Abstract
Free full text

Random and targeted transgene insertion in C. elegans using a modified Mosl transposon
Abstract
We have generated a recombinant Mos1 transposon that can insert up to 45 kb transgenes into the C. elegans genome. The minimal Mos1 transposon (miniMos) is 550 bp long and inserts DNA into the genome at high frequency (~60% of injected animals). Genetic and antibiotic markers can be used for selection and the transposon is active in C. elegans isolates and C. briggsae. We have used the miniMos transposon to generate six universal MosSCI landing sites that allow targeted transgene insertion with a single targeting vector into permissive expression sites on all autosomes. We have also generated two collections of strains: A set of bright fluorescent insertions that are useful as dominant, genetic balancers and a set of lacO insertions to track genome position.
Introduction
Some DNA transposons can carry non-transposon DNA and still retain the ability to insert themselves randomly into chromosomal DNA. For example, the P-element is used extensively to insert transgenes into the fruit fly Drosophila melanogaster1. The P-element has also been used in the fly to generate large-scale gene knock-out libraries, to drive tissue-specific expression using the Gal4 enhancer trap, to study genomic position effects, and to generate targeted transgene insertion sites2-5. Similarly, other DNA-based transposons (for example, Sleeping Beauty, piggyBac and Tol2) have successfully been used for transgenesis in a variety of genetically tractable systems including human tissue culture cells, mice, zebrafish, frogs and flies6.
In C. elegans, transgenic animals are most frequently generated by DNA injection into the syncytial germline to generate extra-chromosomal arrays7. Biolistic transformation can be used for stable, but random, genomic integration of a single or a small number of plasmids8. The fly transposon Mosl is active in C. elegans but has limited cargo capacity (~500 bp) and is therefore not used directly for transgenesis9. Instead excisions of Mosl inserts are used to generate double-strand DNA breaks, which are repaired from injected template DNA10. By using positive and negative selection markers, a single copy of a transgene can be inserted into the genome directly via injection (Mosl-mediated Single Copy Insertion, mosSCI)11,12. An alternative method to modify genomes that does not rely on transposons but on the bacterial Cas9/CRISPR system13 has recently been adapted for C. elegans to allow genome editing at endogenous loci14-16.
Here we demonstrate that a modified Mos1 transposon (miniMos) can carry large fragments of DNA, even 45 kb fosmids, into the C. elegans genome. We show that insertions can be selected using either genetic or antibiotic markers and that the transposon can be mobilized in wild isolates of C. elegans and C. briggsae. We have used miniMos to generate a set of strains with fluorescent markers, that can be used as genetic balancers, and lacO insertions that can track genome position in the nucleus. Furthermore, we have used the miniMos transposon to generate six universal MosSCI landing sites that allow insertion of a single transgene construct into permissive sites on all autosomes.
Results
A recombinant Mos1 element transposes with exogenous DNA
The requirements for transposition of mariner elements (Mos1 and the closely related Peach transposon) vary depending on whether the transposon is embedded in chromatin or is contained within injected plasmid DNA. Mariner transposons within chromosomes require internal sequences to transpose17 and can carry cargo only if the cargo is flanked by intact transposons18. By contrast, transposons injected as plasmids can transpose efficiently even if they contain internal deletions and carry cargo19. Experiments in vitro have further demonstrated that modifications to the inverted terminal repeats improve transposition frequency20. We tested whether modified Mosl elements and plasmid injection protocols11 could overcome previously described limitations for Mosl transposition in C. elegans9. We generated a composite Mosl transposon with a 7.5 kb transgene (containing Ppie-1:GFP:histone and cb-unc-119(+)) and tested transposition by plasmid injection (Fig. 1 and Supplementary Fig. 1). We co-injected the composite Mosl transposon with a helper plasmid expressing the transposase and fluorescent extra-chromosomal array markers. We injected 27 unc-119 animals and identified 17 independent lines with recombinant Mosl insertions (62% P0 insertion frequency). 47% (8/17) of the strains expressed GFP in the germline (Fig. 1). We mapped four GFP expressors and four non-expressors by inverse PCR21 to unique insertion sites. Non-fluorescent insertions were found on autosomal arms, which have high levels of repressive chromatin marks22 or the X-chromosome, which is inactivated in the germline23 (Fig. 1). It is likely that these Ppie-1:GFP:histone insertions are silenced through a combination of small RNAs that detect foreign DNAs and protect endogenous gene expression in the germline24-26 and subsequent modifications to the chromatin environment. We are currently characterizing germline and somatic position effects in detail (unpublished C.F.-J. & E.M.J.).
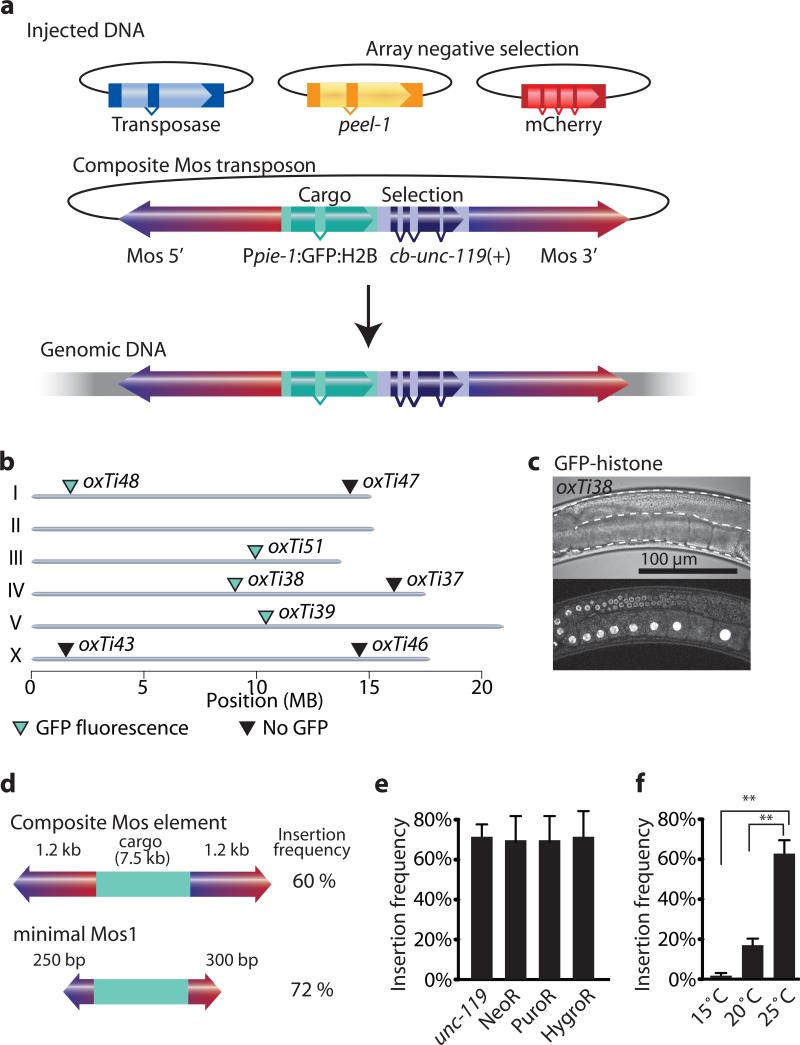
(a) Schematic of recombinant Mosl insertion protocol. Transposon DNA is co-injected with a helper plasmid expressing the transposase (Peft-3:Mos1 transposase). Negative selection markers (Phsp16.41:peel-1, Pmyo-2:mCherry, Prab-3:mCherry and Pmyo-3:mCherry) were used to select against array-bearing transgenic animals. (b) Genomic locations of insertions identified by cb-unc-119(+) rescue of unc-119 mutants. All insertions rescued unc-119, but not all strains expressed GFP-histone in the germline. Germline fluorescence is indicated with turquoise (GFP-positive) or black (no fluorescence) triangles. (c) Fluorescence image of germline expression. Transposon insertion oxTi38 expressed GFP-histone in the germline (Ppie-1:GFP:H2B). Above, differential interference contrast; below confocal fluorescence image. Scale bar = 100 μm. (d) Schematic of the minimal Mos1 transposon (miniMos). 550 bp was enough to retain full insertion frequency. (e) Bar-graph of insertion frequencies with the genetic marker unc-119(+) and antibiotic selection markers G418 (NeoR), puromycin (PuroR) or hygromycin B (HygroR). Values show the average of two independent injections and error bars show the 95% confidence interval (modified Wald method). (f) Bar-graph of insertion frequency at different temperatures. Values shown are averages of three independent replicates (injections) and error bars represent standard error of mean (SEM). Statistics: repeated measures ANOVA (P = 0.0017). Bonferroni post-hoc comparison. **, P < 0.01.
The composite Mosl element was flanked by two essentially full-length Mosl elements (Supplementary Fig. 1). To identify a minimal Mosl element (miniMos) we tested transposition of truncated composite elements. Only 250-300 bp on either side was required for efficient transposition with 7.5 kb cargo with no decrease in efficiency compared to the composite transposon (Supplementary Fig. 1).
The composite transposon could also be mobilized from extra-chromosomal arrays containing the transposon and the transposase under the control of a heat-shock promoter. From one extra-chromosomal line (EG6346) we isolated two insertions from 300 heat-shocked animals (0.7 %) and from a second line (EG6347) we isolated 12 insertions from 410 heat-shocked animals (2.9 %). All insertions generated by mobilization from arrays were independent and mapped to unique genomic locations. It might be possible to generate large-scale transposon collections using a heat-shock protocol, similar to the genome-wide collection of wild-type Mosl inserts27. However, currently it is more efficient to generate insertions directly by plasmid injection.
To determine if composite Mosl insertions can be remobilized from genomic locations, we tried to remobilize the oxTi51 insert by injection of the transposase gene and use of selection markers to detect germline excision and repair (details in Supplementary Note). We were unable to detect remobilization from 48 injections.
Thus, in agreement with experiments in flies18-20,28: (1) composite Mosl elements were able to transpose at high efficiency from injected plasmids and did not require most internal Mosl sequences, (2) composite Mosl elements transposed at lower efficiency from extra-chromosomal arrays, and (3) genomic insertions were not easily remobilized.
Insertion into natural isolates and C. briggsae
We tested other genetic and antibiotic constructs as selectable markers for miniMos insertion. We generated insertions of otherwise identical constructs using unc-119(+)29, G418 (NeoR)30, puromycin (PuroR)31 and hygromycin B (HygroR)32 selection at similar frequencies (Fig. 1). The genetic marker unc-18(+) was also as efficient as unc-119(+) selection (unc-18(+) 38% N=13; unc-119(+) 34% N=32) for a different construct. We were unable to generate insertions with two temperature-sensitive selection markers lin-5 and spd-1 that are necessary in the germline. Insertions were probably not recovered because miniMos transposition was strongly temperature sensitive with insertions occurring only at low frequency at 15°C but at high frequency at 25°C (2% at 15°C N=114; 62% at 25°C N=102) (Fig. 1). Extra-chromosomal arrays are generally silenced in the germline33 and injected DNA therefore cannot rescue lin-5 and spd-1 animals at 25°C. Excision of the native Mos1 element for mosSCI transgenesis at ttTi5605 showed no temperature-dependence (15% at 15°C N=71; 13% at 20°C N=75; 15% at 25°C N=71). It may be possible to use temperature-sensitive genetic markers such as lin-5 or spd-1 by injecting into balanced strains that can be maintained at 25°C.
We tested the P0 insertion frequency into three highly diverged natural C. elegans isolates with NeoR selection: CB4856 (Hawaii), ED3040 (South Africa) and JU345 (France)34. The miniMos element was active in all strains although with variable insertion frequencies (6%, CB4856, N = 17; 68%, ED3040, N = 22; 16%, JU345, N = 19). This variation might be due to differences in genetic backgrounds or differences in susceptibility to antibiotics30. MiniMos could also be mobilized in other species. We successfully inserted a Ppie-1:GFP:histone construct into a C. briggsae strain (6%, N = 90), that was mutant for cb-unc-11935; two of five animals showed stable GFP expression in the germline. In an attempt to improve transposition efficiency in C. briggsae we generated cbr-Peft-3:Mos1 transposase and cbr-Ppie-1:Mos1 transposase constructs; however the insertion frequency did not improve with either construct (0%, cbr-Peft-3, N=137 and 5%, cbr-Ppie-1, N= 43).
Each strain contains a single miniMos insertion
To determine the insertion frequency in F1 animals and the transgene copy-number in each strain, we injected a mix of three different miniMos elements that could be distinguished based on color (red or green) and cellular localization (cytosolic or nuclear) (Table 1). We injected five P0 animals, picked 156 unc-119 rescued F1 animals to individual plates and recovered 20 independent insertions (11.5% F1 insertion frequency). This frequency is comparable to the frequency of generating semi-stable transgenic animals by simple array injection (10%)7. All 20 insertions were fluorescent and expressed only one of the fluorophores from the injection mix (Table 1). Insertions from the same injected animal were independent; we determined all seven insertion sites from animal #5 by inverse PCR and all mapped to unique positions in the genome (oxTi306 - oxTi312, Supplementary Table 1).
We also confirmed that insertion strains contain a single insertion by segregation in crosses (Supplementary Note). How can a single injection generate several independent insertions and yet each strain contains only a single insertion? We determined that this is possible because insertions were generated at relatively low frequency but occurred in the F1 generation when the population has expanded (Supplementary Fig. 2).
To facilitate identification of transposon insertion sites we added new symmetric restriction sites to the miniMos vectors and optimized the inverse PCR protocols (Supplementary Fig. 3 and Supplementary Protocols). We tested the optimized protocol in individual reactions and 96-well reactions on a collection of bright fluorescent Peft-3:tdTomato:H2B inserts, which will be useful as dominant chromosome balancers for C. elegans crosses (Supplementary Fig. 4).
12% of the inverse PCR reactions contained sequences from the injected plasmid backbone, indicating that some transpositions included two adjacent miniMos elements (‘composite transposition’, Supplementary Fig. 1). Sequencing showed that the entire backbone of the injected plasmid had inserted. Incorporating the heat-shock inducible negative peel-1 selection marker11 into the backbone of injected miniMos plasmids effectively selected against these types of complex insertions.
P element transgenesis has been used to generate loss of function mutants in Drosophila3. Although we did not directly screen for mutant phenotypes we noted that several of the Peft-3:tdTomato:H2B insertions were inserted into introns and exons of genes with obvious phenotypes: unc-13 I, unc-22 IV and him-4 X. All three insertions showed the phenotypes expected from loss of function alleles.
To test if expression of insertions was affected by neighboring promoters, we generated strains with promoters driving GFP expression in pharyngeal muscles (Pmyo-2, N = 3) and body wall muscle (Punc-54, N = 3). In this relatively small sample we were unable to detect mis-expression in other tissues (Supplementary Fig. 5).
The insertion frequency and fidelity of insertions is robust enough that miniMos transposition could be a convenient alternative to extra-chromosomal arrays in cases where the unstable and multi-copy nature of arrays is undesirable (Supplementary Note and Supplementary Fig. 6).
Mos1 can transpose with fosmids and lacO repeats
To determine the maximum cargo capacity of recombinant Mos1 elements we generated Mosl-based fosmids (Mosmids) by recombineering36. We inserted a cassette with a 1 kb recombinant Mosl element and cb-unc-119(+) into the backbone of several fosmids with GFP-tagged genes (Fig. 2). We injected five different Mosmids into unc-119 animals and obtained stable integrated lines at P0 frequencies ranging from 2% to 14% (5% ± 2%) of all constructs. The drop in insertion frequency was likely caused by two effects: larger cargo may inhibit transposition and Mosmid injections only inefficiently form extra-chromosomal arrays. Inserted Mosmids expressed GFP in the expected tissues, including the germline (Fig. 2).
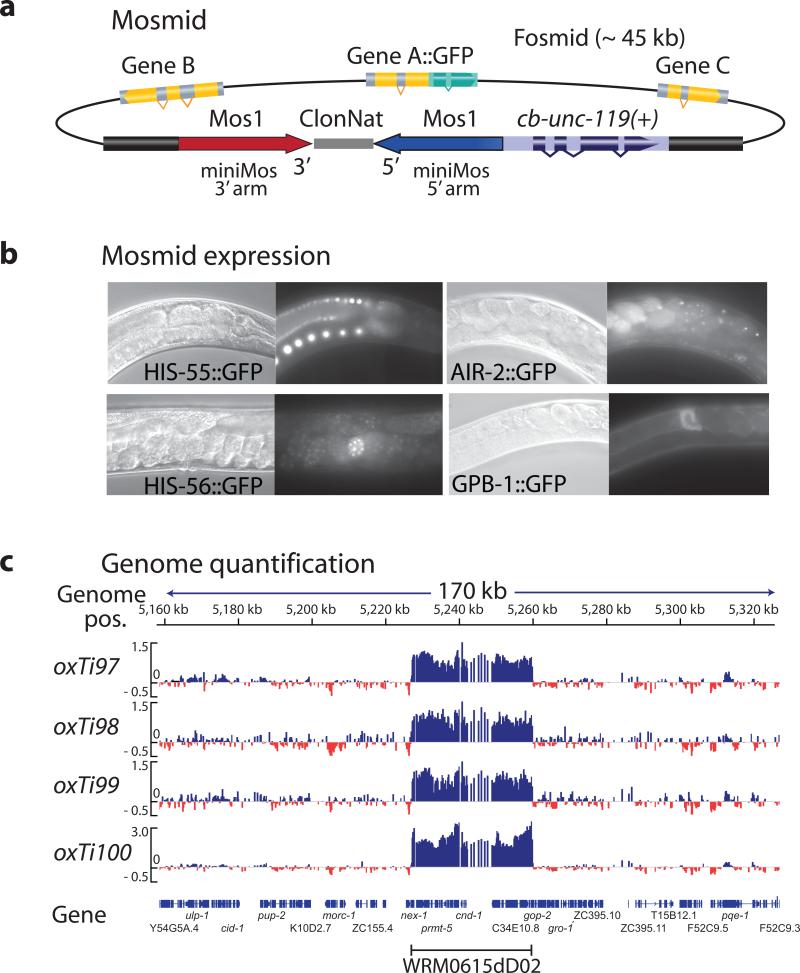
(a) Schematic of Mos1-based fosmids (Mosmids). Mos1 and cb-unc-119(+) selection recombineered into the backbone of a fosmid carrying a GFP-tagged gene. (b) Fluorescence microscopy of Mosmid insertions. Four different Mosmid insertions with GFP show expression from the tagged genes. (c) Comparative genome hybridization (CGH) of genomic DNA from four independent insertions of the Mosmid WRM0615dD02 containing tagged cnd-1. CGH is based on dense oligo arrays tiled against a genome of interest and labeling of sample DNA and control DNA with different fluorophores. Genomic regions that differ between sample and control will show a difference in the ratio between the two color intensities. The Mosmid with cnd-1:eGFP contained an error rendering the fusion protein non-fluorescent.
From one Mosmid (air-2:eGFP) we obtained 18 independent insertions that were all fluorescent, which suggests that Mosmid insertions were generally intact. We verified the integrity of the inserted fosmids by comparative genome hybridization; using this method, deletions, insertions and even single basepair mutations, can be identified with high sensitivity37,38 (Fig. 2 and Supplementary. Fig. 7). In the four lines generated from a tagged cnd-1 gene either a single, fully intact copy or two full copies (into a single location) of the Mosmid were inserted. We observed similar full-length insertions by CGH on lines from gbp-1, his-55 and air-2 inserts (Supplementary Fig. 7).
lacO repeats can be used to visualize chromosome position by binding to a fluorescently tagged LacI repressor39. We tested if a recombinant Mos1 element could insert a large repetitive transgene containing 256x lacO repeats and selection markers. We generated 20 independent insertions (Supplementary Fig. 8). These strains showed two distinct fluorescent dots in embryos when crossed into a line expressing LacI:GFP, corresponding to the two homologous chromosomes containing the lacO repeats (P Meister, University of Bern, personal communication).
These experiments showed that the miniMos element is compatible with a wide variety of transgenic cargo and selection markers. We have generated a set of 16 standardized miniMos cloning vectors to facilitate use of the technique (Supplementary Fig. 9).
A set of universal MosSCI insertion sites
The PhiC31 recombinase has been used in flies to develop universal insertion sites that are compatible with a single targeting vector4,40. We unsuccessfully attempted to adapt the PhiC31 system for C. elegans (M.S. and C.F-J., unpublished observations). As an alternative, we developed a miniMos system that achieves the same goal. We generated a miniMos element containing the ttTi5605 MosSCI site and flanked it with two selection markers, unc-18 and either NeoR or Pmyo-2:GFP:H2B (Fig. 3). The embedded ttTi5605 Mos element within the miniMos transposon can be used as a landing site for single copy insertion using MosSCI12 and is compatible with previously published targeting vectors (pCFJ150 or pCFJ350) (Fig. 3). Furthermore, mosSCI insertions can be followed in crosses by the adjacent selection marker (NeoR or Pmyo-2:GFP:H2B). We generated a set of validated single-copy, full-length mosSCI universal insertion sites that were permissive for germline expression (Fig. 3). Additionally, we targeted the insertion of a universal landing site into the ttTi25545 Mos1 site at the center of Chr. III by mosSCI because no insertion site on Chr. III was compatible with germline expression (data not shown). All universal landing sites were validated: we could generate single copy inserts at frequencies similar to insertions into the native ttTi5605 site and a Pdpy-30:GFP:H2B transgene was expressed in the germline (Supplementary Table 1).
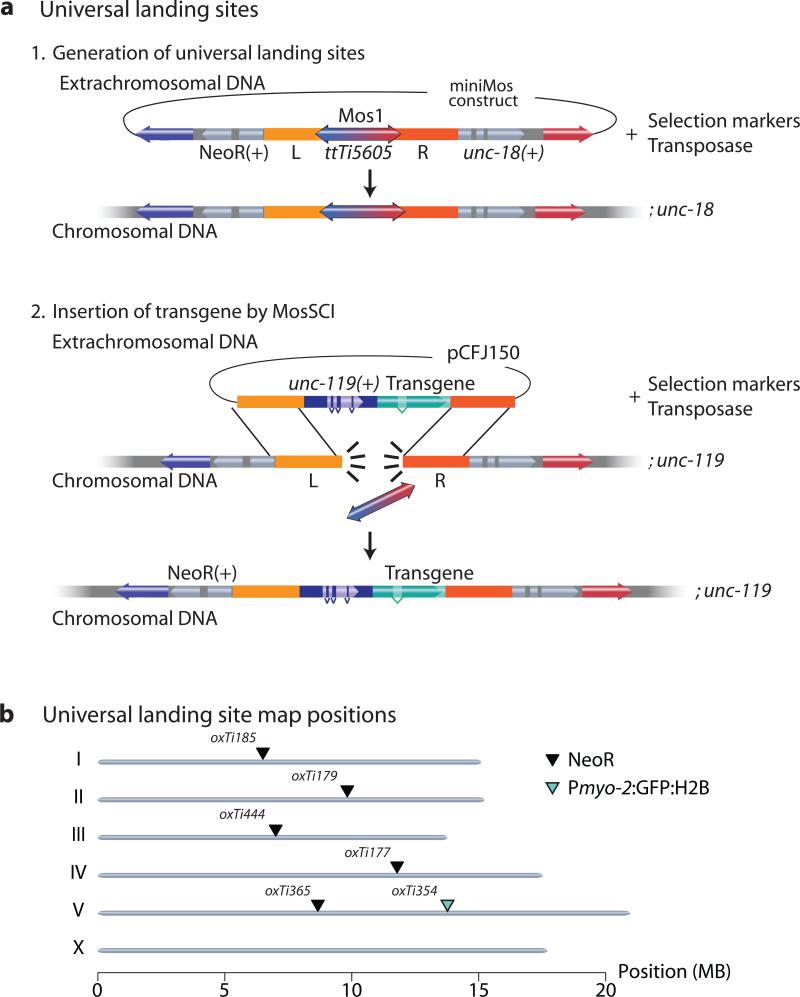
(a) Schematic of method to generate universal mosSCI insertion sites. Step 1: Insert miniMos with the ttTi5605 genomic region (including the native Mos1 element) into unc-18 mutants. Cross inserts to unc-119. Step 2: Inject pCFJ150-based targeting vector to insert transgene by mosSCI. All insertions were verified as functional, single-copy insertions. (b) Genomic location of universal mosSCI insertion sites with verified germline expression. Black arrowhead: NeoR marker. Green arrowhead: Pmyo-2:GFP:H2B marker. NeoR = Neomycin Resistance gene.
Discussion
Random insertion of transgenes with the miniMos element has several advantages relative to biolistic transformation8. First, the exact insertion site can be determined by PCR. Knowledge of the exact insertion site ensures that mutations caused by miniMos insertion, or effects on expression of the transgene by the genomic environment, can be assessed. Second, a single intact copy of the transgene with well-defined endpoints in the genome is inserted. Third, the miniMos element can insert intact fosmids41 and is active in other species and natural C. elegans isolates42. Finally, the insertion frequency of the miniMos element is high enough that several insertions are frequently generated from a single injection. Redundant inserts improve the chance of identifying insertions that do not disrupt endogenous genes and that are appropriately expressed.
We imagine miniMos transgenesis will mostly be used to insert single copies of transgenes, but there are at least four additional uses for the miniMos resources described here: (1) The set of dominant chromosome balancers is comprised of 158 inserts that express red or green fluorescent proteins in somatic nuclei spaced about every 2-5 map units (Supplementary Fig. 4). These balancers can be used to generate strains with complicated genotypes. (2) We generated two mapping strains that contain three distinguishable fluorescent markers that cover all six chromosomes in high-incidence of male (Him) mutant backgrounds. These strains are useful for mapping new mutations to chromosomes. (3) The lacO insertions mark twenty different genomic sites and can be used to locate chromosome positions in the nucleus, for example during meiosis or differentiation43. (4) We generated a set of universal mosSCI insertion sites that are compatible with a single targeting vector. These strains can be used to insert single-copy transgenes at multiple positions in the genome.
In the future, two compelling uses for miniMos will be to probe the genome on a global scale for chromatin effects and to determine expression patterns using gene trap constructs. First, the preliminary experiments with the composite Mos inserts demonstrate that transgene expression in both soma and germline of C. elegans is position dependent, with high degrees of silencing on the X-chromosome and on autosomal arms. For example, almost all of the non-fluorescent Ppie-1:GFP insertions were inserted into the X-chromosome, which is inactivated in the germline23 or into autosomal arms containing a high incidence of repressive histone marks22. Second, miniMos constructs can be used to generate enhancer-trap and gene-trap constructs. For determining the expression pattern of a single gene, it will be much more efficient to specifically target the gene with the CAS9/CRISPR system15,16,44. But to determine the expression patterns of all genes, random insertions using miniMos will be useful, as has been done in Drosophila using P elements2. The miniMos element could be combined with the Q-system45 to generate strong, inducible driver lines for most tissues. In particular, it may be possible to identify promoters or enhancers that target expression individually to many of the 302 neurons of the adult nervous system.
Online Methods
Reagents
Please see the webpage www.wormbuilder.org for annotated plasmid sequences, protocols and a searchable lists of strains. Plasmids are available from Addgene (Cambridge, MA) as a single kit (#1000000031) or as individual plasmids. Strains were maintained using standard methods46. Temperature sensitive strains lin-5 and spd-1 were grown at 15°C. All other strains were grown at room temperature on OP50 or HB101 bacteria. Fluorescent balancer strains, including the two mapping strains, have been deposited with the Caenorhabditis Genetics Center (CGC).
Molecular biology
Plasmids were designed with ApE (A plasmid Editor, M.W. Davis) which is freely available at http://www.biology.utah.edu/jorgensen/wayned/ape/.
All plasmids were generated by standard molecular techniques, including Isothermal Assembly47 and Three-fragment Gateway Cloning (Life Technologies Corporation). PCR amplification was performed using a high quality DNA polymerase Phusion (New England Biolabs).
Please see Supplementary Table 1 for Genbank-formatted plasmid sequences of all plasmids used in this study.
Reproducibility
All injections were performed at least in duplicate and usually in triplicate on different days. Only injections with DNA isolated by the same preparation method were compared. The number of injections and the sample size were selected to reach statistical significance in tests that correct for multiple comparisons. Overall, the reproducibility on different days was high. This is particular apparent in the experiment to identify the minimal Mos1 element (miniMos), where all truncated constructs larger than the miniMos transposon show reproducible insertion frequencies (Supplementary Fig. 1).
Exclusion criteria
Plates that did not contain any transgenic F1 progeny based on phenotypic rescue (unc-119 injections) or by the presence of fluorescent co-injection markers (antibiotic injections) were not counted towards the number of injected animals. This exclusion criteria excluded approximately 5-10% of all injected animals and served to reduce the variability caused by differences in injection needles between separate injections.
Blinding
No blinding was performed.
Randomization
No randomization was performed.
Recombinant Mos1 insertions
miniMos insertions
Insertions were generated and mapped as described in detail in Supplementary Protocols In brief, injection strains were maintained on HB101 bacteria at 15°C -20°C. An injection mix containing the miniMos transgene at 10-15 ng/ul, red fluorescent co-injection markers pGH8 at 10 ng/ul, pCFJ90 at 2.5 ng/ul, pCFJ104 at 10 ng/ul, a helper plasmid expressing the Mos1 transposase pCFJ601 at 50 ng/ul and the negative, heat-shock inducible peel-1 selection marker pMA122 at 10 ng/ul. The remaining volume was made up of milliQ purified water. Injected worms were placed at room temperature for 1-2 hours, transferred to individual plates and incubated at 25°C until starvation (approximately 1 week). For experiments aimed at quantifying insertion frequency, plates were screened for F1 rescue three days after injection and plates with no F1 rescue were discarded. Once starved, plates were heat-shocked for two hours at 34°C or for 1 hour at 37°C in an air incubator to kill animals with extra-chromosomal arrays. All plates were screened for miniMos insertions the day after heat-shock on a fluorescence microscope based on rescue and the absence of red co-injection markers. Because of obvious visual differences (state of animals at 25°C vs 15°C or the fluorescence of injected plasmids), the investigator was not systematically blinded to the injected constructs. A single animal from each plate containing insertions was picked for further analysis. The location of miniMos elements was determined by an inverse PCR protocol modified from Boulin and Bessereau (2007) on genomic DNA isolated with the kits “ZR Tissue and Insect DNA miniprep” or “ZR-96 Genomic DNA Tissue miniprep” (Zymo Research). The DNA was digested with restriction enzymes (New England Biolabs) for 3 hours to overnight, ligated with T4 ligase (Enzymatics) and PCR amplified twice with oligos that anneal in the miniMos transposon with Phusion DNA Polymerase. The PCR product was electrophoresed on an 1% agarose gel and single bands gel purified with the “Zymoclean Gel DNA Recovery Kit” (Zymo Research). The gel-purified product was Sanger sequenced at the University of Utah Sequencing Core.
We performed two or three independent injections for each set of conditions tested (for example, temperature or length of composite miniMos transposon) to minimize effects of a single bad injection needle. Generally, we observed very little variability between independent injections. After advice from M. Maduro, we determined that the largest source of variability was in the quality of injected DNA. We isolated DNA with Spin Miniprep (cat. no. 27106) and Plasmid Plus Midiprep (cat. no. 12943) kits from Qiagen and with a PureLink HQ Mini Plasmid kit from Invitrogen (cat. no. K2100-01). The higher quality DNA kits (Qiagen Midi and Invitrogen Mini kits) resulted in a four-fold increase in F1-rescued animals (20 vs 5 rescued animals per injection) and a 50% (Qiagen Midi) to 100% (Invitrogen mini) increase in mosSCI insertion frequency (Supplementary Fig. 6). Although we have not tested the effect of DNA purity on miniMos insertion frequency, we generally recommend using DNA of higher purity DNA for injection than what is isolated with the standard Qiagen Miniprep Kit. At the time of injections performed to quantify the insertion frequency of the miniMos transposon we were not aware of the increased frequency resulting from higher DNA quality and these injections were therefore all done with the Qiagen miniprep kit.
Quantification of insertions per injection (Table 1)
We injected a mix of three different miniMos plasmids carrying Peft-3:GFP:H2B, Peft-3:mCherry or Peft-3:tdTomato:H2B with the cb-unc-119(+) selection together with the Mos1 transposase and the negative PEEL-1 selection plasmid into unc-119 mutant animals. We picked rescued animals in the F1 generation to individual plates and allowed these plates to starve out at 25°C. We heat-shocked plates with rescued F2-F3 animals to kill animals with extra-chromosomal arrays and screened for insertions the following day. We screened each plate containing an insertion for the presence of multiple different fluorescent patterns and picked a single animal from each plate for further analysis. We isolated genomic DNA and performed inverse PCR on all seven different insertions (oxTi306-oxTi312) that originated from injection into P0 animal #5. All seven insertions mapped to different genomic locations.
Universal insertion sites
The universal insertion sites were generated by injection into unc-18(md299) animals following the protocol for miniMos insertions. The internal Mos1 element depressed miniMos insertion frequency from approximately 60% to 12% (N = 180) and resulted in a high frequency of complex insertions (56%, N=23). Strains with a putative insertion were tested for antibiotic resistance to G418 (NeoR) and ability to be homozygozed. Genomic DNA was isolated from homozygous, G418 resistant strains and tested by PCR for the presence of the ttTi5605 Mos1 element and the absence of backbone fragments from the cloning vector. Inverse PCR was performed on strains with intact universal insertion sites with oligos that specifically detect the miniMos element and not the wild-type (internal) Mos1 element. The genomic location was determined by Sanger sequencing and verified by oligos designed for each individual insertion (Supplementary Table 1). Strains with universal insertion sites were out-crossed five times against an 11x outcrossed unc-119(ed3) strain EG6207 derived from PS6038 (a kind gift from Amir Sapir and Paul Sternberg) by following Neomycin resistance. The universal insertion sites were homozygozed in the unc-119 background and verified by PCR. The ability to insert transgenes into all universal landing sites was verified by insertion of pCFJ150 derived constructs with Peft-3:GFP:H2B:tbb-2 UTR, Pdpy-30:GFP:H2B:tbb-2 UTR or Ppie-1:GFP:H2B:pie-1 UTR transgenes.
In one case, oxTi444, a universal insertion site was generated by targeted insertion of the universal landing site into a pre-existing mosSCI site ttTi25545. In this case, the miniMos element was exchanged for left and right homology regions adjacent to ttTi25545 and inserted by the standard mosSCI protocol11.
Antibiotic selection protocol
We used antibiotic selection protocols modified from Giordano-Santini et al. (2010), Semple et al. (2010) and Radman et al. (2013). For G418 selection, we made a 25 mg/ml (Gold Biotechnology) solution in water and filter-sterilized the solution with a 0.2 um filter. For puromycin selection we purchased a 10 mg/ml solution (InvivoGen) and added 0.1% Triton 100 (Sigma). For hygromycin B we made a 20 mg/ml (Gold Biotechnology) solution in water and filter-sterilized the solution with a 0.2 um filter. To use for antibiotic selections, 500 ul of the stock solutions were added directly to plates containing wild-type worms that had been injected one or two days before. Plates were allowed to dry with the lid off. Dry plates were returned to the 25°C incubator and allowed to starve. The animals were heat-shocked to remove animals with extra-chromosomal arrays and screened for insertions the next day based on survival on antibiotic plates, lack of fluorescent co-injection markers and fluorescence from the miniMos construct carrying Peft-3:GFP(NLS). At least 10 animals from each antibiotic selection were propagated and homozygosed by fluorescence to verify true insertions. We note that the antibiotic selection markers are very convenient for injecting into healthier strains, such as wild-type animals, but suffer from the disadvantage that they are harder to homozygose, especially in the absence of a fluorescent insertion marker. In our hands, G418 and hygromycin B kill almost all non-transgenic animals within two days, whereas puromycin typically takes three to four days to kill non-transgenic animals.
Composite Mosl remobilization
To determine if composite Mosl insertions can be remobilized from genomic locations, we generated a strain carrying an insertion (oxTi51, Fig. 1b) and a mutation in the unc-18 gene. A rescuing template containing unc-18(+) was constructed so that a double-strand break generated by transposon excision would be repaired by homologous recombination and copy unc-18(+) into the excision site. From 48 injected animals we did not recover any targeted unc-18(+) insertions. This result is in agreement with similar experiments in Drosophila, where the insertion frequency was intact but genome mobilization was reduced by two orders of magnitude for modified transposons of the same family as Mos18.
Bioinformatic analysis of recombinant Mosl insertions
The locations of transposons was determined by inverse PCR. Genomic location was determined by identifying the junction between the transposon and genomic DNA. A BLAST search at www.wormbase.org against genome version WS190 (ce6) was used to determine the genomic position. Generally only uniquely identified insertions were used; however some insertions that map to several position within a small genomic interval (~10 kb) were included in some figures.
Comparative Genome Hybridization
Genomic DNA from worms was isolated with the ZR Tissue & Insect DNA MiniPrep kit (Zymo Research) following the manufacturer's protocol. DNA labeling, sample hybridization, image acquisition, and determination of fluorescence were all performed as previously described37,38. We used a 3x high-density (HD) chip divided into three 720 K whole genome sections for all experiments. The chip design was based on our original 385 K whole genome chip. All microarrays were manufactured by Roche-NimbleGen with oligonucleotides synthesized at random positions on the arrays. Chip design name is 90420_Cele_RZ_CGH_HX3. Quantile normalization was performed on the intensity ratios for all experiments. Seven strains, EG7784 (oxTi97), EG7785 (oxTi98), EG7786 (oxTi99), EG7787 (oxTil00), EG6840(oxTi109), EG6731 (oxTi114) and EG6788 (oxTi118), were tested against wild-type DNA. All strains showed a duplication of the full genomic region contained within the recombineered fosmid, except for the strain EG7787 which contains a dual insertion. PCR amplification from EG7787 shows the presence of backbone DNA, which is consistent with a duplicate insertion into the same genomic locus. For all analyzed Mosmid insertions, the end-points of genomic duplications identified by CGH closely matched the ends of recombineered fosmids and no second-site duplications were detected.
Fosmid recombineering
The fosmids were engineered essentially as in41, except for the fosmid backbone modification step, where the Mos1 transposon (1000 bp) with inverted repeats (IR) were added to the cb-unc-119-Nat cassette (on each side of the NatR marker). To make the fosmid host bacteria EPI300 (Epicentre) proficient for recombineering the EPI300 cells were transformed with the pRedFlp4 plasmid, which allows for inducible expression of either the lambda Red operon+RecA or the flp Recombinase. For gene tagging, a multipurpose tagging cassette that contains a flexible linker peptide TY1, GFP, FRT-flanked positive selection (NeoR), counterselection (rpsL), and the affinity tag 3xFlag was PCR amplified. The PCR used gene specific primer extensions of 50 bp upstream and downstream of the insertion point that serve as homology arms for recombineering. Recombinants were selected for Kanamycin resistance in liquid culture. The rpsl/neo selection counter-selection marker was removed by Flp/FRT recombination. The homology arms targeting the cb-unc-119/IR NatR IR cassette to the fosmid backbone were the same for all fosmids and were included in the same plasmid (pCFJ496); this cassette was isolated by restriction digest from pCFJ496 and used for recombineering the fosmid containing a GFP-tagged gene. Both the template for the multipurpose tagging cassette and the template for inserting the Mos1 and cb-unc-119 genes were cloned in plasmids with the R6K origin of replication, which is nonfunctional in the fosmid host strain and thereby removal of the plasmid is not required prior to recombineering. The fosmid modification cassette pCFJ496 is available from Addgene (Plasmid #44488).
Mosmids generally integrate into the genome at lower frequencies than miniMos transposons that can be propagated as high-copy plasmids in bacteria. The lower insertion frequency is likely due to (1) lower transposition frequency of the miniMos element with larger cargo, (2) decreased ability of fosmids to form extra-chromosomal arrays due to reduced homology, and (3) toxic sequences present on the fosmid. Some of the Mosmids that we tested were specifically chosen because integrated lines could not be generated by biolistic transformation despite repeated attempts and appear to be toxic (M. S., unpublished data). For example, we injected 48 and 60 unc-119 animals with the his-55:eGFP and his-56:eGFP Mosmids, respectively. From these injections we did not recover a single rescued F1 animal but were able to isolate one his-55:eGFP (2%) and two his-56:eGFP (3%) rescued insertion lines in the F2 progeny. This suggests that these Mosmids are toxic at high copy number and that higher integration efficiencies may be achieved by titrating the Mosmid concentration. In support of this, we did not observe any toxicity from an air-2:eGFP mosmid and recovered 18 independent insertions from 125 injected unc-119 animals (14%).
Statistics
Statistical methods are described in the legend for each figure.
Table 1
Five unc-119 animals were injected with a mix containing three miniMos elements carrying cb-unc-119 and Peft-3:GFP:H2B, Peft-3:mCherry or Peft-3:tdTomato:H2B transgenes. Three days later, rescued F1 animals were singled. A week later, plates were heat-shocked to express PEEL-1 and kill array-bearing animals, and insertions from rescued F1s screened for the presence of single (“single fluorophore”) or multiple (“multiple fluorophores”) transgenes. All 7 insertions from strain #5 mapped to independent genomic locations.
| Injected P0 animals | #1 | #2 | #3 | #4 | #5 | Total |
|---|---|---|---|---|---|---|
| Singled F1s (rescued) | 24 | 45 | 40 | 18 | 29 | 156 |
| Insertions from rescued F1s | 5 | 5 | 1 | 1 | 6 | 18 |
| Insertions from non-rescued F1s | 0 | 1 | 0 | 0 | 1 | 2 |
| Single fluorophore | 5 | 6 | 1 | 1 | 7 | 20 |
| Multiple fluorophores | 0 | 0 | 0 | 0 | 0 | 0 |
| Peft-3:GFP::H2B | 1 | 1 | 1 | 0 | 2 | 5 |
| Peft-3:mCherry | 2 | 3 | 0 | 1 | 2 | 8 |
| Peft-3:tdTomato::H2B | 2 | 2 | 0 | 0 | 3 | 7 |
Acknowledgements
We thank B. Waterston, A. Sapir, P. Sternberg and the NemaGENETAG consortium for strains; B. Meyer and P. Meister for validating LacO insertions; J. Chin, D. Dupuy, B. Lehner and G. Seydoux labs for plasmids; M. Maduro for improving mosSCI insertion frequency; K. Hoe for expert technical assistance. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by US National Institute of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the Carlsberg Foundation (C.F.-J.), US National Institutes of Health grant 1R01GM095817 (E.M.J), US National Science Foundation grant NSF IOS-0920069 (E.M.J), and the Howard Hughes Medical Institute (E.M.J.). The Mosmid engineering work was supported by the German Max Plank (MPG) Initiative “BAC TransgeneOmics” and the US National Institute of Health ModENCODE project. Work in the laboratory of D.G.M. was supported by the Canadian Institute for Health Research. D.G.M. is a Senior Fellow of the Canadian Institute for Advanced Research.
Footnotes
Author Contributions
C.F.-J. designed experiments under the supervision of E.M.J. and M.W.D.. C.F.-J., M.S., A.P., J.T., M.L.B. and S.F. performed the research. C.F.-J. performed molecular biology, injections, imaging and genetics, M.L.B. generated mapping strains, M.S. and A.P. performed fosmid recombineering, J.T., S.F. and D.G.M. performed comparative genome hybridization. C.F.-J. and E.M.J wrote the paper with input from all co-authors.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nmeth.2889
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4126194?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
nlr-1/CNTNAP regulates dopamine circuit structure and foraging behaviors in C. elegans.
Commun Biol, 7(1):1248, 02 Oct 2024
Cited by: 0 articles | PMID: 39358459 | PMCID: PMC11447218
SoMarker: a genetic marker searching tool for Caenorhabditis elegans.
G3 (Bethesda), 14(10):jkae197, 01 Oct 2024
Cited by: 0 articles | PMID: 39152791 | PMCID: PMC11457077
A specific folate activates serotonergic neurons to control C. elegans behavior.
Nat Commun, 15(1):8471, 30 Sep 2024
Cited by: 0 articles | PMID: 39349491 | PMCID: PMC11442744
The Caenorhabditis elegans protein SOC-3 permits an alternative mode of signal transduction by the EGL-15 FGF receptor.
Dev Biol, 516:183-195, 21 Aug 2024
Cited by: 0 articles | PMID: 39173814
UNC-30/PITX coordinates neurotransmitter identity with postsynaptic GABA receptor clustering.
Development, 151(16):dev202733, 27 Aug 2024
Cited by: 1 article | PMID: 39190555 | PMCID: PMC11385328
Go to all (222) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Transposon-Assisted Genetic Engineering with Mos1-Mediated Single-Copy Insertion (MosSCI).
Methods Mol Biol, 1327:49-58, 01 Jan 2015
Cited by: 13 articles | PMID: 26423967
Engineering the Caenorhabditis elegans genome by Mos1-induced transgene-instructed gene conversion.
Methods Mol Biol, 859:189-201, 01 Jan 2012
Cited by: 5 articles | PMID: 22367873
Targeted and Random Transposon-Assisted Single-Copy Transgene Insertion in C. elegans.
Methods Mol Biol, 2468:239-256, 01 Jan 2022
Cited by: 0 articles | PMID: 35320568
Insertional mutagenesis in C. elegans using the Drosophila transposon Mos1: a method for the rapid identification of mutated genes.
Methods Mol Biol, 351:59-73, 01 Jan 2006
Cited by: 10 articles | PMID: 16988426
Review
Funding
Funders who supported this work.
CIHR
Howard Hughes Medical Institute
NIGMS NIH HHS (3)
Grant ID: R01 GM095817
Grant ID: 1R01GM095817
Grant ID: T32 GM007464
NIH HHS (1)
Grant ID: P40 OD010440





