.
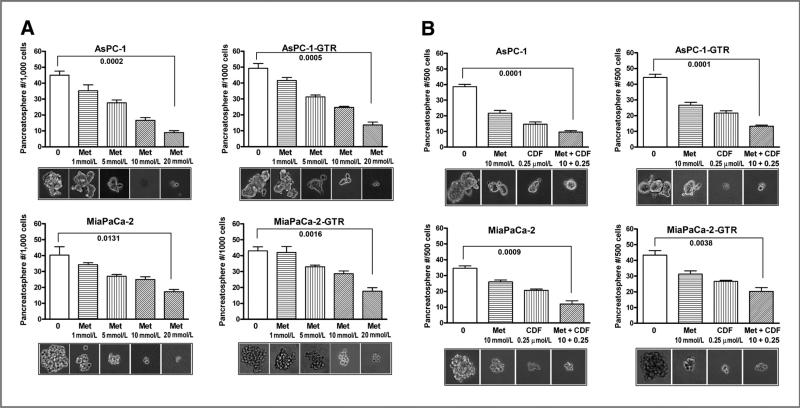
Metformin (Met) and/or its combination with CDF inhibited the capacity of CSC self-renewal in primary (A) and secondary (B) pancreatospheres of AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR cells. The sphere-forming assay was conducted using drug-sensitive and drug-resistant cells incubated with different concentrations of metformin or its combination with CDF (10 mmol/L metformin plus 0.25 μmol/L CDF) for 7 days.
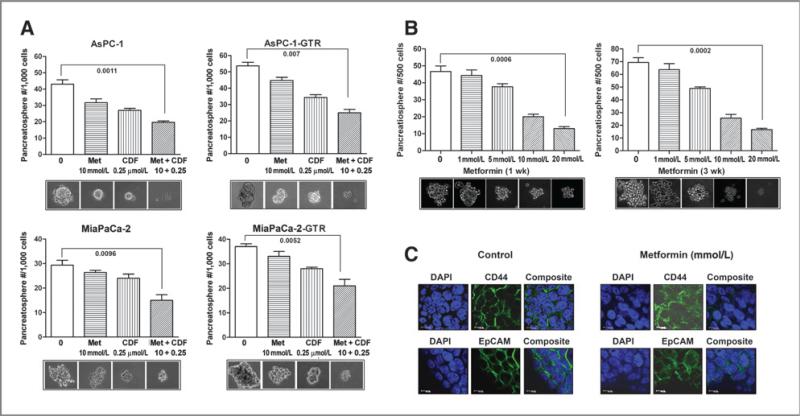
Metformin or/and its combination with CDF increased the disintegration of pancreatospheres (A), decreased CSC self-renewal capacity in the secondary pancreatospheres of mouse xenograft tumor derived from MiaPaCa-2 sphere-forming cells after 1 and 3 weeks of treatment (B). C, metformin (Met; 20 mmol/L) decreased the expression of CD44 and EpCAM in the secondary pancreatospheres of mouse xenograft tumor derived from MiaPaCa-2 sphere-forming cells after 1 week of treatment. DAPI, 4′,6-diamidino-2-phenylindole.
Effect of long-term metformin treatment on the formation of pancreatospheres of tumor cells derived from mouse xenograft tumors established by MiaPaCa-2 sphere-forming cells
To examine the long-term effect of metformin treatment on the formation of pancreatospheres induced by CSC-like cells, we conducted the sphere formation assay in the secondary pancreatospheres of mouse xenograft tumors derived from MiaPaCa-2 sphere-forming cells after 1 or 3 weeks of metformin treatment. The results show that metformin significantly decreased the formation of pancreatospheres in the secondary pancreatospheres assay (Fig. 3B), suggesting that long-term metformin treatment can inhibit the capacity of CSC self-renewal.
Metformin decreased the expression of CD44 and EpCAM in pancreatospheres of mouse xenograft tumors derived from MiaPaCa-2 sphere-forming cells.
We conducted confocal imaging microscopy to examine the effect of metformin on the expression of CSC surface biomarkers, CD44 and EpCAM, in the secondary pancreatospheres of mouse xenograft tumors derived from MiaPaCa-2 sphere-forming cells. We found that metformin inhibited the expression of CD44 and EpCAM in pancreatospheres (Fig. 3C), suggesting that the inhibition of pancreatospheres may be associated with the downregulation of CD44 and EpCAM expression, markers of CSCs. These results are consistent with our in vitro findings.
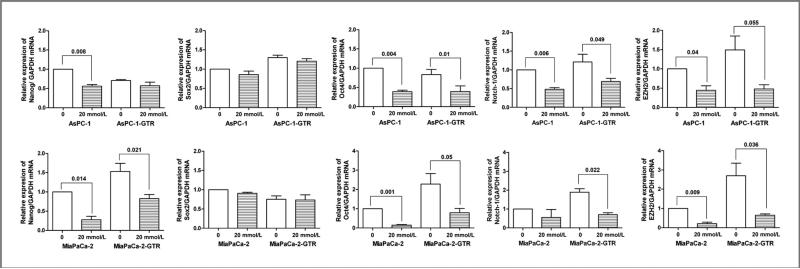
Effect of metformin on the mRNA expression of CSC marker genes in pancreatospheres of drug-resistant pancreatic cancer cells
To examine the effect of metformin on the mRNA expression of CSC marker genes in drug-sensitive and drug-resistant pancreatospheres of pancreatic cancer cells, we measured the relative mRNA levels of CSC marker genes, Nanog, Sox2, Oct4, Notch-1, and EZH2, in AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR cells by quantitative RT-PCR (qRT-PCR). We found that metformin significantly decreased the expressions of Oct4, Notch-1, and EZH2 mRNAs in pancreatospheres of AsPC-1, AsPC-1-GTR, MiaPaCa-2, and MiaPaCa-2-GTR cells (Fig. 4). Metformin only decreased the relative expression of Nanog mRNA in pancreatospheres of MiaPaCa-2 and MiaPaCa-2-GTR cells (Fig. 4). These results strongly suggest that metformin elicits pronounced inhibitory effects on the expression of CSC marker genes in pancreatospheres of pancreatic cancer cells.
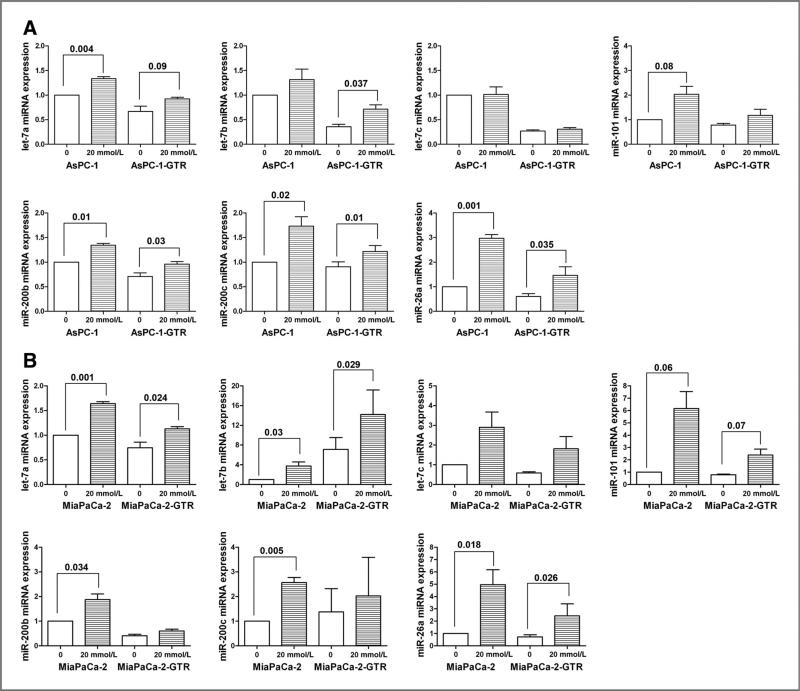
Effect of metformin on the expression of miRNAs (let-7, miR-26a, miR-101, and miR-200) in pancreatospheres of pancreatic cancer cells
We examined the effect of metformin on the expression of miRNAs (let-7, miR-26a, miR-101, and miR-200) in pancreatospheres of pancreatic cancer cells by qRT-PCR. We found that metformin treatment increased the relative expressions of let-7a, let-7b, miR-26a, miR-101, miR-200b, and miR-200c in pancreatospheres of AsPC-1, AsPC-1-GTR (Fig. 5A), MiaPaCa-2, and MiaPaCa-2-GTR cells (Fig. 5B). Metformin increased the relative expressions of let-7c miRNA in pancreatospheres of MiaPaCa-2 and MiaPaCa-2-GTR cells but not AsPC-1 and AsPC-1-GTR cells. Metformin increased the relative expression of miR-200b in pancreatospheres of AsPC-1, AsPC-1-GTR, and MiaPaCa-2 cells but not MiaPaCa-2-GTR cells. These results suggest that metformin could cause reexpression of specific miRNAs, although there are certain degrees of selectivity among the pancreatic cancer cells used in our study.
The role of miR-26a in the regulation EZH2 and EpCAM expression in pancreatic cancer cells
We investigated the mechanistic role of miR-26a in the regulation of EZH2 and EpCAM in pancreatic cancer cells by transfection of miR-26a precursor into MiaPaCa-2 cells. We found that the transfection of miR-26a precursor increased the relative level of miR-26a and consequently decreased the relative levels of EZH2 and EpCAM proteins and mRNAs in MiaPaCa-2 cells (Fig. 6A and B). Reexpression of miR-26a significantly decreased the relative mRNA levels of EZH2, Oct4, Notch-1, and EpCAM in MiaPaCa-2 cells (Fig. 6C), which suggest that reexpression of miR-26a is mechanistically associated with the downregulation of EZH2, Oct4, Notch-1, and EpCAM expression in pancreatic cancer cells, and thus metformin-mediated reexpression of miR-26a is mechanistically associated with its biologic effects on CSCs.
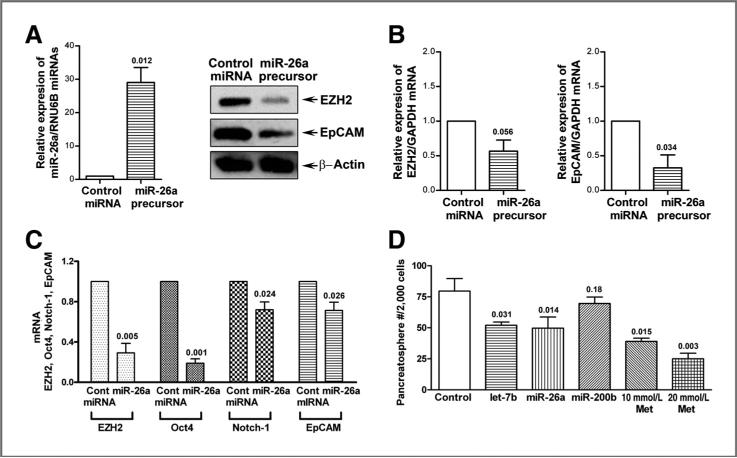
Reexpression of miR-26a decreased the expression of EZH2 and EpCAM proteins (A) and mRNAs (B) in MiaPaCa-2 cells and inhibited the mRNA expressions of EZH2, Oct4, Notch-1, and EpCAM in MiaPaCa-2 tumor sphere cells (C). Reexpression of let-7b and miR-26a decreased the formation of pancreatospheres of MiaPaCa-2 cells (D). The transfection of miR-26a miRNA precursors were conducted in MiaPaCa-2 cells and its tumor sphere cells as described under the Materials and Methods section. Western blot analysis and qRT-PCR were carried out to measure the relative protein levels and mRNA, respectively. The sphere forming assay was conducted as described under the Materials and Methods section. P values represent comparison between controls as calculated by the paired t test. Cont, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Met, metformin.
Effect of reexpression of let-7b, miR-26a, and miR-200b on the formation of pancreatospheres of pancreatic cancer cells
To examine the role of let-7b, miR-26a, and miR-200b in the growth of pancreatospheres, we conducted the sphere forming assay using pancreatic cancer cells transfected with pre-miRNA. The results show that reexpression of let-7b and miR-26a significantly decreased the formation of pancreatospheres (P < 0.05; Fig. 6D), suggesting that the loss of expression of let-7b and miR-26a is mechanistically linked with the regulation of the growth of pancreatospheres of pancreatic cancer cells, which is consistent with the acquisition of CSC function of pancreatic cancer cells. These results clearly suggest that the reexpression of miRNAs that are lost in aggressive pancreatic cancer cells could become a novel approach for the treatment of pancreatic cancer, which requires further in vivo preclinical studies.
Discussion
Accumulating in vitro and in vivo studies have shown that metformin could exert its antitumor effect by targeting multiple pathways such as AMPK/mTOR, anti-inflammatory pathway via inactivation of NF-κB, cell cycle/apoptosis, insulin/IGF-1R, and angiogenesis in cancers (9–11, 26) Moreover, recent studies have shown that metformin can block tumor growth by inactivation of breast CSC–like cells and by targeted killing of EMT phenotypic cells (12, 27–29). It is now well accepted that CSCs posses the ability to self-renew and give rise to differentiated tumor cells, which contribute to resistance to conventional therapeutics, and it is believed to be the root cause of tumor recurrence and metastasis (12). Therefore, targeting CSC self-renewal pathways might provide a new strategy for cancer treatment with better therapeutic outcome.
It has also been shown that metformin can decrease the dose of chemotherapy (doxorubicin) and prolonged tumor remission in mouse xenograft of human breast cancer cells by selectively killing CSCs (30). This effect of metformin is due to inhibition of cell proliferation, migration and invasion, the CSC self-renewal capacity, and the expression of CSC marker genes, such as CD44, EpCAM, EZH2, Notch-1, Nanog, and Oct4, in gemcitabine-resistant human pancreatic cancer cells as documented in our current study. Collectively, these results suggest that metformin could inhibit tumor cell growth via targeted killing of CSCs that are highly resistant to conventional therapeutics especially for pancreatic cancer.
Enhancer of Zeste Homologue 2 (EZH2) is a histone methyltransferase enhancer of polycomb group complexes, and it is a critical part of the cellular machinery involved in the epigenetic regulation of gene transcription (31, 32). The overexpression of EZH2 has been observed in a variety of tumors such as breast cancer, prostate cancer, and pancreatic cancer (33, 34), and the overexpression of EZH2 is believed to directly responsible for de novo suppression of multiple genes in human cancers (35, 36). Emerging evidence also suggest that the overexpression of EZH2 might cause normal cells to dedifferentiate back to the stem cell–like state by epigenetically repressing cell fate–regulating genes and tumor suppressor genes, leading to the development of tumor (35, 37). In this study, we have shown for the first time that metformin decreases the gene expression of EZH2 in pancreatospheres of drug-resistant pancreatic cancer cells, suggesting that metformin may exert its antitumor activity by targeting EZH2 in pancreatic cancer cells. We further investigated how metformin could downregulate EZH2 expression as well as the expression of other CSC marker genes. To that end, it is known that microRNAs are key players which function as endogenous posttranscriptional gene regulators by specific binding to the 3′ untranslated region (3′UTR) of target mRNAs to mediate protein synthesis or mRNA stability (38, 39). A large number of miRNAs have been reported to be associated with tumorigenesis and are suspected to be mechanistically associated with malignant diseases (40–42); however, their exact roles in the pathogenesis of pancreatic cancer are not fully understood.
In the pathogenesis of cancer, let-7 family of miRNAs has been found to be downregulated, thereby increasing the expression of Ras and c-Myc in malignant cells (40, 42, 43). In addition, the downregulation of let-7 has been observed in pancreatic cancer, which was associated with the aggressiveness of tumors (44, 45). Recent studies have shown that the increased expression of CSC marker genes has been found to be associated with the downregulation of let-7 expression (44, 45). In the current study, we have shown that metformin treatment could increase the expression of let-7a and let-7b in pancreatospheres of pancreatic cancer cells, but most importantly in drug-resistant cells, suggesting that inactivation of CSC function would inhibit CSC self-renewal capacity by metformin treatment, which is, in part, due to reexpression of let-7. Emerging evidence also suggest that miR-26a and miR-101, potential tumor suppressors, could modulate the cancer epigenome by repressing the polycomb group protein, EZH2 (46–49) whose expression has been linked with tumor angiogenesis, self-renewal capacity, and regulation of CSC marker genes associated with tumor invasion and metastasis (31, 50). Altered expression of miR-26a and miR-101 has been found in a wide variety of cancers such as gastric cancer, breast cancer, prostate cancer, nasopharyngeal carcinoma, and pancreatic cancer (41, 49). Reexpression of miR-26a or miR-101 in cancer cells caused downregulation of EZH2, resulting in the inhibition of tumor invasion and metastasis (46, 49). Therefore, targeting miR-26a and miR-101 provides a novel therapeutic approach for cancer treatment. Our data showed that reexpression of miR-26a results in the downregulation of EZH2 and EpCAM expression, which was consistent with metformin-mediated reexpression of miR-26a associated with the inhibition of cell proliferation, migration and invasion, and CSC function, all of which was, in part, due to downregulation of EZH2 and other CSC marker genes. Collectively, our results suggest that metformin could be useful for reverting CSC function or the killing of CSCs by reexpression of specific miRNAs and downregulation of CSC-specific genes that are typically overexpressed in pancreatic cancer and thus will result in better treatment outcome of patients diagnosed with pancreatic cancer, which requires further in-depth preclinical experiments using relevant animal models.
Acknowledgments
The authors thank Puschelberg and Guido foundations for their generous financial support.
Grant Support
The study was supported by National Cancer Institute, NIH grants 5R01CA131151, 3R01CA131151-02S1, and 5R01CA132794 (F.H. Sarkar).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/1940-6207.capr-11-0299
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3786260?pdf=render
Citations & impact
Impact metrics
Article citations
Challenges of Managing Type 3c Diabetes in the Context of Pancreatic Resection, Cancer and Trauma.
J Clin Med, 13(10):2993, 19 May 2024
Cited by: 1 article | PMID: 38792534 | PMCID: PMC11122338
Review Free full text in Europe PMC
Metformin: A Dual-Role Player in Cancer Treatment and Prevention.
Int J Mol Sci, 25(7):4083, 06 Apr 2024
Cited by: 6 articles | PMID: 38612893 | PMCID: PMC11012626
Review Free full text in Europe PMC
The Role of MicroRNA, Long Non-Coding RNA and Circular RNA in the Pathogenesis of Polycystic Ovary Syndrome: A Literature Review.
Int J Mol Sci, 25(2):903, 11 Jan 2024
Cited by: 3 articles | PMID: 38255975 | PMCID: PMC10815174
Review Free full text in Europe PMC
Deciphering cellular plasticity in pancreatic cancer for effective treatments.
Cancer Metastasis Rev, 43(1):393-408, 09 Jan 2024
Cited by: 2 articles | PMID: 38194153
Review
Chronic treatment with TNF-α, alone and in combination with Takinib, SB203580 and metformin induce cell death in breast cancer.
Heliyon, 9(11):e21060, 16 Oct 2023
Cited by: 0 articles | PMID: 37964831 | PMCID: PMC10641119
Go to all (220) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MicroRNA-200c overexpression inhibits chemoresistance, invasion and colony formation of human pancreatic cancer stem cells.
Int J Clin Exp Pathol, 8(6):6533-6539, 01 Jun 2015
Cited by: 37 articles | PMID: 26261532 | PMCID: PMC4525866
Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells.
Cancer Lett, 307(1):26-36, 03 Apr 2011
Cited by: 204 articles | PMID: 21463919 | PMCID: PMC3104092
Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression.
Cancer Res, 72(1):335-345, 22 Nov 2011
Cited by: 185 articles | PMID: 22108826 | PMCID: PMC3792589
The Link between Diabetes, Pancreatic Tumors, and miRNAs-New Players for Diagnosis and Therapy?
Int J Mol Sci, 24(12):10252, 16 Jun 2023
Cited by: 4 articles | PMID: 37373398 | PMCID: PMC10299694
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: R01 CA132794
Grant ID: 3R01CA131151-02S1
Grant ID: 5R01CA132794
Grant ID: R01 CA131151
Grant ID: R01 CA154321
Grant ID: 5R01CA131151