Abstract
Objective
Cardiovascular disease is a complex disorder influenced by interactions of genetic variants with environmental factors. However, there is no information from large community-based studies examining the relationship of circulating cell-specific RNA to inflammatory proteins. In light of the associations among inflammatory biomarkers, obesity, platelet function, and cardiovascular disease, we sought to examine the relationships of C-reactive protein (CRP) and interleukin-6 (IL-6) to the expression of key inflammatory transcripts in platelets.Approach and results
We quantified circulating levels of CRP and IL-6 in 1625 participants of the Framingham Heart Study (FHS) Offspring cohort examination 8 (mean age, 66.6 ± 6.6 years; 46% men). We measured the expression of 15 relevant genes by high-throughput quantitative reverse transcriptase polymerase chain reaction from platelet-derived RNA and used multivariable regression to relate serum concentrations of CRP and IL-6 with gene expression. Levels of CRP and IL-6 were associated with 10 of the 15 platelet-derived inflammatory transcripts, ALOX5, CRP, IFIT1, IL6, PTGER2, S100A9, SELENBP1, TLR2, TLR4, and TNFRSF1B (P<0.001). Associations between platelet mRNA expression with CRP and IL-6 persisted after multivariable adjustment for potentially confounding factors. Six genes positively associated with CRP or IL-6 in the FHS sample were also upregulated in megakaryocytes in response to CRP or IL-6 exposure.Conclusions
Our data highlight the strong connection between the circulating inflammatory biomarkers CRP and IL-6 and platelet gene expression, adjusting for cardiovascular disease risk factors. Our results also suggest that body weight may directly influence these associations.Free full text

Relationship Among Circulating Inflammatory Proteins, Platelet Gene Expression, and Cardiovascular Risk
Associated Data
Abstract
Objective
Cardiovascular disease is a complex disorder influenced by interactions of genetic variants with environmental factors. However, there is no information from large community-based studies examining the relationship of circulating cell–specific RNA to inflammatory proteins. In light of the associations among inflammatory biomarkers, obesity, platelet function, and cardiovascular disease, we sought to examine the relationships of C-reactive protein (CRP) and interleukin-6 (IL-6) to the expression of key inflammatory transcripts in platelets.
Approach and Results
We quantified circulating levels of CRP and IL-6 in 1625 participants of the Framingham Heart Study (FHS) Offspring cohort examination 8 (mean age, 66.6±6.6 years; 46% men). We measured the expression of 15 relevant genes by high-throughput quantitative reverse transcriptase polymerase chain reaction from platelet-derived RNA and used multivariable regression to relate serum concentrations of CRP and IL-6 with gene expression. Levels of CRP and IL-6 were associated with 10 of the 15 platelet-derived inflammatory transcripts, ALOX5, CRP, IFIT1, IL6, PTGER2, S100A9, SELENBP1, TLR2, TLR4, and TNFRSF1B (P<0.001). Associations between platelet mRNA expression with CRP and IL-6 persisted after multivariable adjustment for potentially confounding factors. Six genes positively associated with CRP or IL-6 in the FHS sample were also upregulated in megakaryocytes in response to CRP or IL-6 exposure.
Conclusions
Our data highlight the strong connection between the circulating inflammatory biomarkers CRP and IL-6 and platelet gene expression, adjusting for cardiovascular disease risk factors. Our results also suggest that body weight may directly influence these associations.
Human genome-wide association studies have led to better understanding of the genetic basis of complex cardiovascular diseases (CVD) and risk factors.1,2 In contradistinction, few gene expression profiling studies have been reported from large community-based cohorts.3,4 Quantitative differences in the relative abundance of transcripts contribute to susceptibility to, and prognosis of, human diseases, including CVD.5,6 Transcript expression is heritable, and genetic expression analyses provide insights into the pathogenesis of disease based on the strong relationships among DNA, RNA, and proteins.5 The emerging literature suggests that levels of gene expression differ between individuals with and without CVD and its risk factors (eg, obesity, high-density lipoprotein concentrations).7–14
Inflammatory cytokines, most notably C-reactive protein (CRP), have generated considerable interest as biomarkers to risk stratify and guide therapy for individuals at risk for CVD.15–20 In addition to predicting CVD, CRP may directly facilitate atherogenesis and thrombosis.21,22 Interleukin 6 (IL-6), another inflammatory cytokine, through the IL-6 receptor triggers the production of CRP and other acute-phase reactants.22 Importantly, IL-6 production occurs not only in hepatocytes but also in adipose tissue.23 Multiple recent large studies have demonstrated that specific genetic mutations in the IL-6/IL-6 receptor pathway, leading to less IL-6 signaling, were found to lower coronary heart disease risk.24,25
Based on previous in vitro and animal work, we selected 15 platelet-derived genes whose expression may relate to circulating IL-6 or CRP.7,26–37 Our primary hypothesis was that the circulating inflammatory proteins IL-6 and CRP are associated with platelet transcripts and are modulated by obesity.
Materials and Methods
Materials and Methods are available in the online-only Supplement.38,39
Results
Clinical, demographic, and laboratory characteristics of the 1625 FHS Offspring cohort 8 participants in our analysis are shown in Table 1. We studied middle-aged to older adults (67±9 years; 46% men) with a modest prevalence of CVD risk factors.
Table 1
FHS Offspring Study Characteristics
| Variable | n=1625 |
|---|---|
| Male, n (%) | 645 (46) |
| Age, y | 67±9 |
| Body mass index, kg/m2 | 28.1±5.1 |
| Overweight, 25≤BMI<30 kg/m2, n (%) | 679 (42) |
| Obese, BMI ≥30 kg/m2, n (%) | 489 (30) |
| Diabetes mellitus, n (%) | 232 (14) |
| Total cholesterol/HDL ratio, mg/100 mL | 3.5±1.1 |
| Triglyceride, mg/100 mL | 117±6 |
| Systolic blood pressure, mm Hg | 129±17 |
| Diastolic blood pressure, mm Hg | 73±10 |
| Prevalent CHD, n (%) | 171 (11) |
| Smoker, n (%) | 129 (8) |
| Antihypertensive treatment, n (%) | 815 (50) |
| Lipid treatment, n (%) | 715 (44) |
| Aspirin within the past 3 weeks, n (%) | 743 (46) |
| Current hormone replacement therapy, n (%) | 94 (6%) |
| CRP, mg/L | 2.1±1.9 |
| ln-CRP | 0.3±0.9 |
| IL-6, pg/mL | 2.1±1.5 |
| ln-IL-6 | 0.6±0.6 |
Data are presented as means ± SD or number (percentage) for dichotomous traits. BMI indicates body mass index; CHD, coronary heart disease; CRP, C-reactive protein; FHS, Framingham Heart Study; HDL, high-density lipoprotein; and IL-6, interleukin 6.
After adjustment for potentially confounding variables, circulating levels of ln-transformed CRP and IL-6 were significantly associated with 8 of 15 platelet-derived genes (Table 2). Regression models including all covariates in adjusted models and fitted with CRP or IL-6 as the dependent variable, as well as platelet gene expression, are shown in Table I in the online-only Data Supplement. Arachidonate 5-lipoxygenase (ALOX5), PTGER2, S100A9, SELENBP1, TLR4, and TNFRSF1B expression was higher with greater mean levels of ln-transformed CRP and IL-6. In addition, higher mean levels of circulating CRP were also associated with greater CD163 and NFKB1 expression. Increased circulating IL-6 was negatively associated with CRP gene expression but positively associated with TLR2 gene expression.
Table 2
Association Between Gene Expression and CRP or IL-6 Levels
| Gene | Gene Name | Function | IL-6, pg/mL
| CRP, mg/L
| ||
|---|---|---|---|---|---|---|
| Coeff ± SE | P Value | Coeff ± SE | P Value | |||
| ALOX5 | Arachidonate 5-Lipoxygenase | Inflammation | 0.012±0.005 | 0.01* | 0.020±0.008 | 0.01* |
| CD163 | Hemoglobin scavenger receptor | Inflammation | 0.008±0.005 | 0.11 | 0.018±0.008 | 0.02* |
| CRP | C-reactive protein | Inflammation | −0.009±0.004 | 0.01* | −0.012±0.006 | 0.05 |
| ICAM1 | Intercellular adhesion molecule 1 | Adhesion | 0.009±0.005 | 0.06 | 0.010±0.008 | 0.20 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | N/A | 0.008±0.004 | 0.06 | 0.008±0.007 | 0.24 |
| IL6 | Interleukin 6 | Inflammation | −0.006±0.004 | 0.15 | −0.009±0.007 | 0.20 |
| NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 1 | Inflammation | 0.010±0.005 | 0.05 | 0.020±0.008 | 0.01* |
| NFKBIA | Nuclear factor -κB inhibitor α | Inflammation | 0.006±0.005 | 0.22 | 0.005±0.007 | 0.46 |
| PTGER2 | Prostaglandin E receptor 2 (subtype EP2) | Inflammation | 0.012±0.005 | 0.01* | 0.022±0.008 | 0.006* |
| S100A9 | S100 calcium-binding protein A9 | Inflammation | 0.020±0.005 | <0.001* | 0.029±0.008 | <0.001* |
| SELENBP1 | Selenium-binding protein 1 | N/A | 0.013±0.004 | 0.003* | 0.022±0.007 | 0.003* |
| SIRPA | Signal-regulatory protein α | Inflammation | 0.006±0.004 | 0.13 | 0.011±0.007 | 0.13 |
| TLR2 | Toll-like receptor 2 | Inflammation | 0.011±0.004 | 0.006* | 0.012±0.007 | 0.06 |
| TLR4 | Toll-like receptor 4 | Inflammation | 0.017±0.005 | <0.001* | 0.019±0.008 | 0.02* |
| TNFRSF1B | Tumor necrosis factor receptor superfamily 1B | Inflammation | 0.012±0.005 | <0.001* | 0.022±0.008 | 0.004* |
These are from models correcting for the clinical confounders listed in the Materials and Methods section using the log-transformed assay levels. Coeff indicates estimated regression coefficient.
We found evidence of an interaction between obesity (as defined by a body mass index ≥30 kg/m2) and ALOX5, NFKBIA, SELENBP1, and SIRPA gene expression for IL-6 and NFKBIA and SIRPA for CRP (Figure 1). We did not observe an interaction between gene expression and inflammatory biomarkers in overweight or normal weight individuals. In an effort to better understand the associations among obesity, inflammatory cytokines, and platelet gene expression, we related levels of several circulating biomarkers associated with either body mass index or circulating adipocytokines, including intercellular adhesion molecule 1, monocyte chemoattractant protein-1, P-selectin, and tumor necrosis factor receptor.40,41 Adjustment for circulating adipose-related factors did not substantively alter observed associations between CRP and IL-6 and platelet gene expression (Table II in the online-only Data Supplement).
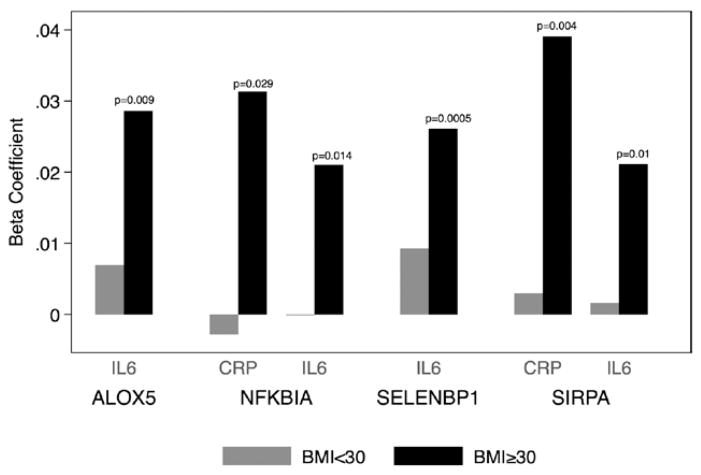
Effect of obesity on the associations between platelet gene expression and circulating C-reactive protein (CRP) or inter-leukin-6 (IL-6) levels. β coefficients are from models correcting for the clinical factors listed in the Materials and Methods section using the log-transformed assay levels and the normalized (ΔCt) RNA expression values. Results are only presented from models in which there was a statistically significant interaction with obesity status. In each case, the association between platelet gene expression was statistically greater (all P<0.05) in subjects with body mass index (BMI) ≥30 kg/m2 (obese) than in subjects with BMI <30 kg/m2.
Gene expression was measured in human megakaryocytes after exposure to IL-6, CRP, or both, using concentrations reported in obese patients and individuals with CVD based on basic42–44 and clinical reports.45–47 Treatment of human megakaryocytes with 20 μg/mL of CRP resulted in a reduction in ALOX5 expression (Table 3). Exposure of these cells to 10 pg/mL of IL-6 resulted in an increase in CD163, IFIT1, IL-6, NFKB1, and TBFRSF1B expression. Combined treatment with IL-6 and CRP did not significantly alter any of the transcripts examined.
Table 3
Effects of CRP and IL-6 on Meg-01 (Human Megakaryocyte) Gene Expression
| Gene | Change Compared With No Treatment
| |||||
|---|---|---|---|---|---|---|
| 20 μg/mL CRP
| 10 pg/mL IL-6
| CRP and IL-6
| ||||
| ΔCt ± SE | Fold Change | ΔCt ± SE | Fold Change | ΔCt ± SE | Fold Change | |
| ALOX5 | 2.69±1.11* | 0.15 | 1.11±1.11 | 0.46 | −0.84±1.22 | 1.79 |
| CD163 | 1.45±0.67 | 0.37 | −2.77±0.82** | 6.82 | −1.47±0.67 | 2.78 |
| CRP | 1.19±0.87 | 0.43 | −1.60±0.97 | 3.03 | −0.77±0.79 | 1.69 |
| ICAM1 | 0.17±1.21 | 0.89 | −1.06±1.21 | 2.09 | −1.62±0.99 | 3.07 |
| IFIT1 | −0.43±1.19 | 1.35 | −3.05±1.19* | 8.32 | −1.28±0.97 | 2.43 |
| IL6 | 0.03±0.58 | 0.97 | −2.92±0.58*** | 7.59 | −0.59±0.47 | 1.51 |
| NFKB1 | −0.06±0.95 | 1.04 | −2.51±0.95** | 5.69 | −0.56±0.77 | 1.48 |
| NFKBIA | −0.86±0.89 | 1.82 | −2.11±0.89 | 4.32 | 0.56±0.89 | 0.67 |
| PTGER2 | 0.01±1.19 | 0.99 | −2.18±1.19 | 3.54 | −0.61±0.98 | 1.52 |
| S100A9 | 1.81±1.24 | 0.28 | −0.48±1.24 | 1.40 | −0.37±1.24 | 1.29 |
| SELENBP1 | 1.01±2.64 | 0.49 | 2.62±2.15 | 0.16 | −1.17±2.64 | 2.25 |
| SIRPA | 1.63±1.31 | 0.32 | −1.20±1.31 | 2.30 | −1.04±1.31 | 2.06 |
| TLR2 | 2.42±1.46 | 0.19 | 0.24±1.46 | 0.85 | −1.09±1.46 | 2.12 |
| TLR4 | −1.22±1.35 | 2.34 | −2.39±1.35 | 5.26 | −1.69±1.10 | 3.22 |
| TNFRSF1B | −2.5±1.32 | 5.69 | −3.43±1.33* | 10.82 | −0.82±1.08 | 1.77 |
CRP indicates C-reactive protein; and IL-6, interleukin 6.
To explore how megakaryocytes responded to different concentrations of IL-6 and CRP exposure, we measured platelet mRNA levels in wild-type mouse megakaryocyte cells treated with 3 different concentrations of IL-6 or CRP. As seen in human megakaryocytes, levels of NFKB1 were significantly increased in mouse megakaryocytes exposed to IL-6 (Table IIIA and IIIB in the online-only Data Supplement). The overall response of mouse megakaryocytes to CRP and IL-6 exposure was distinct from that observed in human cells. Notably, immunofluorescence staining of mouse megakaryocytes for NFκBIA, a transcript upregulated by both IL-6 and CRP in these cells, showed little to no change in NFκBIA protein levels (Figure 2 and Figure IA and IB in the online-only Data Supplement).
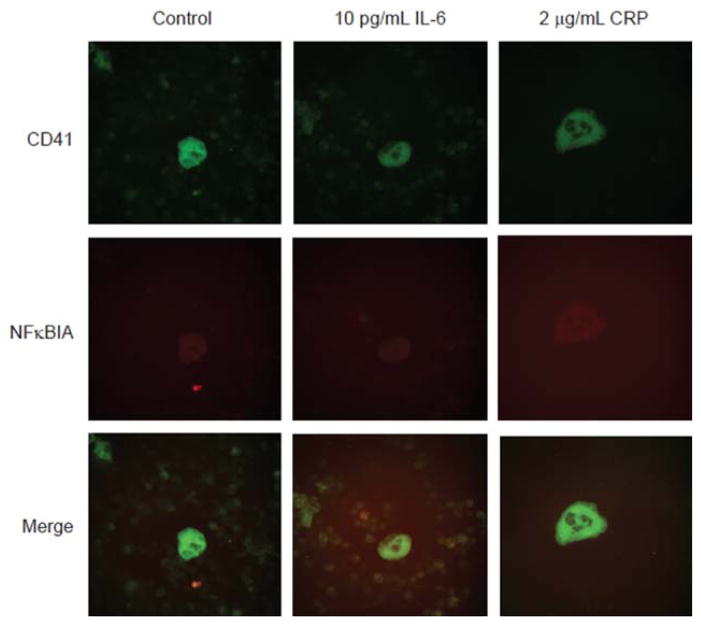
Effects of interleukin-6 (IL-6) and C-reactive protein (CRP) on NFκBIA protein levels in mouse megakaryocytes. Mouse bone marrow was treated with IL-6 or CRP for 24 hours. Permeabilized samples were stained for CD41-fluorescein isothiocyanate (a megakaryocyte marker) and NFκB1A with a Texas Red–labeled secondary antibody. Isotype controls are shown in Figure IA in the online-only Data Supplement.
Recent work has shown that interleukin-1β exposure alters protein levels in mature human platelets.48 We saw no statistically significant changes to levels of key inflammatory transcripts in human platelets treated with IL-6 and CRP versus untreated platelets (Table IVA and IVB in the online-only Data Supplement). To explore the possibility that protein levels are altered without changes to mRNA levels,48 we used Western blots and immunofluorescence to quantify levels of a representative inflammatory cytokine, TNFRSF1B, in human platelets exposed to CRP and IL-6. As seen in Figure 3A and 3B, the total protein levels of TNFRSF1B were increased in treated platelets versus referent platelets. The increase in TNFRSF1B was not caused by any contamination of immune cells because there was no signal for CD45 (Figure IC in the online-only Data Supplement), a transmembrane glycoprotein expressed by multiple immune cells. TNFRSF1B is expressed on the cell surface, and the increase in protein seen in the Western blot is reflected in immunofluorescent staining of intact platelets compared with control (Figure 3C and 3D and Figure ID in the online-only Data Supplement).
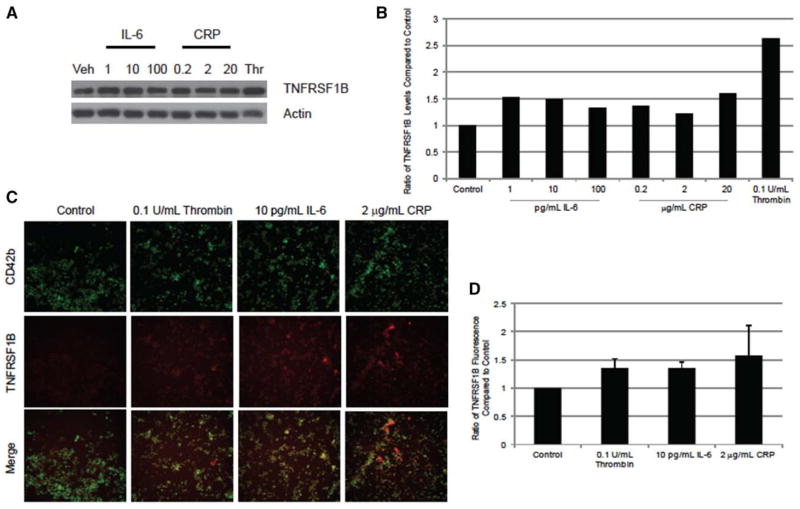
Effects of interleukin-6 (IL-6) and C-reactive protein (CRP) on TNFRSF1B protein levels in human platelets. Platelets were treated with increasing concentrations of IL-6 or CRP or 0.1 U/mL of thrombin for 1 hour. A, Protein levels of TNFRSF1B were measured by Western blot from a representative healthy donor. B, Densitometry was performed on the represented Western blot. Actin served as a loading control. Values were compared with control (resting with and without buffer). C, Confirmation of the Western blot was performed by staining isolated, intact platelets with CD42b-fluorescein isothiocyanate (a platelet marker) and TNFRSF1B-PE antibodies. Isotype controls are shown in Figure ID in the online-only Data Supplement. Representative photographs are shown. D, Fluorescence was determined using ImageJ (n=3), compared with control (with and without buffer).
Discussion
Although studies have previously shown associations between CVD and inflammatory biomarkers, the interaction with blood-derived gene expression in a large population has never been reported. In this community-based study involving 1625 FHS Offspring cohort 8 participants, we sought to examine the relationship between inflammatory cytokines CRP and IL-6 with inflammatory-related transcripts in platelets. We chose to study platelets both because of our previous data demonstrating an association with obesity and the increased relevance of specifically studying a cell that directly contributes to CVD. Of the 15 transcripts, 6 positively correlated with both CRP and IL-6 and 6 were upregulated or down-regulated significantly in megakaryocytes exposed to 1 of the 2 cytokines. Previously, all of these platelet transcripts were associated with higher mean body mass index.7 Two platelet transcripts were specific to each cytokine. TLR2 expression was positively correlated with IL-6, whereas the level of platelet-derived TLR2 was inversely related to circulating levels of CRP. The expression of CD163 and NFκB1 was positively related to CRP levels but not IL-6 levels.
Previously, cohort studies have demonstrated associations among IL-6, CRP, and CVD, including coronary heart disease and sudden death in men and women.49–52 Relevant to our platelet findings, high plasma IL-6 also has been associated with increased coronary stent thrombosis, suggesting that it may be involved in clinically relevant CVD outcomes.53 Despite these observations, little is known about the mechanisms linking circulating inflammatory markers and platelet function.
Platelets are anucleated and receive all their RNA from megakaryocytes, their precursor cells. Megakaryocytes are found primarily in the bone marrow and may respond to circulating inflammatory cytokines, thereby altering the RNA profile of platelets. As seen our in vitro work, megakaryocytes exposed to CRP and IL-6 resulted in altered expression of genes examined in our community-based investigation. We observed that 6 genes associated with circulating IL-6 and CRP levels in our FHS analyses were also significantly upregulated or downregulated in the human megakaryocytes exposed to these factors. Although treatment of mouse megakaryocytes with IL-6 and CRP did not result in statistically significant changes in all 6 of these genes (with the exception of NFKB1), we did observe a dose-dependent and directionally similar effect on levels of 4 of the 6 genes altered in human megakaryocytes. From these data, we cannot deduce whether this difference is as a result of species or some other factor.
There were, however, some inconsistencies between our observations in the platelet and megakaryocyte in vitro work and our FHS data. The discrepant findings may be because of the fact that the intensity and duration of exposure to CRP or IL-6 could not be quantified in vivo and thus may have differed from our in vitro experiment. This hypothesis is supported by the findings of our mouse experiments, showing that different levels of CRP and IL-6 elicit differential transcriptional responses. There are also inherent differences between native megakaryocytes and megakaryoblastic cell lines. As has been previously shown54 and was observed in our experiments, differences in platelet transcripts between humans and mice could have also contributed to discrepant findings. Another possibility, supported by previous work and our finding that levels of TNFRSF1B were altered in human platelets in response to CRP and IL-6, is that mature platelets, not megakaryocytes, may directly respond to circulating IL-6 and CRP. Nevertheless, our findings overall support the hypothesis that inflammatory cytokines, such as IL-6 and CRP, may alter the course of megakaryocyte development and thereby affect platelet gene expression. Levels of platelet-derived proinflammatory transcripts may directly influence platelet function or may transfer RNA to other cell types, including endothelial cells.34,55
We observed an interaction between obesity status and the associations of IL-6 and CRP with levels of 4 platelet-derived gene transcripts. One of the genes, ALOX5, encodes a lipoxygenase responsible for transforming essential fatty acids into leukotrienes, which exert proinflammatory effects, including induction of nuclear factor (NF)-κB and IL-6 secretion by adipose tissue.56–58 We observed that platelet ALOX5 expression was higher among participants with elevated IL-6 and CRP levels after adjustment for several potential confounding factors. Obesity did not seem to modulate the interaction between ALOX5 and CRP, but the association between circulating IL-6 and ALOX5 expression was influenced by obesity status. In vitro, megakaryocyte production of ALOX5 was significantly decreased in the presence of CRP at concentrations reported in obese individuals. The discrepancy may be the result of the amount of time the megakaryocytes were exposed to these inflammatory cytokines or to the concentrations used. Taken together, these data suggest that obesity plays a role in influencing ALOX5 transcript levels, perhaps via circulating CRP and IL-6.
NFKB1 encodes NF-κB, a transcription factor that is activated by intracellular and extracellular cytokines.59 Activated NF-κB stimulates the expression of genes involved in a wide variety of biological functions, including inflammation and thrombosis.60 NFKB1 expression was positively related to CRP but not IL-6 levels, and there was no interaction between NFKB1 expression and obesity status. However, expression of the gene NFKB1A, which encodes a member of a family of proteins that inhibit NF-κB, was significantly higher in obese individuals in relation to both IL-6 and CRP levels. In vitro, it was only significantly upregulated by IL-6 in megakaryocytes. NFKB1A would result in sequestration of NF-κB in an inactivated state and would block the ability of NF-κB to bind to DNA. Increased expression of this inhibitory protein in response to IL-6 and CRP in obese individuals is notable but of unclear biological significance.
SELENBP1 encodes selenium-binding protein 1. Selenium is an essential nutrient, and selenium deficiency has been associated with certain malignancies.61 Although the role of SELENBP1 and its significance to CVD remain unclear, we observed that SELENBP1 was strongly and directly related to circulating IL-6 and CRP. We also noted that obesity significantly modulated the interaction between SELENBP1 and IL-6. There was an increase in the expression of SELENBP1 in the megakaryocytes in the presence of both IL-6 and CRP, but it was not significant. Further studies are warranted to define the effects of selenium and selenium-binding protein 1 on platelet function and atherogenesis.
SIRPA encodes signal-regulatory protein-α, a protein involved in regulating monocyte adhesion, migration of monocytes across the endothelial wall, and phagocytosis.62 Signal-regulatory protein-α may serve a key role in preventing excess monocyte collection and activation of monocytes in the arterial wall during the formation of atherosclerotic plaques.62 As such, it is notable that, although SIRPA expression was not significantly associated with IL-6 or CRP levels, platelet expression of SIRPA was markedly higher in patients with obesity in response to these inflammatory biomarkers. Our in vitro experiment also showed that megakaryocytes coincubated with CRP and IL-6 or IL-6 alone expressed higher levels of SIRPA than megakaryocytes not exposed to these inflammatory cytokines; however, this increase was not significant. Although the biological significance of these findings is unclear, our findings suggest a possible interaction between level of adiposity and SIRPA expression in platelets.
Our findings raise questions including causality and directionality. Although the cell culture experiment suggests alteration in gene expression consistent with the observed associations, it remains possible that levels of circulating cytokines are attributable to platelet gene expression or other unmeasured factor with residual confounding. Furthermore, our results do not completely answer the question as to whether obesity or inflammatory changes alter organ-based protein release and whether these inflammatory modulators alter gene expression in megakaryocytes and platelets (Figure 4). Although the studies are small, there are clinical observations that support this paradigm. Weight loss in 34 individuals led to decreased inflammatory cytokine gene expression including IL-6,51 and another had similar findings with severe calorie restriction.52
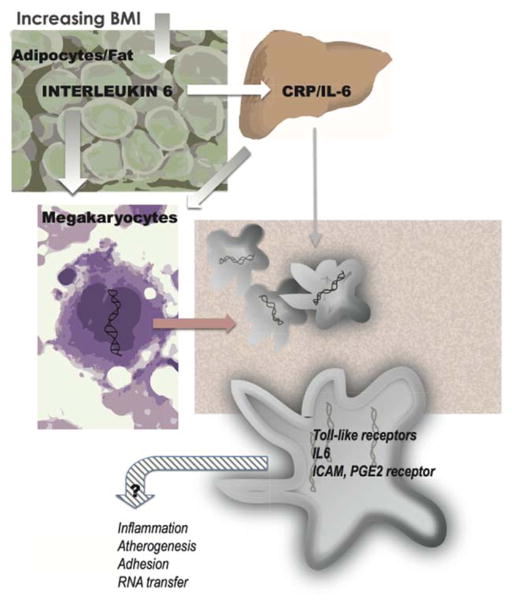
Relationship among circulating interleukin 6 (IL-6), C-reactive protein (CRP), and gene expression impacted by obesity and mechanisms of cardiovascular risk. BMI indicates body mass index; ICAM, intracellular adhesion molecule; IL-6, interleukin 6; and PGE2, prostaglandin E2.
Although there have been other studies examining transcripts in blood, this study is unique in several respects. It reports individual genes from isolated cells using the more robust quantitative platform of quantitative reverse transcriptase polymerase chain reaction and, unlike microarray, does not use an unbiased approach. Although we have proposed some mechanisms for the observed effects, the cross-sectional nature of this analysis impairs our ability to discern the directionality of the association between circulating biomarkers and gene transcripts. In addition, the present study was conducted in middle-aged to elderly individuals of European descent; the generalizability to younger individuals or those of other ethnicity/race is unknown. In addition, our data will need to be replicated in an external cohort. Another limitation of this study is that platelet function testing, although performed during a prior FHS Offspring examination, was not performed during the eighth visit, thus preventing us from relating thrombotic markers, circulating inflammatory biomarker levels, and platelet gene expression.
Acknowledgments
Sources of Funding
This work was supported by N01-HC 25195, RFA-HL-12-008 (to J.E. Freedman, E. Mick), RO1 HL087201A (to J.E. Freedman, K. Tanriverdi), RFA-HL-12-008 (to J.E. Freedman), and 1RO1 HL64753, R01 HL076784, and 1 R01 AG028321 (to E.J. Benjamin) from the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH). Partial salary support is additionally provided by NIH grants 1U01HL105268-01 (to D.D. McManus) and KL2RR031981 (to D.D. McManus). L.M. Beaulieu is supported by P01 AI078894 from the National Institute of Allergy and Infectious Disease.
Nonstandard Abbreviations and Acronyms
| CRP | C-reactive protein |
| CVD | cardiovascular disease |
| FHS | Framingham Heart Study |
| IL-6 | interleukin-6 |
Footnotes
The online-only Data Supplement is available with this article at http://atvb.ahajournals.org/lookup/suppl/10.1161/ATVBAHA.112.301112/-/DC1.
Disclosures
None.
References
Full text links
Read article at publisher's site: https://doi.org/10.1161/atvbaha.112.301112
Read article for free, from open access legal sources, via Unpaywall:
https://www.ahajournals.org/doi/pdf/10.1161/ATVBAHA.112.301112
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1161/atvbaha.112.301112
Article citations
Mean platelet volume might be an effective indicator of poor semen quality in varicocele patients.
Int Urol Nephrol, 56(11):3437-3446, 24 May 2024
Cited by: 0 articles | PMID: 38789870 | PMCID: PMC11464651
Platelet and Monocyte Activation After Transcatheter Aortic Valve Replacement (POTENT-TAVR): A Mechanistic Randomized Trial of Ticagrelor Versus Clopidogrel.
Struct Heart, 7(4):100182, 28 Apr 2023
Cited by: 0 articles | PMID: 37520136 | PMCID: PMC10382989
Platelet response to influenza vaccination reflects effects of aging.
Aging Cell, 22(2):e13749, 19 Jan 2023
Cited by: 3 articles | PMID: 36656789 | PMCID: PMC9924941
Single-cell analysis of platelets from patients with periodontitis and diabetes.
Res Pract Thromb Haemost, 7(2):100099, 01 Feb 2023
Cited by: 4 articles | PMID: 37063767 | PMCID: PMC10099319
Identification of potential biomarkers and immune cell infiltration in acute myocardial infarction (AMI) using bioinformatics strategy.
Bioengineered, 12(1):2890-2905, 01 Dec 2021
Cited by: 7 articles | PMID: 34227921 | PMCID: PMC8806781
Go to all (35) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sex differences in platelet toll-like receptors and their association with cardiovascular risk factors.
Arterioscler Thromb Vasc Biol, 35(4):1030-1037, 05 Feb 2015
Cited by: 70 articles | PMID: 25657311 | PMCID: PMC4376646
Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study.
Circulation, 122(2):119-129, 06 Jul 2010
Cited by: 85 articles | PMID: 20606121 | PMCID: PMC2910759
Clinical correlates of change in inflammatory biomarkers: The Framingham Heart Study.
Atherosclerosis, 228(1):217-223, 04 Feb 2013
Cited by: 36 articles | PMID: 23489346 | PMCID: PMC3650714
Prognostic role of C-reactive protein and interleukin-6 in dialysis patients: a systematic review and meta-analysis.
J Nephrol, 26(2):243-253, 08 Jun 2012
Cited by: 48 articles | PMID: 22684644
Review
Funding
Funders who supported this work.
NCATS NIH HHS (2)
Grant ID: KL2 TR000160
Grant ID: UL1 TR000161
NCRR NIH HHS (2)
Grant ID: KL2RR031981
Grant ID: KL2 RR031981
NHLBI NIH HHS (13)
Grant ID: 1U01HL105268-01
Grant ID: R01 HL098407
Grant ID: 1R01 HL64753
Grant ID: R01 HL076784
Grant ID: RFA-HL-12-008
Grant ID: N01 HC025195
Grant ID: N01-HC 25195
Grant ID: R01 HL087201A
Grant ID: R01 HL092122
Grant ID: N01HC25195
Grant ID: R01 HL064753
Grant ID: R01 HL087201
Grant ID: U01 HL105268
NIA NIH HHS (2)
Grant ID: 1 R01 AG028321
Grant ID: R01 AG028321
NIAID NIH HHS (1)
Grant ID: P01 AI078894





