Abstract
Free full text
Distinguishable Epidemics Within Different Hosts of the Multidrug Resistant Zoonotic Pathogen Salmonella Typhimurium DT104
Abstract
The global epidemic of multidrug resistant Salmonella Typhimurium DT104 provides an important example, both in terms of the agent and its resistance, of a widely disseminated zoonotic pathogen. Here, with an unprecedented national collection of isolates collected contemporaneously from humans and animals, and including a sample of internationally derived isolates, we have used whole genome sequencing to dissect the phylogenetic relationships of the bacterium and its antimicrobial resistance genes through the course of an epidemic. Contrary to current tenets supporting a single homogeneous epidemic, we demonstrate that the bacterium and its resistance genes were largely maintained within animal and human populations separately, and that there was limited transmission, in either direction. We also show considerable variation in the resistance profiles, in contrast to the largely stable bacterial core genome, further emphasizing the critical importance of integrated genotypic datasets in understanding the ecology of bacterial zoonoses and antimicrobial resistance.
S. enterica subspecies enterica is one of the most common bacterial pathogens of humans and other animals (1, 2). The global burden of disease caused by Salmonella infections is substantial, with over 90 million human cases of gastroenteritis alone occurring each year (3). The annual cost of these infections is estimated to be approximately €3 billion in the European Union (4), and approximately $2.7 billion in the United States (5). The public health impact is exacerbated by antimicrobial resistance (AMR), which leads to increased morbidity, mortality, and treatment costs (6, 7). In our study, the inter-related epidemiologies of Salmonella and AMR were examined at the level of the genome. Our results challenge the established view that the human and animal epidemics are synonymous (8-11), and show that the phylogenetic relationships both within and between the resistance determinants and the host bacteria are different.
Throughout the 1990s, there was a global epidemic of multidrug resistant S. Typhimurium Definitive Type 104 (DT104) in animals and humans (12, 13). The DT104 epidemic was important because of its widespread prevalence and perceived zoonotic nature, and the high frequency of resistance to a wide range of commonly used antimicrobials, particularly ampicillin, chloramphenicol, streptomycin, sulphonamides and tetracycline. The primary source of human DT104 infections is thought to be the animal population, particularly local animals, through both direct contact and food-borne transmission (9, 10, 14). Using whole genome sequencing, we have studied the course of the epidemic in Scotland, a restricted geographical location that has a well-characterized collection of DT104 bacteria isolated from both humans and other animals over a 22-year period. By interrogating isolates collected from sympatric and contemporaneous hosts, and using isolates from geographically diverse hosts from other countries to put them in phylogenetic context, we have an unprecedented opportunity to profile the evolution and spread of a local epidemic of this globally important zoonotic pathogen.
In order to investigate the evolutionary changes that characterize the long-term epidemic, and to assess the roles of animals and humans in the spread of both the bacterium and AMR, we sequenced 142 isolates from humans and 120 isolates from other animals in Scotland sampled from 1990 – 2011. To set the Scottish isolates within a broader international context, 111 isolates were included from other locations around the world (Table S1). Sequence reads were mapped against our reference chromosome of DT104 and its virulence plasmid, and genomes were assembled de novo to identify unique regions (15). Maximum likelihood methods were used to produce a phylogenetic tree based on single nucleotide polymorphisms (SNPs) in the core genome (15). This identified a main clade of related DT104 isolates (29 – 123 SNPs compared to the reference core genome) and a separate subset of 14 atypical isolates (620 – 709 SNPs compared to the reference core genome; Fig. S1). Within the main clade there were 2,765 variable sites across the 359 isolates, with pairwise differences between isolates ranging from 0 to 167 SNPs (Fig. 1). Most SNPs were sporadically distributed in the core genome throughout the phylogenetic tree, with relatively few genes showing non-synonymous SNPs in >5 isolates (Fig. S2). These data indicate that the rate of change within the core genome over the 22-year epidemic period, 3.4 × 10−7 substitutions/site/year (95% highest posterior density [HPD] interval: 3.1 × 10−7 – 3.7 × 10−7), is in line with that seen in other S. Typhimurium (16). Although confirmed phenotypically as DT104 by standard phage typing, the 14 atypical isolates are more similar genetically to the non-DT104 S. Typhimurium reference strain LT2, demonstrating the lack of resolution of traditional typing techniques. Isolates from all geographical locations can be found near the root of the tree, consistent with the rapid global spread of DT104 (12). However, many of these isolates, particularly those from England and Wales, can also be found interspersed with Scottish isolates throughout the rest of the tree. The most deeply rooted are primarily derived from humans, and do not have Salmonella Genomic Island 1 (SGI1), a 43kb genomic island with a 13kb multiple drug resistance region (17) that is characteristic of the epidemic strain.
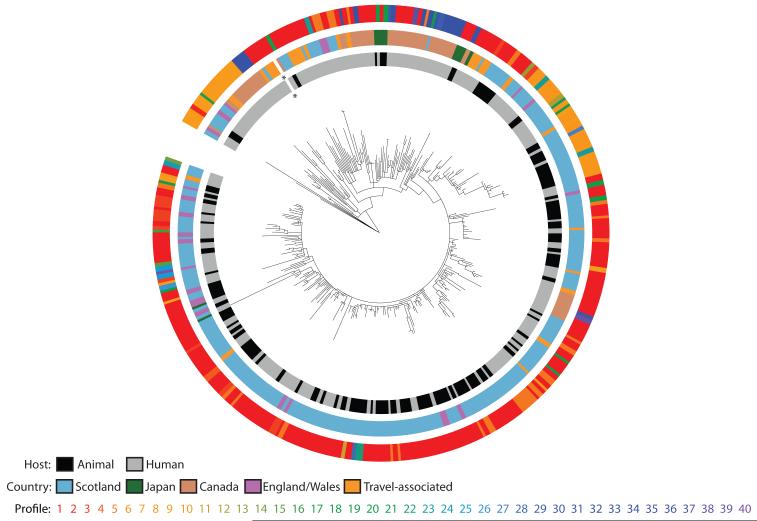
Colored rings indicate, from the center out, the host and country of origin, and the unique genotypic antimicrobial resistance profiles of each isolate (Table S6); the asterisk indicates the location of the reference isolate HF937208. The same tree with bootstrap values added is shown in Fig. S5.
To evaluate interspecies transmission and how DT104 circulated in Scotland, Bayesian phylogenetic diffusion models (18, 19) were used to reconstruct the host population of each branch of the phylogenetic tree (Fig. 2A), using the 135 human and 113 animal Scottish DT104 isolates. The most recent common ancestor of DT104 in Scotland is predicted to date to 1968 (95% HPD interval: 1962 – 1976), consistent with reports of antimicrobial-susceptible DT104 strains isolated from human cases of infection since the 1960s in England and Wales (13), with the first report of multidrug resistant DT104 in Scotland in the mid 1980s (13). What can be observed in Figs. 1 and and2A2A is that while there are isolates from a single host population that cluster together, isolates from animals and humans are also interspersed throughout the tree. However, quantification of the association between phylogeny and host population shows that the clustering is not fully randomized (15). Furthermore, similar to the maximum likelihood tree (Fig. 1), the most deeply rooted branches are estimated to be human (Fig. 2A) but it is difficult to infer with certainty the direction of transmission from these observations alone. Overall, the reconstructed host populations of the branches within the tree suggest that there were relatively few animal-to-human or human-to-animal transitions (Fig. 2B). Using Markov jumps to estimate the number of unobserved human-to-animal and animal-to-human transitions along each branch (15), we show the overall numbers of animal-to-human and human-to-animal transitions were similar, indicating that the majority of human infections in Scotland were unlikely to have been sourced from the local animal population. Markov rewards, from which inference may be made with respect to which populations might act as sources and which might act as sinks, showed that while the human isolates represented just 54% of the sample, 62% (666 years, 95% HPD: 545 – 771) of the total phylogenetic tree length was in the human state (15). Conversely, 38% (400 years, 95% HPD: 318 – 521) of the total tree length was spent in the animal state. Our conclusions are consistent when we perform these analyses on animal isolates by species, and with only cattle isolates, the main animal reservoir (15, 20). These results support two novel hypotheses: 1) DT104 was predominantly circulating separately within each population in Scotland, with a low frequency of spill-over in both directions, and/or 2) that animals and humans were each sinks for a different and separate source of infection, also with a low level of spill-over. Scotland is a net importer of food (21); for example, by value, 58% of all red meat and 38% of raw beef are of non-Scottish origin (22). However, given a lack of surveillance data on food, attribution to this potential source cannot be quantified. Given the likely importance of imported food to the disease burden in humans, the greater global mobility of the human population and the recognition of this as a risk factor for bacterial infections (23, 24), we suggest that while these two hypotheses are not mutually exclusive, the second scenario is the more likely.
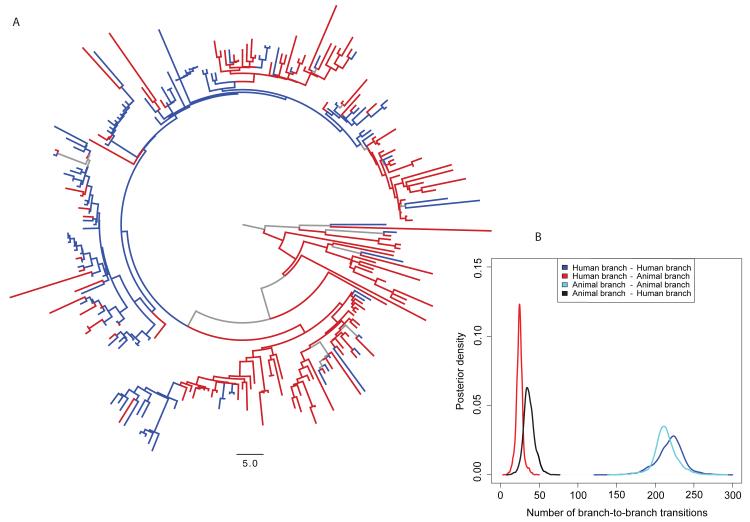
A) Branches with a reconstructed state (host population) posterior probability > 0.75 are colored red for human, blue for animal; branches with a state probability < 0.75 are colored grey. The same tree is shown in Fig. S6, with branch width scaled by the posterior probabilities. B) Posterior density plot of the numbers of human branches ancestral to human branches, human branches ancestral to animal branches, animal branches ancestral to human branches, and animal branches ancestral to animal branches integrated over the subsample of 3,600 phylogenetic trees with reconstructed host population states along branches obtained using BEAST. These results suggest 1) circulation of DT104 predominantly within animals and humans separately, with only a low frequency of spill-over in both directions, and/or or 2) animals and humans were each sinks for different and separate sources of infection, with only a low frequency of spill-over in both directions.
With respect to AMR, our results show that the ecological diversity of antimicrobial resistance, at the level of the genome, is greater in the isolates from the human population than in those from animals, and is not tightly coupled to the bacterial evolutionary history. Examining the distribution of the phenotypic AMR profiles throughout the molecular phylogenetic tree (Fig. S3) (15), allowed us to investigate the degree to which the evolutionary history of the organism is linked to that of the resistance determinants it carries, regardless of whether they are genes or SNPs known to confer resistance. We show that isolates demonstrating the same phenotypic resistance profile, even those putatively conferred by SGI1 (17) (Fig. S3A), do not cluster together exclusively.
The diversity of AMR was assessed further by interrogating the presence or absence of resistance determinants and profiles in the 147 Scottish isolates selected to cover the range of phenotypic resistance profiles observed in the epidemic period (Fig. 3) (15). A greater number of both resistance determinants and genotypic resistance profiles are found in the human isolates. As demonstrated by rarefaction analysis (Fig 3D), these results cannot be accounted for by differential sampling intensity. Multiple, independent recombination events in the multidrug resistance region occurred, as variants of SGI1 (25) are found throughout the tree (Fig. S4). Similarly, while there are some clades with a single AMR gene profile, in many instances profiles can be found in multiple clades and may be due to the gain or loss by horizontal gene transfer of accessory AMR determinants and their plasmid vectors (Tables S2-S5). Although the propensity for horizontal transmission is well recognized (26), these results provide both a unique insight into the genetic flux occurring across a defined 22 year epidemic and a baseline for the concomitant genetic flux in the DT104 core genome. In contrast to the relatively stable core genome with an expected substitution rate, the AMR features of these isolates varied appreciably throughout the time period.
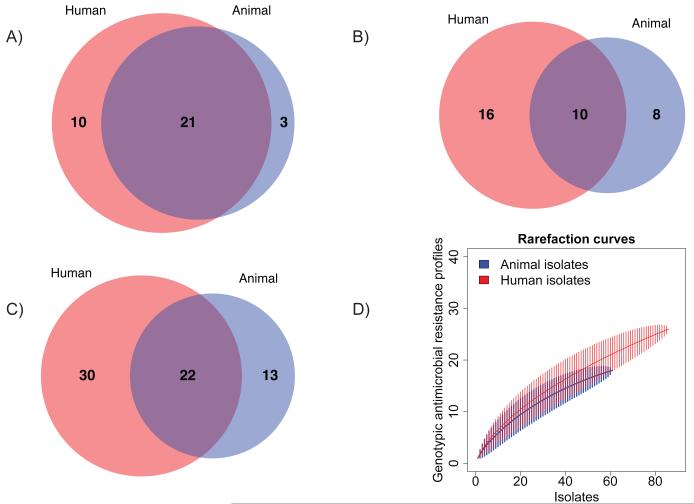
A) The numbers of shared and unique AMR determinants (acquired resistance genes or single nucleotide polymorphisms known to confer resistance) in the 147 Scottish human and animal DT104 isolates investigated for AMR diversity; B) The numbers of shared and unique AMR profiles (unique combinations of AMR determinants) in the 147 isolates; C) The numbers of shared and unique AMR phenotypic profiles (unique combinations of AMR phenotypes) in the original 5,200 surveillance isolates of DT104, 1990-2004, Scotland (11); D) Rarefaction curves, with 95% confidence intervals (vertical lines), of the number of genotypic AMR profiles in the 147 isolates investigated for AMR diversity, demonstrating similar sampling intensity.
Most transmission of DT104 has hitherto been considered to be from animals to humans (27-29), but this view has been based primarily on outbreak investigations. However, on the basis of whole genome sequence analysis, we provide a historical perspective on the epidemic that leads to a different inference. This is the largest study of its kind, utilizing tools with the finest resolution available to investigate the relationships of Salmonella Typhimurium DT104 and AMR between and within co-located human and animal populations, and enabled us to address two separate issues with this dataset, those of the origins and dissemination of AMR and also of Salmonella. First, the greater diversity of AMR genes and profiles in the human DT104 isolates suggests that there were other sources contributing to the diversity of the human resistance burden than the local animals; it is noteworthy that this analysis of molecular data leads to identical conclusions to those based on phenotype (11). Second, we show that a large proportion of transmission of the bacterium appears to have occurred within each host population, and/or that there were separate sources of DT104 for the humans and local animals, with a small degree of spill-over in both directions. If the majority of human infections were obtained from locally produced food or direct contact with animals, the human isolates should be a subset of the animal isolates, but we do not observe this.
With humans having a more diverse source of infections than the local animal population, it is probable that imported food, foreign travel and environmental reservoirs are significant sources of Salmonella and AMR in humans in our study (23, 30), but surveillance data simply do not exist in any coherent form locally, nor consistently across the international community, to quantify the importance of these different sources. If we are to address this deficit, there is an urgent need to construct relevant, robust, and harmonized surveillance protocols to capture this information. Similarly, although the Scottish isolates are similar to those from other countries, this investigation needs to be conducted in other organisms and environments to determine the generality of our conclusions across different agricultural, political, and sociological conditions. This study challenges current views on the contribution of the local animal reservoir as a source of Salmonella and AMR in humans. It demonstrates the critical importance of acquiring targeted genotypic datasets integrated across the veterinary and public health sectors in order to understand the ecology of bacterial zoonoses, identify the sources of antimicrobial resistance, and formulate responsible evidence-based control strategies.
Acknowledgements
AEM was funded by the William Stewart Fellowship while at the University of Glasgow. AEM, SRH, MCF, TRC, JP and NRT are supported by Wellcome Trust grant 098051. PL and MAS acknowledge funding from the European Union Seventh Framework Programme [FP7/2007-2013] under ERC Grant agreement no. 260864. MAS is supported in part by National Institutes of Health R01 grants AI107034 and HG006139 and National Science Foundation grant DMS-1264153. The authors would like to thank James Cotton (Wellcome Trust Sanger Institute) for helpful discussions and assistance. Genome sequences are deposited with the European Nucleotide Archive (study accessions ERP000244, ERP000270, ERP000994) and DNA Data Bank of Japan (accession DRA000942). The finished chromosome and plasmid for Salmonella Typhimurium DT104 are submitted to the European Molecular Biology Laboratory, accessions HF937208 and HF937209, respectively.
References and Notes
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.1240578
Read article for free, from open access legal sources, via Unpaywall:
https://rvc-repository.worktribe.com/preview/1707558/Equine%20Veterinary%20Journal%20-%202023%20-%20Barnab%C3%A9%20-%20Sakiva%20Paper.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Antimicrobial resistance and clonal relationships of Salmonella enterica Serovar Gallinarum biovar pullorum strains isolated in China based on whole genome sequencing.
BMC Microbiol, 24(1):414, 18 Oct 2024
Cited by: 0 articles | PMID: 39425016 | PMCID: PMC11487782
The integrated genomic surveillance system of Andalusia (SIEGA) provides a One Health regional resource connected with the clinic.
Sci Rep, 14(1):19200, 19 Aug 2024
Cited by: 1 article | PMID: 39160186 | PMCID: PMC11333592
Characterization of a Salmonella enterica serovar Typhimurium lineage with rough colony morphology and multidrug resistance.
Nat Commun, 15(1):6123, 20 Jul 2024
Cited by: 1 article | PMID: 39033143 | PMCID: PMC11271444
Comparative Gene Expression Analysis of Salmonella Typhimurium DT104 in Ground Chicken Extract and Brain Heart Infusion Broth.
Microorganisms, 12(7):1461, 18 Jul 2024
Cited by: 0 articles | PMID: 39065229 | PMCID: PMC11279075
A Retrospective Analysis of Salmonella Isolates across 11 Animal Species (1982-1999) Led to the First Identification of Chromosomally Encoded blaSCO-1 in the USA.
Microorganisms, 12(3):528, 06 Mar 2024
Cited by: 0 articles | PMID: 38543579 | PMCID: PMC10974302
Go to all (201) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 6 of 6)
- (1 citation) ENA - ERP000270
- (1 citation) ENA - HF937209
- (1 citation) ENA - DRA000942
- (1 citation) ENA - ERP000244
- (1 citation) ENA - HF937208
- (1 citation) ENA - ERP000994
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
New Variant of Multidrug-Resistant Salmonella enterica Serovar Typhimurium Associated with Invasive Disease in Immunocompromised Patients in Vietnam.
mBio, 9(5):e01056-18, 04 Sep 2018
Cited by: 40 articles | PMID: 30181247 | PMCID: PMC6123440
Microbiology. Sources of antimicrobial resistance.
Science, 341(6153):1460-1461, 12 Sep 2013
Cited by: 61 articles | PMID: 24030495
Genomic relationship of Salmonella enterica serovar typhimurium DT104 isolates from Korea and the United States.
J Microbiol, 42(1):14-19, 01 Mar 2004
Cited by: 10 articles | PMID: 15357286
The genetics of Salmonella genomic island 1.
Microbes Infect, 8(7):1915-1922, 27 Mar 2006
Cited by: 122 articles | PMID: 16713724
Review
Funding
Funders who supported this work.
European Research Council (1)
Evolutionary reconstruction of viral spread in time and space (ViralPhylogeography)
Prof Philippe LEMEY, University of Leuven
Grant ID: 260864
NHGRI NIH HHS (2)
Grant ID: R01 HG006139
Grant ID: HG006139
NIAID NIH HHS (2)
Grant ID: R01 AI107034
Grant ID: AI107034
NIGMS NIH HHS (1)
Grant ID: R01 GM086887
Wellcome Trust (1)
Wellcome Trust Sanger Institute - generic account for deposition of all core- funded research papers
Prof Sir Michael Stratton, Wellcome Trust Sanger Institute
Grant ID: 098051





