Abstract
Free full text

Pure and syndromic optic atrophy explained by deep intronic OPA1 mutations and an intralocus modifier
Abstract
The genetic diagnosis in inherited optic neuropathies often remains challenging, and the emergence of complex neurological phenotypes that involve optic neuropathy is puzzling. Here we unravel two novel principles of genetic mechanisms in optic neuropathies: deep intronic OPA1 mutations, which explain the disease in several so far unsolved cases; and an intralocus OPA1 modifier, which explains the emergence of syndromic ‘optic atrophy plus’ phenotypes in several families. First, we unravelled a deep intronic mutation 364 base pairs 3’ of exon 4b in OPA1 by in-depth investigation of a family with severe optic atrophy plus syndrome in which conventional OPA1 diagnostics including gene dosage analyses were normal. The mutation creates a new splice acceptor site resulting in aberrant OPA1 transcripts with retained intronic sequence and subsequent translational frameshift as shown by complementary DNA analysis. In patient fibroblasts we demonstrate nonsense mediated messenger RNA decay, reduced levels of OPA1 protein, and impairment of mitochondrial dynamics. Subsequent site-specific screening of >360 subjects with unexplained inherited optic neuropathy revealed three additional families carrying this deep intronic mutation and a base exchange four nucleotides upstream, respectively, thus confirming the clinical significance of this mutational mechanism. Second, in all severely affected patients of the index family, the deep intronic mutation occurred in compound heterozygous state with an exonic OPA1 missense variant (p.I382M; NM_015560.2). The variant alone did not cause a phenotype, even in homozygous state indicating that this long debated OPA1 variant is not pathogenic per se, but acts as a phenotypic modifier if it encounters in trans with an OPA1 mutation. Subsequent screening of whole exomes from >600 index patients identified a second family with severe optic atrophy plus syndrome due to compound heterozygous p.I382M, thus confirming this mechanism. In summary, we provide genetic and functional evidence that deep intronic mutations in OPA1 can cause optic atrophy and explain disease in a substantial share of families with unsolved inherited optic neuropathies. Moreover, we show that an OPA1 modifier variant explains the emergence of optic atrophy plus phenotypes if combined in trans with another OPA1 mutation. Both mutational mechanisms identified in this study—deep intronic mutations and intragenic modifiers—might represent more generalizable mechanisms that could be found also in a wide range of other neurodegenerative and optic neuropathy diseases.
Introduction
Optic neuropathies are a substantial cause for blindness in European populations (Munier et al., 1998). The most frequent cause of inherited optic neuropathies are dominant mutations in OPA1, a nuclear encoded mitochondrial dynamin-like protein (OMIM: #165500) (Yu-Wai-Man et al., 2010a). However, the genetic cause of inherited optic neuropathies still remains unsolved in ~55% of patients, including absence of OPA1 mutations by routine diagnostics (Ferre et al., 2009).
Here we addressed the question to what extent and by what mode of action hidden mutations contribute to the share of unsolved cases with inherited optic neuropathy. Moreover, we aimed to elucidate a genetic mechanism why some patients with inherited optic neuropathies develop a complex ‘optic atrophy plus’ syndrome, which includes extraocular features such as deafness, ataxia and peripheral neuropathy (OMIM: #125250). Although this severe phenotype has been acknowledged in ~20% of all OPA1 cases (Amati-Bonneau et al., 2005; Yu-Wai-Man et al., 2010b), mechanisms explaining the emergence of this phenotype remain to be elucidated.
We here report for the first time the presence of disease-causing deep intronic mutations in OPA1, thus introducing a new mutational mechanism for inherited optic neuropathies. Their consequences have been studied by in-depth clinical, genetic and functional investigation of a family with early-onset optic atrophy plus syndrome. The significance of this new type of mutation is emphasized by the identification of other, independent families with inherited optic neuropathy through screening of a large cohort of so far unexplained patients with inherited optic neuropathy. Secondly, we provide evidence for a modifier effect of a certain OPA1 variant (c.1311A>G/p.I437M, NM_130837.1; = c.1146A>G/p.I382M on transcript NM_015560), which causes optic atrophy plus phenotypes by a combined mutational effect. This is exemplified in several families with optic atrophy plus who carry the modifier variant in trans with a pathogenic (exonic or deep intronic) OPA1 mutation. The modifying effect of p.I437M is emphasized by the finding that the variant does not lead to disease even in a homozygous state.
Materials and methods
Phenotypic characterization of the first index family
All available members of Family OAK 587—including the elderly parents (at age 70 and 75 years, respectively)—were systematically phenotyped clinically by a neurologist (M.S.) and an ophthalmologist (F.T.), including funduscopy and optical coherence tomography (for detailed methods, see Supplementary material; for pedigree, see Fig. 3). In addition, all affected members were investigated by nerve conduction studies, motor evoked potentials, MRI, and blood testing for creatine kinase and lactate. Subjects’ consent was obtained according to the Declaration of Helsinki and the study was approved by the ethical committee of the University of Tübingen (Az 598/2011BO1).
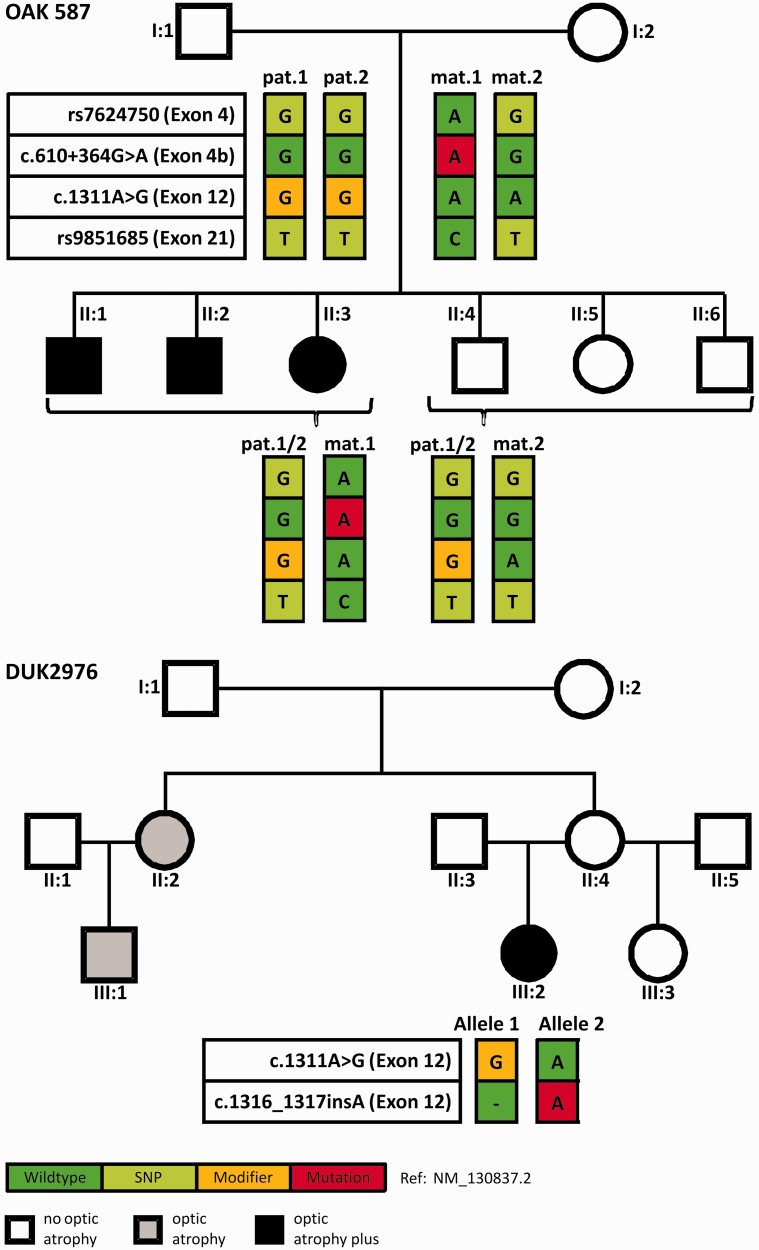
Pedigrees of the Families OAK 587 and DUK2976 including the segregation of OPA1 mutations and coding SNPs. Upper pedigree: All affected siblings of Family OAK 587 inherited the same maternal OPA1 allele (mat.1), comprising the c.610+364G>A deep intronic mutation (red square), whereas all unaffected children inherited the non-mutant maternal OPA1 allele (mat.2). The father, Subject I:1, is homozygous for p.I437M (yellow square), therefore all children are heterozygous for this variant. Note that Subject I:1 is homozygous for all analysed variants in OPA1, most likely due to identity-by-descent. Coding SNPs in exon 4 and exon 21 were used to perform pyrosequencing for discrimination of allelic representation on complementary DNA level. Affected family members are indicated by black symbols and healthy family members by white symbols. Lower pedigree: In Family DUK2976 Subject III:2 is compound heterozygous for p.I437M and c.1316_1317insA/p.N440Kfs*14, and presents with a severe optic atrophy plus phenotype. In contrast, her parents—likely heterozygous for only one of the two OPA1 variants—present only with mild visual disturbances and no symptoms at all, respectively. However, the sister II:2 of the unaffected mother II:4 as well as her daughter III:1 present with non-syndromic optic atrophy (grey).
Genomic screening of candidate genes in Family OAK 587
Exonic mutations in OPA1, OPA3, PANK2, POLG, C10orf2 and in the complete mitochondrial genome (16.569 bp encoding 37 genes and the control region, NC_012920.1) as well as repeat expansions in FXN (Friedreich’s ataxia) and ATXN7 (spinocerebellar ataxia 7) were excluded. Gene rearrangements, including deletions and duplications at the OPA1 locus were excluded by multiplex ligation-dependent probe amplification in two affected subjects and the father (SALSA MLPA KIT P229-B1 OPA1, MRC-Holland). Larger chromosomal rearrangements were excluded by comparative genome hybridization applying a Nimblegen chromosome 3 specific 385K probe array (processed by ImaGenes) in one of the affected patients. The transcript NM_130837.1 is the most complete of the currently 12 protein coding OPA1 transcripts listed in Ensembl release 74, coding for a protein of 1015 amino acids and it is recommended by the OPA1 nomenclature working group of Mitodyn.org. The known OPA1 variant c.1146A>G, p.I382M (NM_015560.2) (Schaaf et al., 2011) corresponds to c.1311A>G, p.I437M based on transcript NM_130837.1. The nomenclature of all following variants refers to transcript NM_130837.1.
Molecular genetic investigation of OPA1 transcripts from patient-derived primary fibroblasts
For standard cell culture conditions of primary fibroblasts from pedigree members of Family OAK 587 as well as Puromycin treatment for detection of nonsense mediated messenger RNA decay, see the online Supplementary material. After complementary DNA synthesis of RNA from puromycin-treated fibroblasts, screening for mutations on the OPA1 transcript level was performed with primer pairs that amplify eight overlapping fragments covering the entire coding sequence of transcript NM_130837 (Supplementary material). Average fragment length was ~500 bp. Primer pairs that flank the alternative exons 4, 4b and 5b amplify multiple complementary DNA fragments of different lengths. These fragments were separated on an agarose gel, excised and cloned for mutation analysis as well as for verification of intact exon–exon boundaries.
Pyrosequencing assays were designed to discriminate between the OPA1 alleles and perform quantitative analysis of allelic expression for the common single nucleotide polymorphisms (SNP) rs7624750 in exon 4 and rs9851685 in exon 21. For primer sequences see the Supplementary material. The primers have been designed using the PyroMark Assay Design 2.0 software (Qiagen). Pyrosequencing was performed using the PyroMark Q96 ID instrument, the sample preparation and the data analysis according to the manufacturer’s instructions.
Western blot and assessment of mitochondrial morphology
Semi-quantitative analysis of OPA1 protein bands was performed from primary fibroblast cell lines of patients and a control (for details, see Supplementary material). Mitochondrial morphology was assessed in fibroblasts of all affected subjects of Family OAK 587 and their two parents as well as an age- and gender-matched control for each individual of this family. Their fibroblasts were cultivated and analysed in parallel (Supplementary material).
Screening for the deep intronic OPA1 mutation in large patient and control cohorts
Three hundred and sixty-nine patients with unexplained bilateral optic atrophy negative for exonic OPA1 mutations and 241 normal controls were screened for the c.610+364G>A by amplification of a 1166 bp fragment containing the entire intron 4b and a subsequent restriction fragment length polymorphism (RFLP) assay (Supplementary material).
Screening for the OPA1 p.I437M variant (rs143319805) in a large exome data set of additional cohorts
The frequency of the proposed phenotypic modifier variant c.1311A>G/p.I437M (rs143319805) was analysed in 6499 whole exome sequencing data sets from the normal population (Exome Variant Server, NHLBI GO Exome Sequencing Project) (URL: http://evs.gs.washington.edu/EVS/) (December 2013). Moreover, we analysed the frequency in 743 exomes from 618 families presenting with at least one of the central neurological features of syndromic OPA1 disease, namely ataxia, pure or complicated hereditary spastic paraplegia or peripheral neuropathy. These exomes were collected in the Genomes Management Application (GEM.app) (Gonzalez et al., 2013).
Results
A family with severe early-onset ‘optic atrophy plus’ syndrome and asymptomatic parents
In Family OAK 587 three of six children from an Italian father and a German mother presented with a relatively uniform phenotype of an early-onset multisystemic neurodegeneration (Subjects II:1, II:2 and II:3; for pedigree see Fig. 3; for video illustrating the marked severity of the phenotype and for clinical examination, see the Supplementary material). This consisted of optic atrophy (onset at age 2 years) with thinning of the peripapillary retinal nerve fibre layer (Fig. 1A), near-blind vision reduced to light perception (20/1000), cerebellar ataxia [onset between age 4 to 6 years, leading to wheelchair-dependency within 10–15 years of age and high ataxia scores (Scale for the Rating and Assessment of Ataxia > 29/40 points) at the current age of 46 to 50 years; Table 1], external ophthalmoplegia, peripheral neuropathy, muscle atrophy, ptosis and spasticity (Table 1). None of the subjects showed evidence of hypacusis. Electrophysiological studies revealed severe axonal sensorimotor neuropathy as well as pyramidal tract damage to both upper and lower extremities. MRI demonstrated mild to moderate cerebellar atrophy and thinning of the cervical spinal cord (Fig. 2). Neither the other three siblings nor the two parents showed any abnormalities on neurological and ophthalmological examination. In particular, neither of the two parents revealed evidence for even subclinical optic atrophy on funduscopy or optical coherence tomography (Fig. 1B).
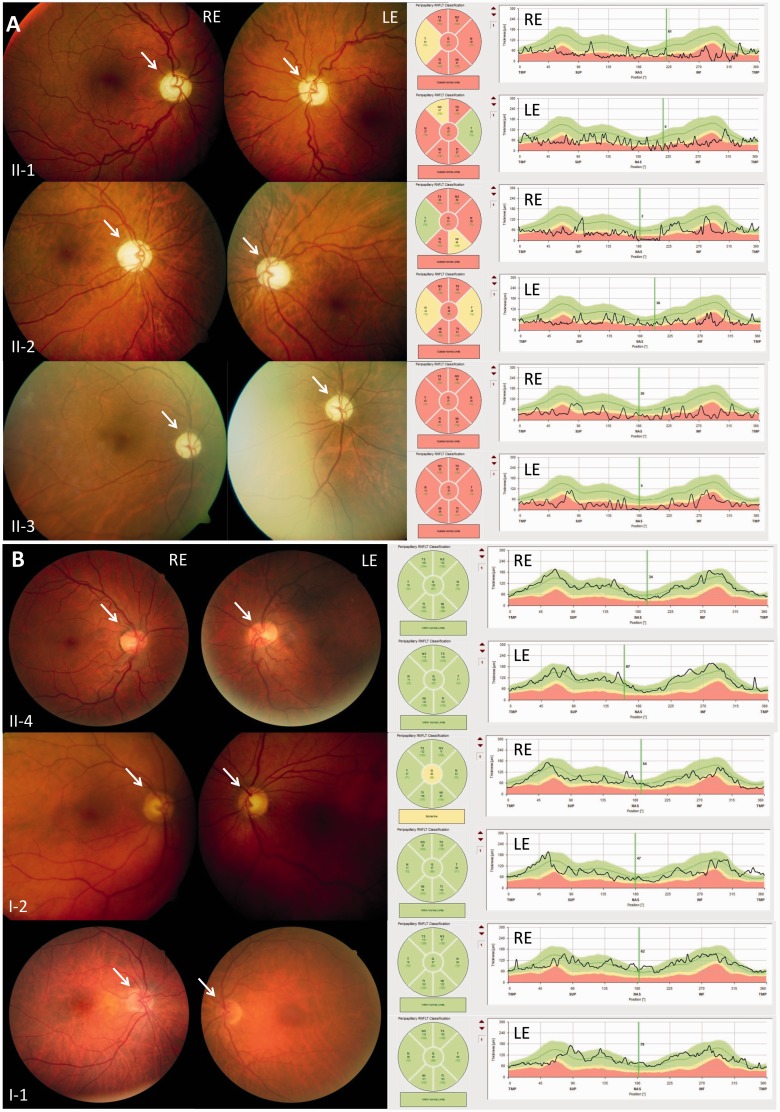
Funduscopy and optical coherence tomography (OCT) in affected and non-affected members of Family OAK 587. (A) Affected family members (Subjects II:1, II:2 and II:3): paleness of the optic nerve heads (white arrow) indicating atrophy of the optic nerve (left column), the optical coherence tomography scan demonstrates a thinning of the peripapillary retinal nerve fibre layer (right column). Both findings are most severe in the subject with the longest disease duration (Subject II:3). LE = left eye; RE = right eye. (B) Family members without clinical manifestation (Subjects II:4, I:2 and I:1): optical nerve heads showing the normal orange-pink appearance, thickness of the peripapillary retinal nerve fibre layer within normal limits.
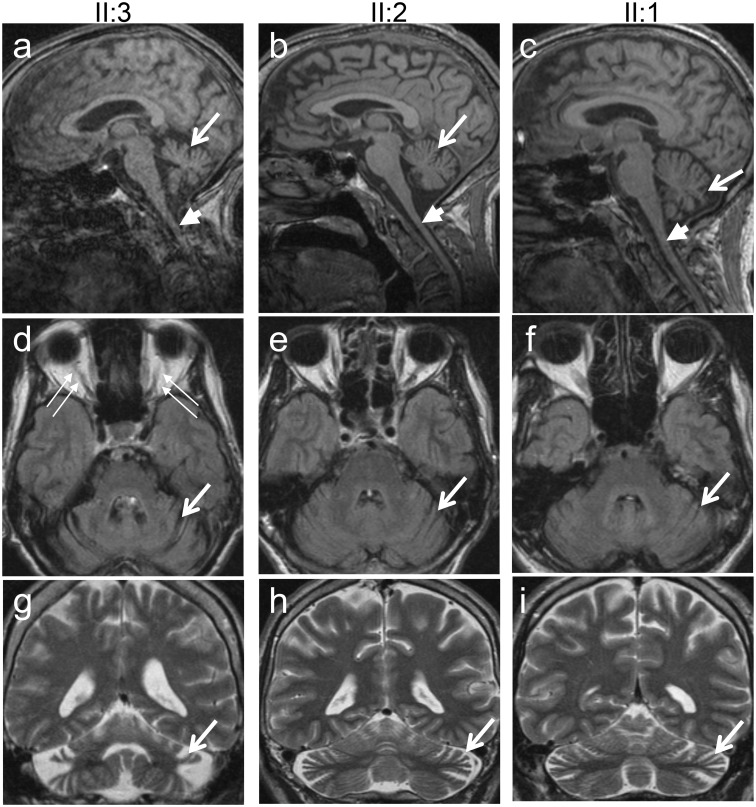
MRI of the brain and cervical spinal cord of affected members of Family OAK 587. MRI of the cerebrum and the upper cervical spinal cord (A–C, sagittal T1; D–F axial FLAIR; G–I coronal T2) demonstrates cerebellar atrophy in all three affected subjects (long arrows), most severely in the subject with the longest disease duration and highest ataxia score (Subject II:3; A, D and G). All three subjects also show, to a variable degree, atrophy of the upper cervical spinal cord (short arrows in A–C).
Table 1
Clinical and electrophysiological characteristics of affected members of the OPA1 index Families OAK587 and DUK2976
| Subject | Family OAK 587 | Family DUK2976 | ||
|---|---|---|---|---|
| II:3 | II:2 | II:1 | III:2 | |
| Age | 50 | 48 | 46 | 13 |
| Clinics | ||||
 Age of onset Age of onset | ||||
    Optic atrophy Optic atrophy | 2 | 2 | 2 | 1 |
    Cerebellar ataxia Cerebellar ataxia | 6 | 6 | 4 | 2 |
    Wheel chair bound Wheel chair bound | 10 | 15 | 14 | n.a. |
    External ophthalmoplegia External ophthalmoplegia | + | + | + | - |
| Vibration sense | Severely reduced | Reduced | Reduced | Reduced |
| Reflexes | ||||
    ATR ATR | - | - | - | - |
    PTR PTR | +++ | +++ | +++ | - |
    BTR BTR | + | + | + | . |
| Spasticity | ||||
    Upper extremity Upper extremity | + | - | - | - |
    Lower extremity Lower extremity | + | + | + | - |
    Babinski sign Babinski sign | +/+ | +/+ | +/+ | -/- |
| SARA score | 34/40 | 30/40 | 29/40 | n.d. |
| Muscle atrophy extremities | + | + | + | + |
| Bilateral ptosis | + | + | + | - |
| Hypacusis | - | - | - | - |
| Blood testing | ||||
| Lactate | Normal (<2.2 mmol/l) | Normal (<2.2 mmol/l) | Normal (<2.2 mmol/l) | Normal |
| Creatine kinase | Normal (<170 U/l) | Normal (<170 U/l) | Normal (<170 U/l) | Normal |
| Electrophysiology | ||||
| Sensory nerve conduction | ||||
    Sural nerve Sural nerve | No SNAP | No SNAP | No SNAP | No SNAP |
    Ulnar nerve Ulnar nerve | No SNAP | No SNAP | No SNAP | No SNAP |
    Medianus nerve Medianus nerve | No SNAP | No SNAP | No SNAP | No SNAP |
| Motor nerve conduction | ||||
    Peroneal nerve Peroneal nerve | No CMAP | No CMAP | No CMAP | |
| Motor evoked potentials | Not evoked UE +LE | Not evoked UE, prolonged LE | Not evoked UE +LE | |
ATR = Achilles tendon reflex; PTR = Patellar tendon reflex; BTR = biceps tendon reflex; SARA = Scale for Assessment and Rating of Ataxia; SNAP = sensory nerve action potential; CMAP = compound muscle action potential; UE = upper extremity; LE = lower extremity; + present; - absent.
Reference values for nerve conduction studies are given in brackets.
Age and age of onset is given in years.
Exon sequencing of candidate genes reveals a rare missense variant in OPA1
No exonic mutation was detected in OPA1, PANK2, POLG, C10orf2, OPA3, mitochondrial DNA and no repeat expansion in FXN and ATXN7, except for a known rare missense variant c.1311A>G/p.I437M in OPA1. The unaffected father in our family was homozygous for p.I437M, whereas the mother was negative for the variant. Correspondingly, the three affected subjects, as well as their unaffected siblings, were heterozygous for c.1311A>G/p.I437M (henceforth p.I437M), which indicates that this variant per se is not pathogenic, neither in heterozygous nor in homozygous state (Fig. 3). Analogous to a previously described family with the p.I437M variant in a compound heterozygous state and a complex early onset inherited optic neuropathy phenotype (Schaaf et al., 2011), we hypothesized that there is a second OPA1 mutation (in trans) in our family that—in concert with p.I437M—leads to the severe multisystemic neurodegenerative phenotype seen in the affected family members. Supporting this hypothesis we found by microsatellite marker-based haplotype reconstruction that all affected family members inherited the same maternal OPA1 allele, whereas the three unaffected siblings inherited the opposite maternal OPA1 allele (data not shown).
Allelic imbalance of OPA1 transcripts in patient fibroblasts
We therefore extended OPA1 mutation screening to the transcript level. RNA isolated from Patient II:2 fibroblasts was used for reverse transcriptase PCR of overlapping complementary DNA fragments. A first round of qualitative complementary DNA analysis did not reveal point mutations or aberrant splice products. We then performed relative quantification of allelic expression of OPA1 transcripts applying pyrosequencing technology. This assay revealed a strong allelic imbalance with a ratio of ~4:1 in favour of the paternally-derived transcripts, whereas equal allele ratios were found at the genomic DNA level (Table 2), indicating reduced steady-state levels of the maternally-derived OPA1 transcripts. These might be caused by reduced transcript stability due to nonsense-mediated messenger RNA decay of transcripts with premature termination codons. To suppress nonsense-mediated messenger RNA decay-dependent allelic decay we used puromycin treatment in patient derived fibroblasts. After this treatment, allelic balance on RNA level could be nearly restored back to normal, as proven by pyrosequencing. Control fibroblasts of healthy donors showed no allelic imbalance and no effect upon puromycin treatment (Table 2).
Table 2
Allelic imbalance of OPA1 transcripts in patient-derived fibroblasts can be rescued by puromycin treatment
| rs9851685, exon 21, all subjects, heterozygous (C/T) | Proportion of allele C (%) | ||
|---|---|---|---|
| Control subject | Patient OAK 587, II:1 | ||
| DNA | Genomic | 48.5 | 50.6 |
| cDNA | Puromycin − | 48.8 | 17.8 |
| Puromycin + | 47.8 | 43.5 | |
Identification of a deep intronic mutation that activates a cryptic exon
We therefore repeated qualitative transcript analysis with complementary DNA (cDNA) from puromycin-treated patient fibroblasts. Gel electrophoretic analysis of one of these complementary DNA amplicons yielded several additional products (Fig. 4). Gel extraction and analysis of these bands showed insertion of additional sequences between exons 4b and 5 (Fig. 5). These additional sequences derived from the intron between exon 4b and 5 and thus constitute a cryptic exon activated in these aberrant transcripts. Two forms of aberrant cDNA products were found: the larger product contained a 93 bp sequence (exon C+; corresponding to c.610+366 to c.610+458; NM_130837.2) that introduces a UAG stop codon after 87 bp into the open reading frame (Supplementary material), whereas the smaller aberrant cDNA contained a shorter variant of the cryptic exon (exon C) that comprises 61 bp (corresponding to c.610+366 to c.610+426; NM_130837.2). The latter starts at exactly the same nucleotide as exon C+ but lacks its last 32 nucleotides. Insertion of exon C into the transcript induces a frameshift and results in a premature termination codon another 84 bp downstream in exon 5. In both cases translation of the maternal allele is aborted before or within exon 5 supporting the hypothesis that nonsense-mediated messenger RNA decay is the cause for the observed allelic imbalance and that haplo-insufficiency contributes to the molecular pathogenesis in this family.
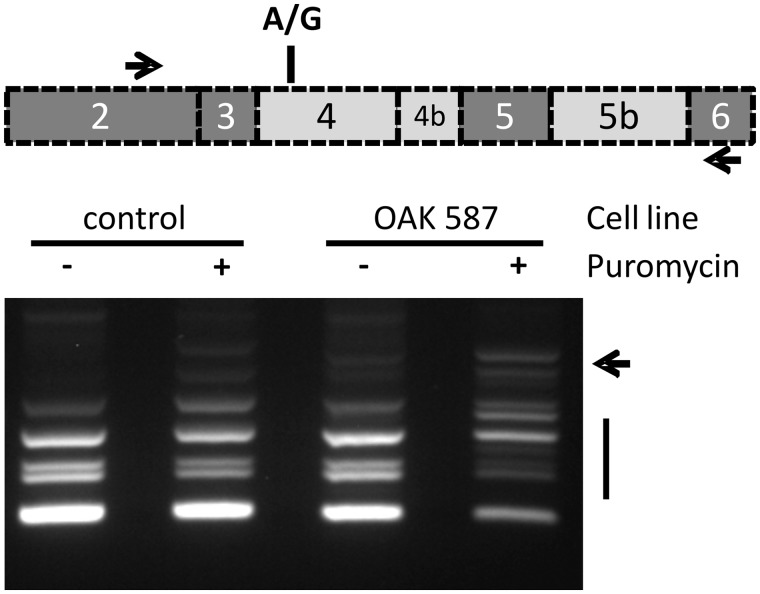
Unmasking of aberrant OPA1 transcripts in puromycin treated patient fibroblasts. Deep intronic mutations in OPA1 reverse transcriptase PCR amplification with primers located in exons 2 and 6 result in multiple cDNA products due to the alternation of constitutively (dark grey in scheme) and facultatively spliced exons (light grey) in this part of the OPA1 gene. The presence of a heterozygous coding SNP in exon 4 (rs7624750; depicted as A/G in the scheme) enabled to differentiate between maternal and paternal-allele derived transcripts in the analysed patient (see text). The pattern of reverse transcriptase PCR products obtained with RNA from untreated fibroblasts of Subject II:1 and the control is virtually identical. However treatment of the patient’s fibroblasts with puromycin before RNA isolation resulted in a drastically altered pattern with additional bands (arrow) and reduced amounts of regular fragments.
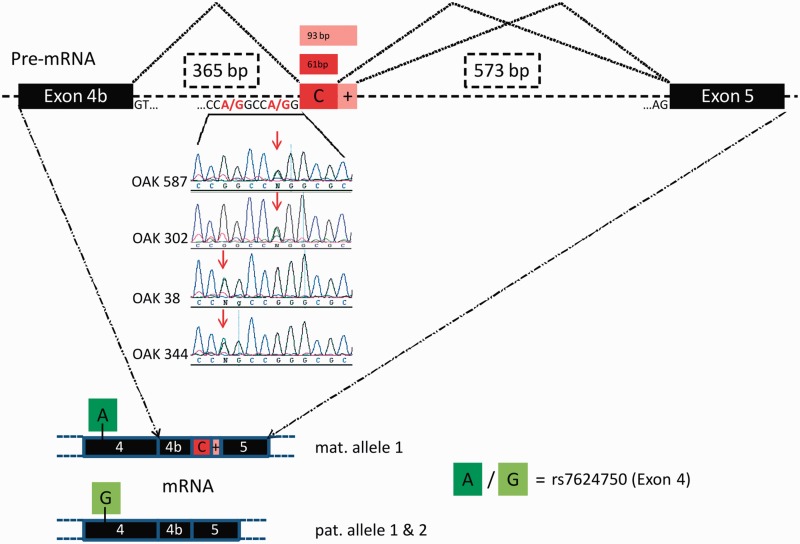
Novel splicing acceptor sites in intron 4b of OPA1 and resulting cryptic exon inclusion. Base transitions of G>A at positions 360 bp or 364 bp downstream of exon 4b create novel splice acceptor sites that exonize downstream sequences of 65 bp / 97 bp or 61 bp/93 bp. Top: Family OAK 587 (index) and Family OAK 302 share the c.610+364G>A mutation, whereas Families OAK 38 and OAK 344 share the mutation c.610+360G>A (centre). The cryptic exon is exclusively found in transcripts of the maternal allele (Family OAK 587) which can be determined e.g. by an SNP in exon 4 (rs7624750, bottom panel). Black bars = exons; dashed lines = introns; red bars = cryptic exons.
Using the heterozygous coding SNP rs7624750 in pyrosequencing as a marker to differentiate between paternal and maternal allele-derived transcripts we found that all maternal allele-derived transcripts were mis-spliced.
Sequencing of the intron between exons 4b and 5 in DNA samples of the affected patients unveiled a single nucleotide exchange G>A at a position 364 bp downstream of exon 4b and adjacent to cryptic exon C (chr3:193335990G>A). The mother and all of the affected siblings were heterozygous for this intronic mutation, whereas the father and the unaffected siblings did not carry the mutation (Fig. 3).
The bioinformatic tool Human Splicing Finder (Desmet et al., 2009) predicts that the deep intronic mutation, henceforth c.610+364G>A (reference: NM_130837.2), creates a novel splice acceptor site with a score of 92.2. The mutation c.610+364G>A was neither observed in 241 healthy Caucasian control samples nor in the 1092 genomes of the 1000 Genome Project (Genomes Project Consortium et al., 2012).
Additional identification of deep intronic mutations in patients with optic atrophy
To further confirm the pathogenicity of the c.610+364G>A OPA1 mutation as well as to determine its frequency in patients with unexplained optic atrophy, we investigated 369 index cases in which conventional OPA1 screening failed to detect the disease-causing mutation. Our RFLP analysis in intron 4b revealed three further index cases (OAK 302, OAK 38 and OAK 344), one carrying the same mutation c.610+364G>A as found in the original family and two other index patients with a G-to-A exchange 4 bp upstream (c.610+360G>A reference: NM_130837.2) (Fig. 5). Subsequent cDNA analysis with blood samples from two of these patients confirmed that both mutations result in aberrant splicing of OPA1 transcripts with pseudoexon activation between exons 4b and 5. Yet in the patients with the c.610+360G>A mutation, the 5’ acceptor site is shifted by 4 nucleotides further upstream whereas the alternative 3’ donor sites of the cryptic exon are conserved for both deep intronic mutations (as seen for exon C+; Supplementary material). Neither of the two newly identified deep intronic mutations were observed in the 1092 genomes of the 1000 Genomes Project Consortium (Genomes Project Consortium et al., 2012), demonstrating that they are not simply frequent SNPs. Each of the three additional deep intronic mutation carriers showed a classic, slowly progressing optic atrophy and no signs of extraocular neurodegenerative impairment.
Reduced OPA1 protein level in affected mutation carriers
We further investigated whether deep intronic mutations result in lower OPA1 protein levels, or whether compensatory mechanisms counteract the genetic defect. Western blot analysis with whole-cell lysates of primary fibroblasts from several members of the OAK 587 and OAK 302 families revealed a reduction of OPA1 protein normalized for ACTB (Fig. 6). This reduction caused by the deep intronic mutation was in the same range as in a patient with a well characterized stop mutation in exon 7 (c.868C>T/p.Arg290stop) (Puomila et al., 2005). We also observed a slight reduction of OPA1 protein in fibroblasts of the unaffected father in OAK 587 (homozygous carrier of p.I437M), suggesting some impairment of this modifier variant on OPA1 protein stability. Comparative immunoblotting with an antibody against the mitochondrial outer membrane protein TOMM40 revealed no significant difference in protein levels compared with the control indicating that reduced amounts of OPA1 do not by large affect mitochondrial mass in fibroblasts (Fig. 6).
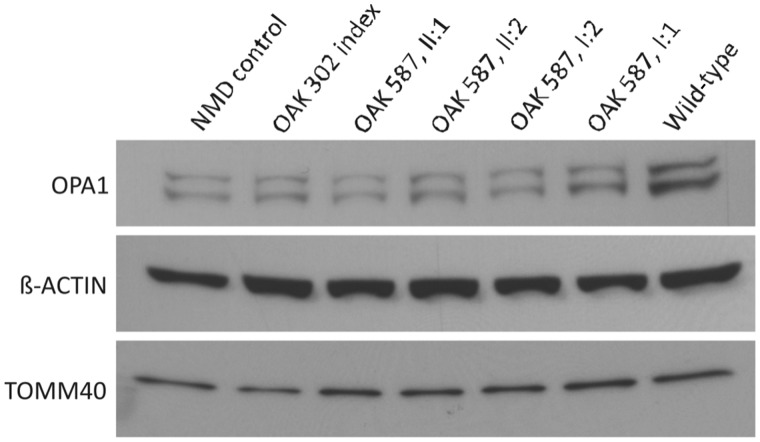
Reduced OPA1 protein amount in patients with deep intronic mutation. Semi-quantitative western blot immunodetection with antibodies against OPA1 (top), ACTB (centre) and TOMM40 (bottom) in whole fibroblast cell lysates from a patient with autosomal dominant optic atrophy with heterozygous c.868C>T/p.Arg290stop in OPA1 (nonsense-mediated messenger RNA decay control), the affected index patient of Family OAK 302, two affected patients (Subjects II:1 and II:2) as well as the parents of Family OAK 587, and a healthy control (wild-type) (from left to right). A clear reduction in OPA1 protein amount can be observed in all samples except the control and the unaffected father (only slight reduction).
Increased mitochondrial fragmentation in mutation carriers
To assess the functional consequences of the two OPA1 variants reported here (c.610+364G>A; and p.I437M), we assessed the morphological characteristics of the mitochondrial network in fibroblasts of the three affected siblings and both parents in Family OAK 587. Compared to age-matched controls, surface (P < 0.05), aspect ratio (P < 0.001), and form factor (P < 0.001) of the mitochondria were significantly reduced in the three affected subjects (Fig. 7A and B). This finding is in line with the well-established effect of OPA1 mutations that lead to reduced mitochondrial fusion and increased mitochondrial fragmentation, respectively (Amati-Bonneau et al., 2005, 2008; Olichon et al., 2007; Zanna et al., 2008). Although clinically asymptomatic, the mother of the affected siblings (Subject I:2) also showed a significantly reduced surface (P < 0.01), aspect ratio (P < 0.05), and form factor (P < 0.01) of the mitochondria (Fig. 7), indicating that even the deep intronic mutation c.610+364G>A per se leads to OPA1-characteristic dysregulation of mitochondrial dynamics. Mitochondria of the father (Subject I:1) were reduced in aspect ratio (P < 0.001) and form factor (P < 0.001), but not in surface (P > 0.05) and volume (P > 0.05) (Fig. 7), compatible with a moderate effect of the homozygous p.I437M modifier variant on mitochondrial dynamics.
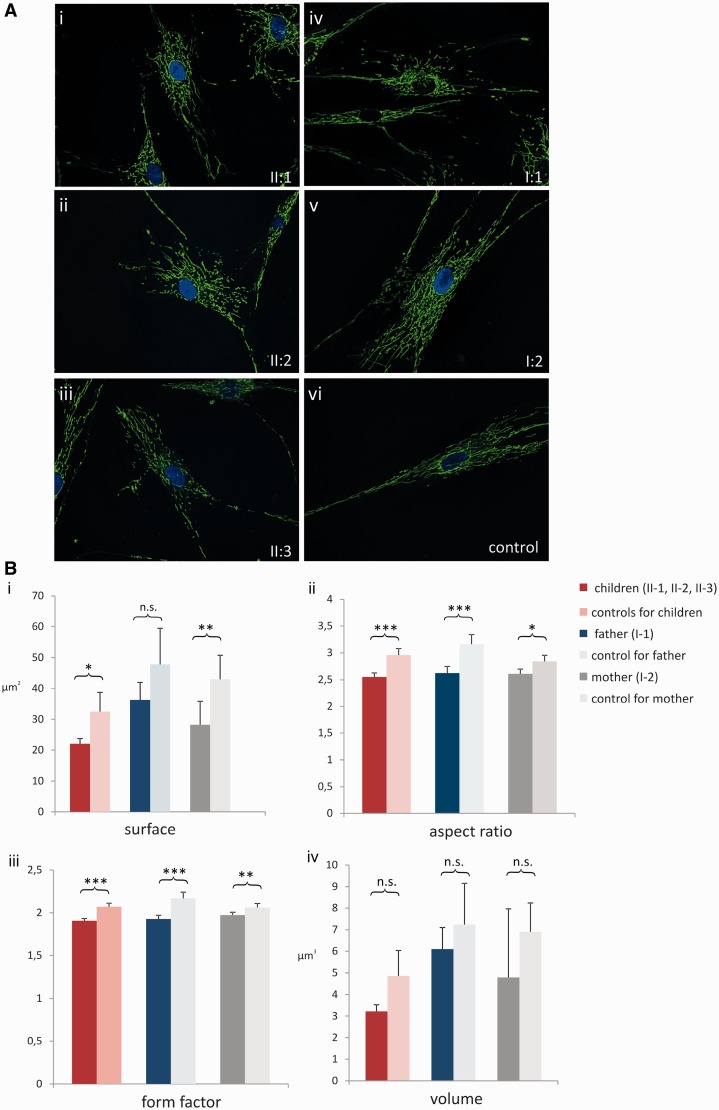
Mitochondrial morphometry. (A) Mitochondrial network stained with MitoTracker Green FM(R) and visualized with a fluorescence microscope. Compared to an exemplary control (i), where mitochondria build a network of lengthy and closely connected mitochondrial alignments, the mitochondrial network is fragmented in all three affected siblings (ii–iv). Similarly, also mitochondria of the mother (vi, Subject I:2) show signs of fragmentation and, to a lesser extent, mitochondria of the father as well (v, Subject I:1). (B) Statistical analysis of mitochondrial morphometry in fibroblasts of the three affected siblings (dark red), the father (dark blue) and the mother (dark grey) of Family OAK 587 compared to age-matched controls. Four aspects of morphology were analysed: (i) surface; (ii) aspect ratio (major axis/minor axis); (iii) form factor (surface2/4 π volume); and (iv) volume. *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant.
Screening for the OPA1 p.I437M variant in additional cohorts
Although the missense variant I437M does not necessarily lead to a clinical manifestation of optic atrophy when present per se at a heterozygous or homozygous state (Family OAK 587), it causes a severe optic atrophy plus syndrome in conjunction with an additional truncating OPA1 mutation, as observed in our Family OAK 587 as well as in a previously published family (Schaaf et al., 2011). To further elucidate the phenotypic effects of the I437M variant we analysed the presence of this variant in two additional cohorts. In the NHLBI Exome Sequencing Project (public data available at data Exome Variant Server), the p.I437M was present at heterozygous state in 5 of 6499 subjects (minor allele frequency 0.077%). In the disease cohort presenting with at least one of the main neurological features of syndromic OPA1 disease (ataxia, spastic paraplegia or peripheral neuropathy), the heterozygous I437M variant was observed three times among 618 families (0.5%).
Among the three carriers of the I437M variant in the disease cohort, only a single index case had an additional compound heterozygous exonic variant in the OPA1 gene, the insertion c.1316_1317insA, p.N440Kfs*14 (Supplementary material).
This 13-year-old patient of US-American origin (Family DUK2976, III:2) presented with a severe optic atrophy plus phenotype consisting of connate bilateral optic atrophy, sensory predominant peripheral neuropathy and truncal as well as limb ataxia with a symptom onset in early childhood (Table 1). The mother of the patient had high arches of her feet at age 31, but was otherwise neurologically asymptomatic. Family history for optic atrophy without further accompanying signs or symptoms was positive on the maternal side (optic atrophy in the maternal aunt and one maternal cousin (Fig. 3, lower pedigree). The father of the index case had myopia, but no further ocular or neurological symptoms in his 30 s.
Discussion
Pathogenic deep intronic mutations cause OPA1 disease
Here we report for the first time deep intronic mutations as a novel genetic mechanism for OPA1-associated neurodegenerative disease. We have identified two deep intronic mutations in intron 4b of the OPA1 gene in a total of four families with optic atrophy. Both deep intronic mutations are novel and absent in healthy controls. They activate a cryptic splice site, leading to exonization of parts of intron 4b and a consecutive shift of the reading frame. We could demonstrate that the mutant transcripts of the deep intronic mutations are subject to nonsense mediated decay and that OPA1 protein levels in fibroblasts of mutation carriers are significantly reduced, leading to mitochondrial fragmentation typical for OPA1-associated disease.
The deep intronic mutations occur at the 5’ end of an AluY repetitive element. Genomic insertion of Alu-sequences into coding regions can cause mis-splicing. This has also been observed for OPA1, where de novo insertion of an Alu-element into intron 7 of OPA1 was shown to cause autosomal dominant optic atrophy by skipping of exon 8 (Gallus et al., 2010). However, this is the first case that shows a point mutation within a pre-existing Alu element to induce mis-splicing in OPA1, although generally, point mutations in Alu elements are a relatively common (though mostly undetected) cause of exonization (Lev-Maor et al., 2003). Considering that we have only targeted a 6 bp sequence in intron 4b by RFLP-based screening, but not systematically screened all OPA1 introns, deep intronic mutations might in fact be a relatively frequent mechanism in OPA1-associated disease. To unravel additional OPA1 deep intronic mutations in hereditary and maybe even apparently sporadic inherited optic neuropathies patients, future studies might use targeted OPA1 genomic sequencing and subsequent functional analysis of the identified intronic variants.
The p.I437M variant acts as a phenotypic modifier to reinforce the pathogenic effect of loss of function mutations
Mechanisms determining severe plus phenotypes rather than pure optic atrophy have been vague so far. Putative dominant negative effects of missense mutations in the GTPase domain, exerting a gain-of-function effect, are frequently associated with a severe phenotype (Amati-Bonneau et al., 2008; Yu-Wai-Man et al., 2010b). An additional mechanism might be a combined mutational effect from a biallelic OPA1 state of two OPA1 variants (which, for example, jointly lower the amount of functional OPA1 protein below a certain critical threshold). For example, it has been shown that, like in Family OAK 587, the I437M variant in compound heterozygous state with another OPA1 mutation (c.2873_2876del/p.V958Gfs*3) presents with a severe optic atrophy plus phenotype (Schaaf et al., 2011). Our findings corroborate and, at the same time, significantly extend this notion. We show that the later scenario does not follow a simple recessive mode of inheritance but rather that the I437M variant acts as an intralocus modifier. This hypomorphic allele causes a severe optic atrophy plus phenotype when combined with a compound heterozygous OPA1 null mutation. That second OPA1 mutation might be either exonic (Family DUK2976) or intronic (Family OAK 587).
The notion of I437M acting as an intralocus modifier is based on the finding that in all three families [OAK 587, DUK2976 and the family described by Schaaf et al. (2011)] the p.I437M variant is associated with a severe ‘optic atrophy plus’ phenotype if occurring with another OPA1 mutation. In contrast, these three truncating OPA1 mutations alone cause a pure optic atrophy phenotype. Compared to the compound heterozygous null mutations, the I437M variant alone does not cause disease per se, neither in heterozygous nor in homozygous state as observed in Family OAK 587. In fact, the homozygous father Subject I:1 is still healthy at the age of 76 years, without evidence for subclinical optic atrophy. In addition to our clinical observation in three independent families, several other lines of evidence also support an adverse effect of the p.I437M on OPA1 function.
First, amino acid sequence alignment of OPA1 and its homologues in diverse species shows that the isoleucine at position 437 (Ile437) localizes to the GTPase domain of OPA1 and is fully conserved in vertebrates, invertebrates (Drosophila melanogaster and Caenorhabditis elegans) as well as in yeast (Supplementary material).
Second, the p.I437M variant is very rare (5/~6500 exomes) in the Exome variant server (http://evs.gs.washington.edu/EVS/), thus demonstrating that it is not simply a frequent SNP.
Finally, Ile437 in OPA1 corresponds to the conserved Ile120 in the dynamin proteins DNM1, DNM2 and DNM3 and is part of a dynamin superfamily-specific β-strand (β2b). This β-strand directly follows the ‘cis-stabilizing loop’ that stabilizes nucleotide-dependent dimerization of the GTPase domains (Chappie et al., 2010). Furthermore, Ile120 points into the hydrophobic core of the GTPase domain and interacts, for example, with Ile101 of the preceding α-helix (Supplementary material). A substitution of Ile120(dynamin)/Ile437(OPA1) to methionine might locally destabilize the GTPase domain and thus partially interfere with its nucleotide-dependent function (Oliver Daumke, personal communication).
OPA1 plus: an autosomal-recessive ataxia mimic with cervical cord atrophy
The clinical presentation of the affected patients in Families OAK 587 and DUK2976—namely, early-onset, multisystemic neurodegenerative disease—mimics a picture well known from a variety of autosomal-recessive neurodegenerative diseases, such as autosomal-recessive ataxias, hereditary spastic paraplegias or lysosomal storage disorders, which might likewise include optic atrophy, spasticity and polyneuropathy [e.g. Friedreich’s ataxia, spastic paraplegia type 7, Gaucher’s disease or SPOAN (spastic paraplegia, optic atrophy and neuropathy) (Macedo-Souza et al., 2009)]. In addition, also the frequently observed reduced penetrance in OPA1 [as seen in Subject I:2 of Family OAK 587 and as described in the literature (Cohn et al., 2007)] might mislead towards an apparently recessive disease in these subjects. Thus, OPA1 mutations should be taken into the differential diagnosis of patients presenting with such seemingly recessive disease, and intronic mutations should be searched for if exonic OPA1 sequencing remains inconclusive.
Although most features of the affected siblings of our index family fit well with the reported phenotype of optic atrophy plus syndromes (Yu-Wai-Man et al., 2010b), our clinical findings modify and extend this phenotypic spectrum. Unlike most patients with optic atrophy plus (Amati-Bonneau et al., 2005; Yu-Wai-Man et al., 2010b; Schaaf et al., 2011), none of our patients was suffering from hearing impairment, demonstrating that this feature is not an obligatory part of the OPA1-plus cluster. Moreover, our MRI results demonstrate that atrophy of the cervical spinal cord can be part of the disease spectrum, whereas cerebellar atrophy might be only mild, despite severe ataxia [SARA (Scale for the Assessment and Rating of Ataxia) scores ≥29 points]. This resembles findings in patients with Friedreich’s ataxia, where atrophy of the spinal cord is also common, whereas cerebellar atrophy is mild even in late disease stages. This parallel might indicate that, as in Friedreich’s ataxia, also OPA1-ataxia might result largely from a spinal degeneration and only to a smaller extent from cerebellar degeneration.
Identification of more generalizable mutational mechanisms?
The mutational mechanisms identified in this study—deep intronic mutations and intragenic modifiers—might also be of relevance beyond the field of OPA1 genetics. This notion is corroborated by the previous identification of deep intronic mutations in another hereditary neurodegenerative disease (ataxia telangiectasia; deep intronic mutation in ATM; Cavalieri et al., 2013) as well as in a retinal degeneration (Leber’s congenital amaurosis, deep intronic mutation in CEP290; den Hollander et al., 2006). Taken together with our findings from OPA1, these observations make it highly likely that deep intronic mutations can be found also in a wide range of other neurodegenerative and optic neuropathy diseases where deep intronic mutations are not yet known and where the relative share of genetically unexplained patients is still high. This is important to acknowledge as these mutations will be missed even if using the current state-of-the-art whole-exome approaches.
The presence and action of genetic modifiers in neurodegenerative disease is frequently postulated, however, genetic mapping of such loci or even the identification of such factors has been rarely achieved. In Leber’s hereditary optic neuropathy, the other major form of inherited optic neuropathy, there is strong evidence that certain mitochondrial haplogroups govern clinical expression in subjects with principle mitochondrial DNA mutations at m.11778 (Hudson et al., 2007; Ji et al., 2008). Moreover, a modifier that may explain the pronounced gender difference in penetrance in this disease has been mapped on Xp21-q21 (Hudson et al., 2005) and Xq25-27.2 (Shankar et al., 2008), respectively, though their identity is still unknown.
Reports on intragenic modifiers that modulate phenotype severity and disease penetrance (as the p.I437M OPA1 variant in this paper) are still limited (Gouya et al., 2002). Nonetheless we suggest that such a mechanism represents an attractive explanation for expression variability of traits that rely on dose-sensitive or threshold dependent functions. As variability in disease severity and disease phenotype within families as well as between subjects carrying the same mutation have been described in a large number of genetic diseases [e.g. for SOD1 mutations in amyotrophic lateral sclerosis (Synofzik et al., 2010) or PRNP mutations in hereditary prion disease (Synofzik et al, 2009)], it is likely that such intragenic modifiers exist also for other neurological/ophthalmological diseases.
Conclusions
We show that deep intronic mutations in OPA1 are a cause of optic atrophy and might be responsible for inherited optic neuropathies in a substantial group of so far unsolved inherited optic neuropathy patients. Furthermore, we demonstrate by example of the I437M variant that certain OPA1 sequence alterations can act as intralocus disease-modifiers. If occurring in a compound-heterozygous state, such a modifier can cause optic atrophy plus disease, following the model of a combined mutational effect. Taken together, we suggest that whole gene sequencing of OPA1 and/or expansion of diagnostics to the transcript level are important tools to solve unexplained cases of optic atrophy and optic atrophy plus syndromes. The mutational mechanisms identified in this study—deep intronic mutations and intragenic modifiers—could represent more generalizable mechanisms that might also be found in a wide range of other neurodegenerative and optic neuropathy diseases.
Acknowledgements
We thank the patients and their family members for participating in our study, Doron Rapaport (Tübingen) for the anti-TOMM40 antibody, Beate Leo-Kottler and Bernhard Jurklies for ophthalmological examinations and recruitment of patients with Autosomal Dominant Optic Atrophy, and in particular Oliver Daumke (Berlin) for the modelling of the 3D structures of the OPA1 GTPase domain and his support in evaluating the putative function of I437M.
Funding
This work was supported by the German Ministry of Education for the E-RARE projects ERMION (to V.C., 01GM1006 to B.W.) and EUROSCAR (01GM1206 to L.S.); by grants of the Fondazione Telethon (GGP06233;, GGP1118; and GPP10005 to V.C.); an FP7 grant for NeurOmics (2012-305121 to L.S.), ‘HSP/CMT genetics’ (PIOF-GA-2012-326681 to R.S.); NIH 5R01NS075764 and MDA (to S.Z.) and mitoNET (01GM1113E to L.S.) and the Interdisciplinary Center for Clinical Research IZKF Tübingen (2191-0-0; to MS and 1970-0-0 to R.S.).
References
- Amati-Bonneau P, Guichet A, Olichon A, Chevrollier A, Viala F, Miot S, et al. OPA1 R445H mutation in optic atrophy associated with sensorineural deafness. Ann Neurol. 2005;58:958–63. [Abstract] [Google Scholar]
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–51. [Abstract] [Google Scholar]
- Cavalieri S, Pozzi E, Gatti RA, Brusco A. Deep-intronic ATM mutation detected by genomic resequencing and corrected in vitro by antisense morpholino oligonucleotide (AMO) Eur J Hum Genet. 2013;21:774–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:435–40. [Europe PMC free article] [Abstract] [Google Scholar]
- Cohn AC, Toomes C, Potter C, Towns KV, Hewitt AW, Inglehearn CF, et al. Autosomal dominant optic atrophy: penetrance and expressivity in patients with OPA1 mutations. Am J Ophthalmol. 2007;143:656–62. [Abstract] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79:556–61. [Europe PMC free article] [Abstract] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. [Europe PMC free article] [Abstract] [Google Scholar]
- Ferre M, Bonneau D, Milea D, Chevrollier A, Verny C, Dollfus H, et al. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 novel OPA1 mutations. Hum Mutat. 2009;30:692–705. [Abstract] [Google Scholar]
- Gallus GN, Cardaioli E, Rufa A, Da Pozzo P, Bianchi S, D'Eramo C, et al. Alu-element insertion in an OPA1 intron sequence associated with autosomal dominant optic atrophy. Mol Vis. 2010;16:178–83. [Europe PMC free article] [Abstract] [Google Scholar]
- Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [Europe PMC free article] [Abstract] [Google Scholar]
- Gonzalez MA, Lebrigio RF, Van Booven D, Ulloa RH, Powell E, Speziani F, et al. GEnomes Management Application (GEM.app): a new software tool for large-scale collaborative genome analysis. Hum Mutat. 2013;34:842–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Gouya L, Puy H, Robreau AM, Bourgeois M, Lamoril J, Da Silva V, et al. The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nat Genet. 2002;30:27–8. [Abstract] [Google Scholar]
- Hudson G, Carelli V, Spruijt L, Gerards M, Mowbray C, Achilli A, et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–33. [Europe PMC free article] [Abstract] [Google Scholar]
- Hudson G, Keers S, Yu Wai Man P, Griffiths P, Huoponen K, Savontaus ML, et al. Identification of an X-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am J Hum Genet. 2005;77:1086–91. [Europe PMC free article] [Abstract] [Google Scholar]
- Ji Y, Zhang AM, Jia X, Zhang YP, Xiao X, Li S, et al. Mitochondrial DNA haplogroups M7b1'2 and M8a affect clinical expression of leber hereditary optic neuropathy in Chinese families with the m.11778G–>a mutation. Am J Hum Genet. 2008;83:760–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons. Science. 2003;300:1288–91. [Abstract] [Google Scholar]
- Macedo-Souza LI, Kok F, Santos S, Licinio L, Lezirovitz K, Cavacana N, et al. Spastic paraplegia, optic atrophy, and neuropathy: new observations, locus refinement, and exclusion of candidate genes. Ann Hum Genet. 2009;73:382–7. [Abstract] [Google Scholar]
- Munier A, Gunning T, Kenny D, O'Keefe M. Causes of blindness in the adult population of the Republic of Ireland. Br J Ophthalmol. 1998;82:630–3. [Europe PMC free article] [Abstract] [Google Scholar]
- Olichon A, Landes T, Arnaune-Pelloquin L, Emorine LJ, Mils V, Guichet A, et al. Effects of OPA1 mutations on mitochondrial morphology and apoptosis: relevance to ADOA pathogenesis. J Cell Physiol. 2007;211:423–30. [Abstract] [Google Scholar]
- Puomila A, Huoponen K, Mantyjarvi M, Hamalainen P, Paananen R, Sankila EM, et al. Dominant optic atrophy: correlation between clinical and molecular genetic studies. Acta Ophthalmol Scand. 2005;83:337–46. [Abstract] [Google Scholar]
- Schaaf CP, Blazo M, Lewis RA, Tonini RE, Takei H, Wang J, et al. Early-onset severe neuromuscular phenotype associated with compound heterozygosity for OPA1 mutations. Mol Genet Metab. 2011;103:383–7. [Abstract] [Google Scholar]
- Shankar SP, Fingert JH, Carelli V, Valentino ML, King TM, Daiger SP, et al. Evidence for a novel x-linked modifier locus for leber hereditary optic neuropathy. Ophthalmic Genet. 2008;29:17–24. [Abstract] [Google Scholar]
- Synofzik M, Bauer P, Schols L. Prion mutation D178N with highly variable disease onset and phenotype. J Neurol Neurosurg Psychiatry. 2009;80:345–6. [Abstract] [Google Scholar]
- Synofzik M, Fernandez-Santiago R, Maetzler W, Schols L, Andersen PM. The human G93A SOD1 phenotype closely resembles sporadic amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2010;81:764–7. [Abstract] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Burke A, Sellar PW, Clarke MP, Gnanaraj L, et al. The prevalence and natural history of dominant optic atrophy due to OPA1 mutations. Ophthalmology. 2010a;117:1538–46. 1546.e1. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Gorman GS, Lourenco CM, Wright AF, Auer-Grumbach M, et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010b;133:771–86. [Europe PMC free article] [Abstract] [Google Scholar]
- Zanna C, Ghelli A, Porcelli AM, Karbowski M, Youle RJ, Schimpf S, et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352–67. [Abstract] [Google Scholar]
Articles from Brain are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/brain/awu165
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/brain/article-pdf/137/8/2164/13798500/awu165.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/brain/awu165
Article citations
<i>Drosophila</i> model to clarify the pathological significance of OPA1 in autosomal dominant optic atrophy.
Elife, 12:RP87880, 23 Aug 2024
Cited by: 0 articles | PMID: 39177028 | PMCID: PMC11343565
Autosomal recessive cerebellar ataxias: a diagnostic classification approach according to ocular features.
Front Integr Neurosci, 17:1275794, 07 Feb 2024
Cited by: 0 articles | PMID: 38390227 | PMCID: PMC10883068
Genetic background modulates phenotypic expressivity in OPA1 mutated mice, relevance to DOA pathogenesis.
Front Mol Neurosci, 16:1241222, 06 Sep 2023
Cited by: 0 articles | PMID: 37736113 | PMCID: PMC10510408
Evaluation of a Less Invasive Cochlear Implant Surgery in OPA1 Mutations Provoking Deafblindness.
Genes (Basel), 14(3):627, 02 Mar 2023
Cited by: 0 articles | PMID: 36980899 | PMCID: PMC10048538
Dynamic features of human mitochondrial DNA maintenance and transcription.
Front Cell Dev Biol, 10:984245, 07 Sep 2022
Cited by: 6 articles | PMID: 36158192 | PMCID: PMC9491825
Review Free full text in Europe PMC
Go to all (43) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases (2)
- (1 citation) OMIM - 165500
- (1 citation) OMIM - 125250
RefSeq - NCBI Reference Sequence Database (2)
- (2 citations) RefSeq - NM_130837.2
- (1 citation) RefSeq - NM_130837
SNPs (3)
- (4 citations) dbSNP - rs7624750
- (2 citations) dbSNP - rs143319805
- (1 citation) dbSNP - rs9851685
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of two novel intronic OPA1 mutations resulting in aberrant pre-mRNA splicing.
BMC Med Genet, 18(1):22, 28 Feb 2017
Cited by: 2 articles | PMID: 28245802 | PMCID: PMC5331656
Mutation survey of the optic atrophy 1 gene in 193 Chinese families with suspected hereditary optic neuropathy.
Mol Vis, 19:292-302, 06 Feb 2013
Cited by: 9 articles | PMID: 23401657 | PMCID: PMC3566897
A comprehensive survey of mutations in the OPA1 gene in patients with autosomal dominant optic atrophy.
Invest Ophthalmol Vis Sci, 43(6):1715-1724, 01 Jun 2002
Cited by: 51 articles | PMID: 12036970
Recessive optic atrophy, sensorimotor neuropathy and cataract associated with novel compound heterozygous mutations in OPA1.
Mol Med Rep, 14(1):33-40, 04 May 2016
Cited by: 10 articles | PMID: 27150940 | PMCID: PMC4918608
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NINDS NIH HHS (2)
Grant ID: R01 NS072248
Grant ID: 5R01NS075764
Telethon (3)
Systematic gene hunting for nuclear modifiers in Leber's hereditary optic neuropathy and their validation in model systems
Prof. Palmiro Cantatore, Università degli Studi di Bari Aldo Moro, Dipartimento di Bioscienze, Biotecnologie e Scienze Farmacologiche
Grant ID: GGP11182
Therapeutic strategies to combat mitochondrial disorders
Prof Rosario Rizzuto, Università di Padova, Dipartimento di Scienze Biomediche
Grant ID: GPP10005
PATHOGENIC MECHANISMS FOR DEGENERATION OF RETINAL GANGLION CELLS IN MITOCHONDRIAL OPTIC NEUROPATHIS
Prof. Palmiro Cantatore, Università degli Studi di Bari Aldo Moro, Dipartimento di Biochimica e Biologia Molecolare - Facoltà di Scienze
Grant ID: GGP06233
 1 and
1 and 




