Abstract
Free full text

Mechanism of Human Antibody-Mediated Neutralization of Marburg Virus
Associated Data
Summary
The mechanisms by which neutralizing antibodies inhibit Marburg virus (MARV) are not known. We isolated a panel of neutralizing antibodies from a human MARV survivor that bind to MARV glycoprotein (GP) and compete for binding to a single major antigenic site. Remarkably, several of the antibodies also bind to Ebola virus (EBOV) GP. Single-particle EM structures of Antibody-GP complexes reveals that all of the neutralizing antibodies bind to MARV GP at or near the predicted region of the receptor-binding site. The presence of the glycan cap or mucin-like domain blocks binding of neutralizing antibodies to EBOV GP but not to MARV GP. The data suggest that MARV neutralizing antibodies inhibit virus by binding to infectious virions at the exposed MARV receptor-binding site, revealing a mechanism of filovirus inhibition.
Introduction
Marburg virus (MARV) and Ebola virus (EBOV) are members of the family Filoviridae, which infect humans and non-human primates causing a hemorrhagic fever with mortality rates up to 90% (Brauburger et al., 2012). There have been a dozen outbreaks of Marburg virus infection in humans reported to date, including the most recent report from Uganda of a 30-year old male health worker who died in September, 2014 (WHO, 2014a). As of January 7, 2015, there have been in excess of 20,000 confirmed, probable, and suspected cases of Ebola virus disease (EVD) in the current EBOV outbreak in nine affected countries (Guinea, Liberia, Mali, Nigeria, Senegal, Sierra Leone, Spain, the United Kingdom and the United States of America) with more than 8,000 deaths (WHO, 2014b).
There is no licensed treatment or vaccine for filovirus infection. Recently, several studies showed that filovirus glycoprotein (GP)-specific neutralizing antibodies (nAbs) can reduce mortality following experimental inoculation of animals with a lethal dose of EBOV (Dye et al., 2012; Marzi et al., 2012; Olinger et al., 2012; Qiu et al., 2012; Pettitt et al., 2013; Qiu et al., 2014) or MARV (Dye et al., 2012). The primary target of these nAbs, the filovirus surface GP, is a trimer composed of three heavily glycosylated GP1-GP2 heterodimers (Figure S1). The GP1 subunit can be divided further into base, head, glycan cap and mucin-like domains (Lee et al., 2008). During viral entry, the mucin-like domain and glycan cap mediate binding to multiple host attachment factors present on the cell membrane. After the virus enters the host cell by macropinocytosis (Nanbo et al., 2010; Saeed et al., 2010), the GP is cleaved by host proteases that remove approximately 80% of the mass of the GP1 subunit, including the mucin-like domain and glycan cap (Chandran et al., 2005; Dube et al., 2009). After cleavage of GP in the endosome, the receptor-binding sites on GP become exposed, and the GP1 head then is able to bind to its receptor, Niemann-Pick C1 (NPC1) protein (Carette et al., 2011; Chandran et al., 2005; Côté et al., 2011). Subsequent conformational changes in GP facilitate fusion between viral and endosomal membranes.
The dense clustering of glycans on the glycan cap and mucin-like domain likely shield much of the surface of EBOV GP from humoral immune surveillance, leaving only a few sites on the EBOV GP protein where nAbs could bind without interference by glycans (Cook and Lee, 2013). Most of our knowledge about humoral response against filovirus infections has come from studies of murine Abs that recognize EBOV GP. From those studies, we learned that mouse nAbs preferentially target peptides exposed in upper, heavily glycosylated domains or lower areas (the GP1 base) where rearrangements occur that drive fusion of viral and host membranes (Saphire, 2013). Abs have not been identified that target protein features of the GP1 head subdomain, where the receptor-binding site to NPC1 protein is located. Ab KZ52, the only reported human EBOV GP-specific mAb, was obtained from a phage display library that was constructed from bone marrow RNA obtained from a survivor (Maruyama et al., 1999). KZ52 binds a site at the base of the GP and neutralizes EBOV, most likely by inhibiting the conformational changes required for fusion of viral and endosomal membranes (Lee et al., 2008). Some murine Abs also have been reported to bind to the base region of Ebola virus GPs (Dias et al., 2011, Murin et al., 2014). In contrast, very little is known about the mechanisms by which Abs neutralize MARV. Two murine Abs that bound the mucin-like domain of MARV GP reduced MARV budding from infected cells in culture, but failed to neutralize virus directly (Kajihara et al., 2012). Polyclonal MARV-specific Abs were shown to protect non-human primates when administrated passively after challenge (Dye et al., 2012). The epitopes recognized by such polyclonal nAbs, and the mechanism of neutralization by which these Abs act, are unknown. In this study, we isolated a large panel of human nAbs from B cells of a human survivor of severe MARV infection and used these Abs to define the molecular basis of MARV neutralization by human Abs. The results show that MARV nAbs recognize the NPC1 receptor-binding domain of MARV GP, and in some cases also recognize conserved structural features in the equivalent receptor-binding domain on EBOV GP.
Results
Isolation of monoclonal antibodies (mAbs)
We tested plasma of a MARV survivor previously infected in Uganda for the 50% neutralization activity against the Uganda strain of MARV and found a serum neutralizing titer of 1:1,010. To generate human hybridoma cell lines secreting mAbs to MARV, we screened supernatants from EBV-transformed B cell lines derived from the survivor for binding to several recombinant forms of MARV GP or to irradiated cell lysates prepared from MARV-infected cell cultures. We fused transformed cells from B cell lines producing MARV-reactive Abs to the MARV antigens with myeloma cells and generated 51 cloned hybridomas secreting MARV-specific human mAbs. Thirty-nine of these mAbs were specific to the MARV GP, while 12 bound to infected-cell lysate but not to GP; these latter mAbs were shown in secondary screens to bind to MARV internal proteins (NP, VP35 or VP40; data not shown). Analysis of the Ab heavy- and light-chain variable domain sequences revealed that all MARV-specific mAbs were encoded by unique Ab genes.
Neutralization activity
To evaluate the inhibitory activity of the mAbs, we first performed in vitro neutralization studies using a chimeric vesicular stomatitis virus with MARV GP from Uganda strain on its surface (VSV/GP-Uganda). Eighteen of the 39 MARV GP-specific mAbs exhibited neutralization activity against VSV/GP-Uganda (Figures 1A, 1C; Figure S2, S4). Of those 18 nAbs, 9 displayed strong (IC50 < 10 μg/mL), 8 nAbs displayed moderate (IC50: 10-99 μg/mL) and one displayed weak (IC50: 100-1,000 μg/mL) neutralizing activity against VSV/GP-Uganda. We also tested the neutralization potency of all nAbs that bound to MARV GP in a plaque reduction assay using live MARV-Uganda virus. Of 18 Abs that neutralized VSV/GP-Uganda, 10 Abs exhibited neutralizing activity against MARV-Uganda (Figure 1A, 1C; Figure S3, S4). These data suggest that VSV/GP, often used to study neutralizing potency of Abs because of its BSL-2 containment level, is more susceptible to Ab-mediated neutralization than live MARV. This difference is likely explained by the significantly lower copy number of MARV GP molecules that incorporate into VSV particles compared with the large number of GP molecules on the surface of filovirus filaments (Beniac et al., 2012; Thomas et al., 1985). Comparison of MARV neutralizing and non-neutralizing antibodies at concentration up to 1.6 mg/mL revealed dose-dependent activity of those mAbs that neutralized. The neutralization activity of nAbs was not enhanced by the presence of complement (data not shown). As expected, we did not detect neutralizing activity for any of the 12 Abs specific to MARV NP, VP35 or VP40 proteins.
(A) Neutralization activity of MR77 (non-neutralizing antibody) or MR213 (neutralizing antibody) against VSV/GP-Uganda (red circles) or MARV-Uganda (black circles). Error bars represent the standard errors of the experiment performed in triplicate.
(B) Binding of representative mAbs from four distinct Binding Groups to the MARV GP (blue squares) or MARV GPΔmuc (green squares). Dotted line indicates 0.5 μg/mL threshold for categorizing Group 3 antibodies as possessing low (3A) or high (3B) EC50 values.
(C) Heatmap showing the neutralization potency of MARV GP-specific mAbs against VSV/GP-Uganda or MARV-Uganda. IC50 value for each virus-mAb combination is shown, with dark red, orange, yellow or white shading indicating high, intermediate, low or no potency. IC50 values greater than 1,000 μg/mL are indicated by >. Neutralization assays were performed in triplicate.
(D) Data from competition binding assays using mAbs from Binding Groups 2, 3A or 3B. Numbers indicate the percent binding of the competing mAb in the presence of the first mAb, compared to binding of competing mAb alone. MAbs were judged to compete for the same site if maximum binding of the competing mAb was reduced to <30% of its un-competed binding (black boxes with white numbers). MAbs were considered non-competing if maximum binding of the competing mAb was >70% of its un-competed binding (white boxes with red numbers). Grey boxes with black numbers indicate an intermediate phenotype (between 30 and 70% of un-competed binding). See also Figures S1 to S5.
Recognition of varying forms of GP
To characterize the binding of isolated Abs to recombinant MARV GPs, we performed binding assays using either a recombinant MARV GP ectodomain containing the mucin-like domain (MARV GP) or a recombinant GP lacking residues 257-425 of the mucin-like domain (MARV GPΔmuc). Based on OD405 values at the highest Ab concentration tested (Emax) and 50% effective concentration (EC50), we divided the MARV-GP-specific Abs into four major groups, based on binding phenotype (designated Binding Groups 1, 2, 3A and 3B; Figure 1B and Figure S5). Binding Group 1 mAbs had an Emax to GP < 2 [i.e., these mAbs never exhibited a maximal binding level to MARV GP]; Binding Group 2 mAbs had an Emax to GP >2, with EC50 for GP < EC50 for GPΔmuc [i.e., these mAbs bound to the mucin-like domain or glycan cap]; Binding Group 3 had an Emax to GP > 2, with EC50 for GP ≈ EC50 for GPΔmuc [i.e., these mAbs bound equally well to full-length and mucin-deleted forms of GP], with the Group 3A mAbs having an EC50 for GP < 0.5 μg/mL and the Group 3B mAbs having an EC50 for GP > 0.5 μg/mL [suggesting that, as a class, the Group 3B mAbs possess a lower steady state KD of binding to GP than Group 3A mAbs].
Abs that lacked neutralization activity against VSV/GP-Uganda or MARV-Uganda fell principally into Binding Groups 1, 2, 3A. Interestingly, all VSV/GP-Uganda nAbs displayed a unique binding pattern and segregated into Binding Group 3B (Figure 1C). It was interesting that while both mAbs from Groups 3A and 3B bound equally well to the full-length MARV GP and to the GPΔmuc, EC50 values for nAbs from Binding Group 3B were higher than those for non-neutralizing Abs from Group 3A.
Competition-binding studies
To determine whether mAbs from distinct binding groups targeted different antigenic regions on the MARV GP surface, we performed a competition-binding assay using a real-time biosensor. We tested 18 MARV nAbs from Binding Group 3B, four Abs from Binding Group 3A and one Ab from Binding Group 2 in a tandem blocking assay in which biotinylated GPΔmuc was attached to a streptavidin biosensor. Abs from Group 1 and the two non-neutralizing Abs from Binding Group 3B did not bind to biotinylated GPΔmuc in the competition assay and were excluded from the analysis. While non-neutralizing Abs from Binding Groups 2 and 3A did not prevent binding of the Binding Group 3B nAbs to GPΔmuc, all nAbs blocked binding of each of the other nAbs to the antigen and segregated into a single competition-binding group (Figure 1D). These data suggested that all of the nAbs target a single major antigenic region on the MARV GP surface.
Electron microscopy studies of antigen-antibody complexes
To determine the location of the antigenic region targeted by MARV nAbs, we performed negative stain single-particle electron microscopy (EM) studies using complexes of GPΔmuc with Fab fragments of seven nAbs from Binding Group 3B. The EM reconstructions clearly showed that Fab fragments for all seven nAbs bind at the top of the GP in or near the NPC1 protein receptor-binding site (Figure 2A,B). The binding pattern of these Abs could be divided further into two major groups based on their relative angle of approach to the GP head domain. MAbs MR72, MR78, MR201 and MR82 bound toward the top and side of GP1 at a shallow angle relative to the central three-fold axis, while mAbs MR191, MR111 and MR198 bound at a steeper angle toward the top of GP1 (Figure 2C, 2D). When we compared IC50 values for nAbs that bound in the two binding poses, we did not detect a significant difference in neutralization potency based on the angle of approach (Figure 1C).
(A) Representative reference-free 2D class averages of the MARV GPΔMuc:MR Fab complexes.
(B) EM reconstructions of seven Fab fragments of neutralizing antibodies bound to MARV GPΔmuc (side views). All seven antibodies target a similar epitope on the top of GP.
(C) These antibodies can be subdivided based on their angles of approach: i) those that bind toward the top and side of GP1 at a shallow angle relative to the central three-fold axis (MR72 in red, MR78 in orange, MR201 in yellow or MR82 in green) and ii) those that bind at a steeper angle toward the top of GP1 (MR191 in cyan, MR111 in blue or MR198 in purple).
(D) The crystal structure of EBOV GPΔmuc (GP1 in white and GP2 in dark grey) is modeled into the MARV GP density (mesh) and the angles of approach of the neutralizing antibodies are indicated with arrows, colored as in (B). The footprint of the antibodies is indicated by a black circle targeting residues in the putative receptor-binding site (RBS) through a variety of approach angles.
Antibody neutralization escape mutant viruses
As an additional strategy to determine residues on MARV GP involved in binding to nAbs, we generated VSV/GP-Uganda variant viruses that escaped neutralization, and then we determined the sequence of the GP of those mAb escape viruses. Vero E6 cells were inoculated with VSV/GP-Uganda in the presence of MR72 or MR78 nAbs. Two escape mutant viruses were isolated: virus variant VSV/GP-72.5 contained three missense mutations in the MARV GP gene (N129S in the putative NPC1 receptor-binding site, S220P in the glycan cap and P455L in the mucin-like domain) and virus variant VSV/GP-78.1 possessed missense mutation C226Y in the glycan cap (Figure 3A). Consistent with the EM data, six out of seven nAbs tested displayed a higher level of neutralization activity against the wild-type VSV/GP-Uganda than to the VSV/GP-72.5 or VSV/GP-78.1 escape mutant viruses, suggesting these nAbs recognize MARV GP in a similar fashion (Figure 3B). MAb MR198 exhibited equal neutralization potency against wild-type VSV/GP-Uganda or the two escape mutant viruses (Figure 3B). As all nAbs segregated into one competition group (Figure 1D), bound the MARV GP at the NPC1 receptor-binding site (Figure 2A-D) and displayed a similar profile of neutralization of escape mutant viruses (Figure 3B), we propose that blocking of MARV GP binding to NPC1 is the principal mechanism of MARV neutralization by these naturally-occurring human Abs. This model is supported by the data in the accompanying paper (Hashiguchi T. et al., 2015) showing that MR78 inhibits binding of NPC1 domain C to MARV GP.
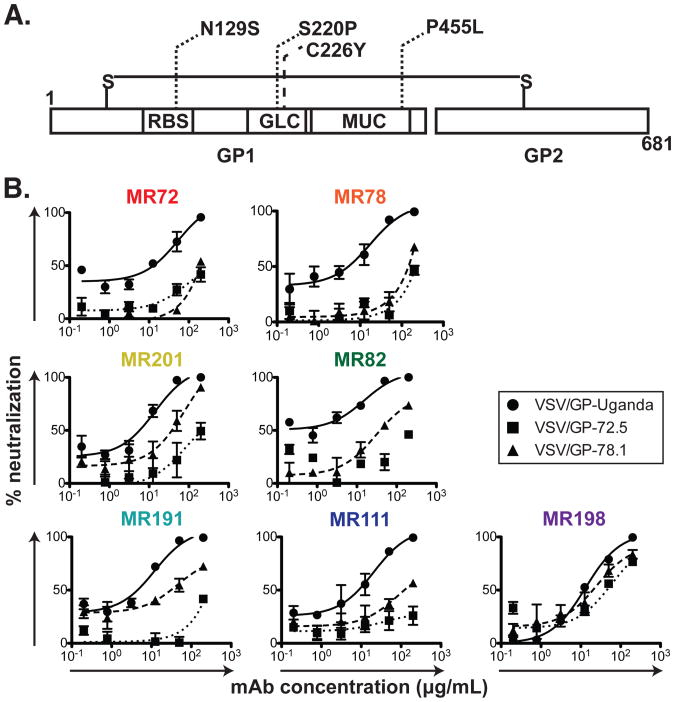
(A) VSV-MARV-72.5 (dotted lines) or VSV-MARV-78.1 (dashed line) escape mutations mapped onto the domain schematic of MARV GP. RBS = Receptor binding site; GLC = glycan cap; MUC = mucin-like domain.
(B) Neutralization activity of antibodies from Binding Group 3B against wild-type VSV/GP-Uganda (circles, straight curves), VSV/GP-72.5 (squares, dotted curves) or VSV/GP-78.1 (triangles, dashed curves) escape mutant viruses.
Cross-reactive binding of MARV antibodies with EBOV GP
It is surprising that human MARV nAbs recognize the putative NPC1 protein receptor-binding site on GP, since previous studies suggested that the NPC1 protein receptor-binding site on EBOV GP may be obscured from Ab binding by the presence of the highly glycosylated glycan cap and mucin-like domain (Lee et al., 2008). To determine whether the MARV nAbs we isolated also could bind in a cross-reactive manner to the EBOV GP receptor-binding site, we performed ELISA binding assays using three recombinant forms of MARV and EBOV GPs: full-length GP ectodomain containing the glycan cap and mucin-like domain (designated MARV or EBOV GP), ectodomains lacking residues 257-425 (MARV) or 314-462 (EBOV) of the mucin-like domain (designated MARV or EBOV GPΔmuc) and cleaved GP ectodomains enzymatically treated to remove the mucin-like domain and glycan cap (designated MARV or EBOV GPcl). Three of the MARV nAbs, designated MR78, MR111 and MR191, recognized the EBOV GPcl that lacked the glycan cap and mucin-like domain (Figure 4A). Remarkably, the MARV nAb MR72 bound all three forms of both EBOV and MARV GPs with similar EC50 and Emax values, indicating that its epitope, and the EBOV receptor-binding site which it likely overlaps, might be partially accessible for Ab binding even in the full-length form (Figure 4A). We tested the breadth of neutralization of MARV nAbs for filoviruses using a panel of different MARV and EBOV isolates. While multiple MARV Abs displayed neutralizing activity towards different MARV strains, MARV nAbs did not exhibit detectable neutralization activity against EBOV or VSV/EBOV (Figure 4B). Structural analysis of MARV and EBOV GP in the accompanying paper (Hashiguchi T. et al., 2015) reveals that the glycan cap and mucin-like domain likely obscure the receptor-binding domain in EBOV but not MARV.
(A) A heat map showing the binding in ELISA of neutralizing mAbs from Binding Group 3B to the MARV and EBOV GPs. EC50 value for each antigen-mAb combination is shown, with dark red shading indicating lower EC50 values and orange or yellow shading indicating intermediate or higher EC50 values. EC50 values greater than 1,000 μg/mL are indicated by >.
(B) A heatmap showing the neutralization breadth of mAbs from Binding Group 3B. The IC50 value for each virus-mAb combination is shown, with dark red shading indicating increased potency and orange or yellow shading indicating intermediate or low potency. IC50 values greater than 1,000 μg/mL are indicated by >. Neutralization assays were performed in triplicate.
In vivo testing
We tested the in vivo protective activity of the mAbs in a murine model using mouse-adapted MARV strain Ci67 (Warfield et al., 2007; Warfield et al., 2009). Inoculation of mice with MARV Ci67 causes clinical disease, and in a proportion of animals causes lethal disease, although typically less than 100% lethality in mice (Warren et al., 2014). We selected four of the mAbs among those with the lowest in vitro neutralization IC50 values: MR72, MR82, MR213, and MR232. The IC50 values in neutralization assays with MARV Uganda or mouse-adapted MARV strain Ci67 were comparable (within two-fold). Seven week-old BALB/c mice were injected with 100 μg of antibody by the IM route and challenged with 1,000 PFU of Ci67. Twenty four hours later, antibody treatment was repeated. By day 6, all five control (untreated) mice developed progressive loss of weight and symptoms of the disease, including dyspnea, recumbency and unresponsiveness, and on days 8 and 9, two animals were found dead and one animal was found moribund and euthanized. The remaining two animals demonstrated recovery by day 11. In contrast, all animals treated with any antibody survived and did not display the elevation of the disease score, with the exception of two animals treated with MR72, which showed a transient marginal loss of weight and increase of the disease score on days 6-9, which did not exceed 1 (Figure 5). The observed level of protection was remarkable given the relatively modest in vitro neutralizing potency of the antibodies.
Groups of mice at 5 animals per group were injected with individual mAbs by the intraperitoneal route twice: 1 h prior and 24 h after MARV challenge at 100 μg per treatment. Untreated animals served as controls.
(A) Kaplan-Meier survival curves.
(B) Body weight
(C) Illness score
Discussion
There is an obvious urgent need for prophylactic and therapeutic interventions for filovirus infections given the recurrence of MARV outbreaks including in October 2014 in Uganda and a massive outbreak of EBOV infections in West Africa in 2014. There is very little information about the structural determinants of neutralization on which to base the rational selection of antibodies, and for MARV there have been no reported human nAbs.
This study reveals that naturally occurring human MARV nAbs isolated from the B cells of a recovered donor principally target the MARV NPC1 protein receptor-binding site, suggesting that a major mechanism of MARV neutralization could be inhibition of binding to receptor. Remarkably, some of the isolated antibodies also bound to the EBOV GP. This mechanism of MARV neutralization was unexpected, because previous studies with EBOV showed that the putative receptor-binding domain on GP is obscured on the surface of virions by the presence of the glycan cap and mucin-like domain, only becoming exposed following cleavage by cathepsin in the endosome. These studies suggest that the configuration of the MARV GP differs significantly from that of EBOV GP because the receptor-binding domain must be accessible for immune recognition on MARV GP. Indeed, determination of the structure of the MARV GP and structural analysis of the interaction of mAb MR78 with MARV and EBOV GP molecules shows this to be the case (see accompanying paper, Hashiguchi et al., 2015).
The information obtained from these studies can be used to inform development of new therapeutics and structure-based vaccine designs against filoviruses. Furthermore, as these nAbs are fully human and exhibit inhibitory activity, they might be useful as a component of a prophylactic or therapeutic approach for filovirus infection and disease. The challenge studies using a murine model here show clear evidence of in vivo activity and suggest additional preclinical studies in other species such as guinea pigs and macaques are warranted. Their ability to bind a broad range of MARV isolates indicates they may offer detection of or efficacy against new viral strains yet to emerge. Although some of these mAbs bind to certain forms of EBOV GP, these antibodies are not likely to be effective against natural Ebola infection because the EBOV receptor-binding site is obscured on the viral surface. However, such mAbs might neutralize EBOV if they could be delivered to the endosome where the EBOV receptor-binding site is exposed following GP cleavage.
Experimental Procedures
Donor
The donor was an otherwise healthy adult woman who contracted Marburg virus (MARV) infection in 2008 following exposure to fruit bats in the Python Cave in Queen Elizabeth National Park, Uganda. The donor's clinical course was documented previously (CDC, 2009). Peripheral blood from the donor was obtained in 2012, four years after the illness, following informed consent. The study was approved by the Vanderbilt University Institutional Review Board.
Viruses
MARV strain 200702854 Uganda (MARV-Uganda) was isolated originally from a subject designated “Patient A” during the outbreak in Uganda in 2007 (CDC, 2009; Towner et al., 2009) and underwent 4 passages in Vero E6 cells. MARV strain Musoke (MARV-Musoke) was isolated during the outbreak in Kenya in 1980 (Smith et al., 1982) and passaged 5 times in Vero E6 cells. MARV strain 200501379 Angola (MARV-Angola) was isolated during the outbreak in Angola in 2005 (Towner et al., 2006) and passaged 3 times in Vero E6 cells. MARV Ravn virus (Ravn) was isolated from a patient in 1987 in Kenya (Johnson et al., 1996) and passaged 4 times in Vero E6 cells. All strains of MARV were obtained originally from the Special Pathogens Branch, U.S. Centers for Disease Control (CDC) and deposited at the World Reference Center of Emerging Viruses and Arboviruses (WRCEVA) housed at UTMB. The recombinant Ebola Zaire strain Mayinga (EBOV) expressing eGFP was generated in our laboratory by reverse genetics (Lubaki et al., 2013; Towner et al., 2005) from plasmids provided by the Special Pathogens Branch at CDC and passaged 3 times in Vero E6 cells. For analysis of antibody binding by ELISA, viruses were gamma-irradiated with the dose of 5 × 106 rad. The recombinant VSV in which the VSV GP protein was replaced with that of MARV strain Musoke (VSV/GP-Musoke) or EBOV strain Mayinga (Garbutt et al., 2004) were provided by Dr. Thomas Geisbert (UTMB) and Dr. Heinz Feldmann (NIH), respectively; a similar virus with GP from MARV (strain 200702854 Uganda) was constructed as described below. All work with EBOV and MARV was performed within the Galveston National Laboratory BSL-4 laboratories.
We used a mouse-adapted strain of MARV for testing the effect of mAbs in vivo. The mouse-adapted Ci67 strain of Marburg virus (Warfield et al., 2007) was provided by Dr. Sina Bavari (U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Maryland) and amplified by a single passage in Vero-E6 cells.
Generation of a chimeric strain of VSV in which VSV G protein was replaced with the GP protein of MARV strain Uganda (VSV/GP-Uganda)
The plasmid pVSV-XN2 carrying cDNA of the full-length VSV anti-genome sequence and the support plasmids pBS-N, pBS-L and pBS-P encoding the internal VSV proteins under control of the T7 promoter were kindly provided by Dr. John Rose (Yale University). The plasmid pC-T7, encoding the T7 polymerase, was kindly provided by Dr. Yoshihiro Kawaoka (University of Wisconsin). For generation of the VSV/GP-Uganda construct, Vero E6 cell monolayers were inoculated with MARV strain 200702854 and total cellular RNA was isolated and reverse-transcribed. MARV GP ORF was PCR-amplified from cDNA using forward primer 5′-CATGTACGACGCGTCAACATGAGGACTA-3′ and reverse primer 5′-TCTAGCAGCTCGAGCTATCCAATATATTTAGTAAAGATACGACAA-3′ (underlined are MluI and XhoI endonuclease sites, respectively; italicized are the start and the end of MARV GP ORF – direct and complementary sequences, respectively). To replace VSV G with MARV GP, the resulting PCR-product was cloned into pVSV-XN2 using the unique MluI and XhoI endonuclease sites located between the VSV G gene-start and gene-end signals and flanking its ORF, resulting in the plasmid pVSV/GP-Uganda. To recover the recombinant virus, 1 × 106 BSR-T7 cells, kindly provided by Dr. Ursula Buchholz (U.S. National Institute of Allergy and Infectious Diseases), were transfected with the following plasmids: pVSV/GP-Uganda, 5 μg; pBS-N, 1.5 μg; pBS-P, 2.5 μg; pBS-L, 1 μg; pC-T7, 5 μg. After 48 hours, transfected BSR-T7 cells were collected with a cell scraper and transferred, along with the supernates, to Vero E6 cell monolayers for amplification of the recovered VSV/GP-Uganda.
Generation of human hybridomas secreting monoclonal antibodies (mAbs)
Peripheral blood mononuclear cells (PBMCs) from the donor were isolated with Ficoll-Histopaque by density gradient centrifugation. The cells were cryopreserved immediately and stored in the vapor phase of liquid nitrogen until use. Previously cryopreserved samples were thawed, and 10 million PBMCs were plated into 384-well plates (Nunc #164688) using: 17 mL of cell culture medium (ClonaCell-HY Medium A, Stemcell Technologies #03801), 8 μg/mL of the TLR agonist CpG (phosphorothioate-modified oligodeoxynucleotide ZOEZOEZZZZZOEEZOEZZZT, Invitrogen), 3 μg/mL Chk2 inhibitor (Sigma #C3742), 1 μg/mL cyclosporin A (Sigma #C1832) and 4.5 mL of clarified supernate from cultures of B95.8 cells (ATCC VR-1492) containing Epstein-Barr virus (EBV). After 7 days, cells from each 384-well culture plate were expanded into four 96-well culture plates (Falcon #353072) using cell culture medium containing 8 μg/mL CpG, 3 μg/mL Chk2i and 10 million irradiated heterologous human PBMCs (Nashville Red Cross) and incubated for an additional four days. Plates were screened for MARV antigen-specific antibody-secreting cell lines using enzyme-linked immunosorbent assays (ELISAs). Cells from wells with supernates reacting in a MARV antigen ELISA were fused with HMMA2.5 myeloma cells using an established electrofusion technique (Yu et al., 2008). After fusion, hybridomas were resuspended in medium containing 100 μM hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine (HAT Media Supplement, Sigma #HO262) and 7 μg/mL ouabain (Sigma #O3125) and incubated for 18 days before screening hybridomas for antibody production by ELISA.
Human mAb and Fab production and purification
After fusion with HMMA2.5 myeloma cells, hybridomas producing MARV-specific antibodies were cloned biologically by two rounds of limiting dilution and by single-cell fluorescence-activated cell sorting. After cloning, hybridomas were expanded in post-fusion medium (ClonaCell-HY Medium E, STEMCELL Technologies #03805) until 50% confluent in 75-cm2 flasks (Corning #430641). For antibody production, cells from one 75-cm2 flask were collected with a cell scraper and expanded to four 225-cm2 flasks (Corning #431082) in serum-free medium (Hybridoma-SFM, Gibco #12045-076). After 21 days, supernates were clarified by centrifugation and sterile filtered using 0.2-μm pore size filter devices. HiTrap Protein G or HiTrap MabSelectSure columns (GE Healthcare Life Sciences #17040501 and #11003494 respectively) were used to purify antibodies from filtered supernates. Fab fragments were generated by papain digestion (Pierce Fab Preparation Kit, Thermo Scientific #44985) and purified by chromatography using a two-column system where the first column contained protein G resin (GE Healthcare Life Sciences #29048581) and the second column contained either anti-kappa or anti-lambda antibody light chain resins (GE Healthcare Life Sciences #17545811 and #17548211 respectively).
Expression and purification of MARV and EBOV GPs
Angola strain MARV GP ectodomains, containing the mucin-like domain (MARV GP) or lacking residues 257-425 of the mucin-like domain (MARV GPΔmuc), were used to screen supernates of transformed B cells and human hybridomas separately. Recombinant proteins for Ravn strain cleaved GP, EBOV Mayinga strain GP, EBOV Mayinga strain GPΔmuc and EBOV Mayinga strain cleaved GP were designed and expressed similarly. Large-scale production of recombinant GP or GPΔmuc was performed by transfection of Drosophila Schneider 2 (S2) cells with modified pMTpuro vectors, followed by stable selection of transfected cells with 6 μg/mL puromycin. Secreted GP ectodomain expression was induced with 0.5 mM CuSO4 for 4 days. Proteins were engineered with a modified double strep tag at the C terminus (enterokinase cleavage site followed by a strep tag/linker/strep tag) to facilitate purification using Strep-Tactin resin (Qiagen #2-1201). Proteins were purified further by Superdex 200 size exclusion chromatography in 10 mM Tris, 150 mM NaCl, pH 7.5 (1× TBS).
Lysates of MARV-infected cells
Lysates were prepared as previously described (Ksiazek et al., 1999). Briefly, Vero E6 cell monolayers in 850 cm2 roller bottles were inoculated with approximately 106 PFU MARV or EBOV and incubated at 37 °C until partial destruction of monolayer occurred (approximately 9-10 days). Cell monolayers were detached using 3 mm glass beads, and cell suspensions were centrifuged at 16,000 × g for 10 min at 4 °C. Supernates were discarded, cell pellets were resuspended in 10× excess of borate buffer saline (10 mM Na2B4O7, 150 mM NaCl, pH 9.0), and centrifuged at 16,000 × g for 10 min at 4 °C. Supernates were discarded, cell pellets were resuspended in cold 1% Triton X-100 (Fisher Scientific) in borate buffer saline, vortexed and gamma-irradiated on dry ice at 5 × 106 rad. The lysates were sonicated with a 600 W Tekmar Sonic Disruptor TM600 (Tekmar) using a cuphorn sonicator at maximum power setting and 50% duty cycle for 10 min, centrifuged at 16,000 × g and the supernates aliquoted.
Screening ELISA
ELISA plates were coated with lysates of MARV infected cells (diluted 1:1,000 in Dulbecco phosphate buffered saline, DPBS) or recombinant MARV GP or MARV GPΔmuc proteins (20 μg in 10 mL DPBS per plate) and incubated at 4 °C overnight. Plates were blocked with 100 μL of blocking solution/well for 1 h. Blocking solution consisted of 10 g powdered milk, 10 mL of goat serum, 100 mL of 10× DPBS, and 0.5 mL of Tween-20 mixed to a 1 L final volume with distilled water. The presence of antibodies bound to the GP was determined using goat anti-human IgG horseradish peroxidase conjugated secondary antibodies (Southern Biotech #2040-05, 1:4,000 dilution) and 1-Step Ultra TMB-ELISA substrate (Thermo Scientific #34029), with optical density read at 450 nM after stopping the reaction with 1M HCl.
Half maximal effective concentration (EC50) binding analysis
MARV or EBOV GPs, MARV or EBOV GPΔmuc, or Ravn or EBOV cathepsin-cleaved GPs were coated onto 384-well plates (Thermo Scientific Nunc #265203) in DPBS at 2 μg/mL overnight, then antigen was removed and plates were blocked with blocking solution made as above. Antibodies were applied to the plates at a concentration range of 1.5 μg/mL to 270 ng/mL (Binding Groups #1, #2 and 3A) and 0.1 μg/mL to 10 ng/mL (Binding Group #3B) using three-fold serial dilutions. The presence of antibodies bound to the GP was determined using goat anti-human IgG alkaline phosphatase conjugate (Meridian Life Science #W99008A, 1:4,000 dilution) and p-nitrophenol phosphate substrate tablets (Sigma #S0942), with optical density read at 405 nM after 120 minutes. A non-linear regression analysis was performed on the resulting curves using Prism version 5 (GraphPad) to calculate EC50 values.
MARV and EBOV neutralization experiments
Dilutions of mAbs in triplicate were mixed with 150 PFU of MARV or EBOV expressing eGFP in MEM containing 10% FBS (HyClone), 50 μg/mL gentamicin (Cellgro #30-005-CR) with or without 5% guinea pig complement (MP Biomedicals #642836) in a total volume of 0.1 mL, and incubated for 1 hour at 37 °C for virus neutralization. Following neutralization, virus-antibody mixtures were placed on monolayers of Vero E6 cells in 24-well plates, incubated for 1 hour at 37 °C for virus adsorption, and overlayed with MEM containing 2% FBS and 0.8% methylcellulose (Sigma-Aldrich #M0512-1KG). After incubation for 5 days, medium was removed, cells were fixed with 10% formalin (Fisher Scientific #245-684), plates were sealed in plastic bags and incubated for 24 hours at room temperature. Sealed plates were taken out of the BSL-4 laboratory according to approved SOPs, and monolayers were washed three times with phosphate buffered saline. Viral plaques were immunostained with the serum of rabbits that had been hyperimmunized with MARV, or with a mAb against EBOV, clone 15H10 (BEI Resources #NR-12184). Alternatively, following virus adsorption, monolayers were covered with MEM containing 10% FBS and 1.6% tragacanth (Sigma-Aldrich #G1128). After incubation for 14 days, medium was removed, cells were fixed with 10% formalin, plates were sealed in plastic bags, incubated for 24 hours at room temperature, and taken out of the BSL-4 laboratory as above. Fixed monolayers were stained with 10% formalin containing 0.25% crystal violet (Fisher Scientific #C581-100), and plaques were counted.
VSV-MARV and VSV-EBOV neutralization tests
Neutralization assays were performed in triplicate, as described above for MARV and EBOV. Following neutralization, virus-antibody mixtures were placed on monolayers of Vero E6 cells in duplicate, incubated for 1 hour at 37 °C for virus adsorption, and overlayed with MEM containing 2% FBS containing 0.9% methylcellulose. After incubation for 3 days, medium was removed, monolayers were fixed and stained with 10% formalin containing 0.25% crystal violet, and plaques were counted.
Generation and sequencing of VSV/GP-Uganda escape mutants
Vero E6 cell monolayers with two-fold dilutions of mAbs (12.5 – 200 μg/mL) added to the medium were inoculated with 200 PFU of recombinant VSV/GP-Uganda and incubated at 37 °C for 2-4 days. To determine which samples contained live virus, supernates were collected, virus was titrated in Vero E6 cell monolayers under methylcellulose overlay, monolayers were incubated at 37 °C for 3-4 days, and plaques were counted. Supernates with the highest concentrations of mAbs, which were found to contain live virus by plaque titration, were incubated in presence of serially diluted mAbs followed by titration of virus as above. The procedure was performed a total of three times. Escape mutant viruses harvested after the third passage were cloned biologically by plaque purification. For biological cloning, Vero E6 cell monolayers in 24-well plates were inoculated with dilutions of the escape mutant viruses in the presence of the corresponding mAbs (200 μg/mL of MR72 or 100 μg/mL of MR78) and covered with 0.7% low melting temperature SeaPlaque agarose (Lonza #50100). Monolayers were incubated at 37 °C for 6 days, plaques were visualized with 0.01% neutral red aqueous solution (Electron Microscopy Sciences), picked, resuspended in medium and transferred to Vero E6 cell monolayers in 24-well plates in presence of the corresponding mAbs (200 μg/mL of MR72 or 100 μg/mL of MR78) for virus propagation. In 2-5 days, based on the extent of CPE observed, virus was harvested, and cells were dissolved in Trizol reagent (Life Technologies 315596018). Total cellular RNA was extracted and reverse-transcribed and amplified by PCR with the primers described above for generation of a chimeric strain of VSV. Two overlapping fragments covering MARV GP ORF were PCR-amplified from cDNA using forward primer 5′-CATGTACGACGCGTCAACATGAGGACTA-3′ and reverse primer 5′-ACTAAGCCCTGCTGCCAGGT-3′ or forward primer 5′-ACAACAATGTACCGAGGCAA-3′ and reverse primer 5′-TCTAGCAGCTCGAGCTATCCAATATATTTAGTAAAGATACGACAA-3′, and the nucleotide sequences of the GP ORFs were determined using standard procedures.
Analysis of growth kinetics of VSV/GP-Uganda escape mutant viruses
Vero E6 cell monolayers in 24-well plates were inoculated in triplicate with VSV/GP-Uganda escape mutants or non-mutated virus at an MOI of 0.00025 PFU/cell in the presence of varying concentrations of the corresponding mAbs. Aliquots of medium were collected every 12 hours and frozen for titration at a later time. Titration of virus in aliquots was performed as above, without adding antibodies to the culture medium.
Biolayer interferometry competition binding assay
Biotinylated GP or GPΔmuc (EZ-link® Micro NHS-PEG4-Biotinylation Kit, Thermo Scientific #21955) (1 μg/mL) was immobilized onto streptavidin-coated biosensor tips (ForteBio #18-5019) for 2 minutes. After measuring the baseline signal in kinetics buffer (KB: 1× PBS, 0.01% BSA and 0.002% Tween 20) for two minutes, biosensor tips were immersed into the wells containing primary antibody at a concentration of 100 μg/mL for 10 minutes. Biosensors then were immersed into wells containing competing mAbs at a concentration of 100 μg/mL for 5 minutes. The percent binding of the competing mAb in the presence of the first mAb was determined by comparing the maximal signal of competing mAb applied after the first mAb complex to the maximal signal of competing mAb alone. MAbs were judged to compete for binding to the same site if maximum binding of the competing mAb was reduced to <30% of its un-competed binding. MAbs were considered non-competing if maximum binding of the competing mAb was >70% of its un-competed binding. A level of 30-70% of its un-competed binding was considered intermediate competition.
Sequence analysis of antibody variable region genes
Total cellular RNA was extracted from clonal hybridomas that produced MARV antibodies, and RT-PCR reaction was performed using mixtures of primers designed to amplify all heavy chain or light chain antibody variable regions. The generated PCR products were purified and cloned into the pJet 1.2 plasmid vector (Thermo Scientific, #K1231) for sequence analysis. The nucleotide sequences of plasmid DNAs were determined using an ABI3700 automated DNA sequencer. Heavy chain or light chain antibody variable region sequences were analyzed using the IMGT/V-Quest program (Brochet et al., 2008; Giudicelli et al., 2011). The analysis involved the identification of germline genes that were used for antibody production, location of complementary determining regions (CDRs) and framework regions (FRs) as well as the number and location of somatic mutations that occurred during affinity maturation.
Statistical analysis
EC50 values for neutralization were determined by finding the concentration of mAb at which a 50% reduction in plaque counts occurred after incubation of virus with neutralizing antibody. A logistic curve was fit to the data using the count as the outcome and the log-concentration as the predictor variable. The results of the model then were transformed back to the concentration scale. Results are presented as the concentration at the dilution that achieve a 50% reduction from challenge control with accompanying 95% confidence intervals. Each antibody was treated as a distinct analysis in a Bayesian non-linear regression model.
Sample preparation for EM studies
A Ravn strain MARV GP mucin-deleted construct (GPΔmuc) was produced by stable cell line expression in Drosophila S2 cells, as described above. Human Fab proteins for MARV-specific antibodies were generated as described above. Fabs were added in molar excess to GPΔmuc and allowed to incubate overnight at 4 °C. Complexes then were purified by Superdex 200 size exclusion chromatography in TBS.
Electron microscopy and sample preparation
A 4 μL aliquot of each complex that had been diluted to a concentration of ~0.03 μg/mL with TBS buffer was placed for 15 seconds onto carbon-coated 400 Cu mesh grids that had been plasma cleaned for 20 s (Gatan), blotted off on the edge of the grid, then immediately stained for 30 s with 4 μL of 2% uranyl formate. The stain was blotted off on the edge of the grid and the grid was allowed to dry. Data were automatically collected with Leginon (Carragher et al., 2000; Potter et al., 1999; Suloway et al., 2005) using a FEI Tecnai F20 electron microscope operating at 120 keV with an electron dose of 30 e-/Å2 and a magnification of 52,000X that resulted in a pixel size of 2.65 Å at the specimen plane when collected with Tietz CMOS 4k × 4k CCD camera. Particle orientations appeared to be generally isotropic and images were acquired at a constant defocus value of -1.0 μ m at 0° stage tilt.
Image processing of protein complexes
Particles were picked automatically using DoG Picker (34) and placed into a particle stack using the Appion software (Lander et al., 2009). Reference-free 2D class averages were generated with the Xmipp clustering 2D alignment software (van Heel et al., 1996) and sorted into an initial 300 classes. Non-GP particles were removed and the stack was further sub-classified into classes with ~100 particles per class in order to generate the final particle stack used for the reconstruction. Various numbers of class averages were chosen to create initial models using EMAN2 common lines software (Tang et al., 2007). A model that best matched its projected classes was then used for refinement against the raw particle stack, imposing C3 symmetry, and the reconstruction was generated with 10 rounds of refinement and increasingly smaller angular sampling rates with EMAN2 (Tang et al., 2007). All model fitting and manipulation was completed using UCSF Chimera (Pettersen et al., 2004).
In vivo testing
The animal protocol for testing of mAbs in mice was approved by the Institutional Animal Care and Use Committee of the University of Texas Medical Branch at Galveston. Seven-week-old BALB/c mice (Harlan) were placed in the ABSL-4 facility of the Galveston National Laboratory. Groups of mice at 5 animals per group were injected with individual mAbs by the intraperitoneal route twice: one h prior and 24 h after MARV challenge, using 100 μg per treatment. Untreated animals served as controls. For the challenge, mice were injected with 1,000 PFU of the mouse-adapted MARV strain Ci67 by the intraperitoneal route. Animals were weighed and monitored daily over the three-week period after challenge. Once animals were symptomatic, they were examined twice per day. The disease was scored using the following parameters: dyspnea (possible scores 0-5), recumbency (0-9), unresponsiveness (0-5), and bleeding/hemorrhage (0-5); the individual scores for each animal were summarized.
Supplementary Material
1
2
3
4
5
6
Acknowledgments
This project received support from the Defense Threat Reduction Agency (grant HDTRA1-13-1-0034 to JEC), U.S. NIH (grant 1U19AI109711 to JEC and AB, and U19AI109762 to EOS and ABW, R01AI089498 to EOS and U01AI082156 to EOS). EOS is an Investigator in the Pathogenesis of Infectious Disease of the Burroughs Wellcome Fund. The project was supported by NCRR Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. TH received support from MEXT and JSPS Postdoctoral Fellowships for Research Abroad and a Research Fellowship of The Uehara Memorial Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Dr. Eugene Agapov (Washington University in St. Louis) for useful suggestions concerning the generation of VSV/GP-Uganda escape mutants. We thank Dr. the donor, Dr. Norman Fujita (Wheat Ridge, CO) and the Vanderbilt Clinical Trials Center for help in sample acquisition. We thank Frances Smith-House, Gloria Fritz, Vidisha Singh and Leland Brown for excellent technical support, and Dr. Scott A. Smith for thoughtful comments and discussions. Flow cytometry experiments were performed in the VMC Flow Cytometry Shared Resource, supported by NIH P30 CA68485 and DK058404. The EM work was conducted at the National Resource for Automated Molecular Microscopy at The Scripps Research Institute, which is supported by the Biomedical Technology Research Center program (GM103310) of the National Institute of General Medical Sciences. EM reconstructions have been deposited in the Electron Microscopy Data Bank under the accession codes EMD-6232 through 6238.
Footnotes
Author Contributions: AIF, PAI, CDM planned, performed, and analyzed experiments and wrote the paper. TG, XS, CK, MLF, TH, ZAB and GS performed and analyzed experiments. JCS performed statistical analysis. TGK and ABW planned and analyzed experiments. EOS, AB, and JEC planned and analyzed experiments and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PloS One. 2012;7:e29608. [Europe PMC free article] [Abstract] [Google Scholar]
- Brauburger K, Hume AJ, Mühlberger E, Olejnik J. Forty-five years of Marburg virus research. Viruses. 2012;4:1878–1927. [Europe PMC free article] [Abstract] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. [Europe PMC free article] [Abstract] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. [Europe PMC free article] [Abstract] [Google Scholar]
- Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A. Leginon: An automated system for acquisition of images from vitreous ice specimens. J Struct Biol. 2000;132:33–45. [Abstract] [Google Scholar]
- CDC. Imported case of Marburg hemorrhagic fever - Colorado, 2008. MMWR. 2009;58:1377–1381. [Abstract] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science (New York, NY) 2005;308:1643–1645. [Europe PMC free article] [Abstract] [Google Scholar]
- Cook JD, Lee JE. The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Path. 2013;9:e1003258. [Europe PMC free article] [Abstract] [Google Scholar]
- Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. [Europe PMC free article] [Abstract] [Google Scholar]
- Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, Kawaoka Y, Chandran K, Dye JM, Saphire EO. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J Virol. 2009;83:2883–2891. [Europe PMC free article] [Abstract] [Google Scholar]
- Dye JM, Herbert AS, Kuehne AI, Barth JF, Muhammad MA, Zak SE, Ortiz RA, Prugar LI, Pratt WD. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc Natl Acad Sci U S A. 2012;109:5034–5039. [Europe PMC free article] [Abstract] [Google Scholar]
- Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk HD, Feldmann H, Stroher U. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol. 2004;78:5458–5465. [Europe PMC free article] [Abstract] [Google Scholar]
- Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011:695–715. [Abstract] [Google Scholar]
- Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, Matsuoka R, Kohda D, Yanagi Y, Hammel M, Crowe JE, Jr, Saphire EO. Cell. 2015 in press. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson ED, Johnson BK, Silverstein D, Tukei P, Geisbert TW, Sanchez AN, Jahrling PB. Characterization of a new Marburg virus isolated from a 1987 fatal case in Kenya. Arch Virol Suppl. 1996;11:101–114. [Abstract] [Google Scholar]
- Kajihara M, Marzi A, Nakayama E, Noda T, Kuroda M, Manzoor R, Matsuno K, Feldmann H, Yoshida R, Kawaoka Y, et al. Inhibition of Marburg virus budding by nonneutralizing antibodies to the envelope glycoprotein. J Virol. 2012;86:13467–13474. [Europe PMC free article] [Abstract] [Google Scholar]
- Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–198. [Abstract] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Bio. 2009;166:95–102. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. [Europe PMC free article] [Abstract] [Google Scholar]
- Lubaki NM, Ilinykh P, Pietzsch C, Tigabu B, Freiberg AN, Koup RA, Bukreyev A. The lack of maturation of Ebola virus-infected dendritic cells results from the cooperative effect of at least two viral domains. J Virol. 2013;87:7471–7485. [Europe PMC free article] [Abstract] [Google Scholar]
- Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PW, Burton DR. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol. 1999;73:6024–6030. [Europe PMC free article] [Abstract] [Google Scholar]
- Marzi A, Yoshida R, Miyamoto H, Ishijima M, Suzuki Y, Higuchi M, Matsuyama Y, Igarashi M, Nakayama E, Kuroda M, et al. Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of Ebola hemorrhagic fever. PloS One. 2012;7:e36192. [Europe PMC free article] [Abstract] [Google Scholar]
- Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A. 2014;111:17182–17187. [Europe PMC free article] [Abstract] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. [Europe PMC free article] [Abstract] [Google Scholar]
- Olinger GG, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A. 2012;109:18030–18035. [Europe PMC free article] [Abstract] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. [Abstract] [Google Scholar]
- Pettitt J, Zeitlin L, Kim DH, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly MH, Whaley KJ, Ingram MF, Zovanyi A, Heinrich M, Piper A, Zelko J, Olinger GG. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med. 2013;5:199ra113. [Abstract] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, et al. Leginon: A system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. [Abstract] [Google Scholar]
- Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB, et al. Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Trans Med. 2012;4:138ra181. 138ra181. [Abstract] [Google Scholar]
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. [Europe PMC free article] [Abstract] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. [Europe PMC free article] [Abstract] [Google Scholar]
- Saphire EO. An update on the use of antibodies against the filoviruses. Immunotherapy. 2013;5:1221–1233. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith DH, Johnson BK, Isaacson M, Swanapoel R, Johnson KM, Killey M, Bagshawe A, Siongok T, Keruga WK. Marburg-virus disease in Kenya. Lancet. 1982;1:816–820. [Abstract] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Bio. 2005;151:41–60. [Abstract] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Bio. 2007;157:38–46. [Abstract] [Google Scholar]
- Thomas D, Newcomb WW, Brown JC, Wall JS, Hainfeld JF, Trus BL, Steven AC. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. [Europe PMC free article] [Abstract] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, Carroll SAR, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Path. 2009;5:e1000536. [Europe PMC free article] [Abstract] [Google Scholar]
- Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, Hartman AL, Comer JA, Zaki SR, Ströher U, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J Virol. 2006;80:6497–6516. [Europe PMC free article] [Abstract] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. [Abstract] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Bio. 1996;116:17–24. [Abstract] [Google Scholar]
- Warfield KL, Alves DA, Bradfute SB, Reed DK, VanTongeren S, Kalina WV, Olinger GG, Bavari S. Development of a model for marburgvirus based on severe-combined immunodeficiency mice. Virol J. 2007;4:108. [Europe PMC free article] [Abstract] [Google Scholar]
- Warfield KL, Bradfute SB, Wells J, Lofts L, Cooper MT, Alves DA, Reed DK, VanTongeren SA, Mech CA, Bavari S. Development and characterization of a mouse model for Marburg hemorrhagic fever. J Virol. 2009;83:6404–6415. [Europe PMC free article] [Abstract] [Google Scholar]
- Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. [Europe PMC free article] [Abstract] [Google Scholar]
- World Health Organization. Ebola Situation Report. WHO Global Alert and Response. 2014a 7 January 2015. [Google Scholar]
- World Health Organization. Marburg virus disease - Uganda, 10 October 2014. WHO Global Alert and Response 2014b [Google Scholar]
- Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Met. 2008;336:142–151. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2015.01.031
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867415000744/pdf
Citations & impact
Impact metrics
Article citations
Emerging and reemerging infectious diseases: global trends and new strategies for their prevention and control.
Signal Transduct Target Ther, 9(1):223, 11 Sep 2024
Cited by: 0 articles | PMID: 39256346 | PMCID: PMC11412324
Review Free full text in Europe PMC
Longitudinal proteome-wide antibody profiling in Marburg virus survivors identifies wing domain immunogen for vaccine design.
Nat Commun, 15(1):8133, 17 Sep 2024
Cited by: 0 articles | PMID: 39285186 | PMCID: PMC11405854
Antibodies targeting the glycan cap of Ebola virus glycoprotein are potent inducers of the complement system
Commun Biol, 7(1):871, 17 Jul 2024
Cited by: 0 articles | PMID: 39020082 | PMCID: PMC11255267
Non-Ebola Filoviruses: Potential Threats to Global Health Security.
Viruses, 16(8):1179, 23 Jul 2024
Cited by: 0 articles | PMID: 39205153 | PMCID: PMC11359311
Review Free full text in Europe PMC
Macaque antibodies targeting Marburg virus glycoprotein induced by multivalent immunization.
J Virol, 98(7):e0015524, 04 Jun 2024
Cited by: 0 articles | PMID: 38832790 | PMCID: PMC11329191
Go to all (106) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Electron Microscopy Data Bank
- (1 citation) Electron Microscopy Data Bank - EMD-6232
Electron Microscopy Data Bank (EMDB) at PDBe (Showing 7 of 7)
Lay summaries
Plain language description
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural basis for Marburg virus neutralization by a cross-reactive human antibody.
Cell, 160(5):904-912, 01 Feb 2015
Cited by: 87 articles | PMID: 25723165 | PMCID: PMC4344967
Non-neutralizing Antibodies from a Marburg Infection Survivor Mediate Protection by Fc-Effector Functions and by Enhancing Efficacy of Other Antibodies.
Cell Host Microbe, 27(6):976-991.e11, 21 Apr 2020
Cited by: 42 articles | PMID: 32320678 | PMCID: PMC7292764
Discovery of Marburg virus neutralizing antibodies from virus-naïve human antibody repertoires using large-scale structural predictions.
Proc Natl Acad Sci U S A, 117(49):31142-31148, 23 Nov 2020
Cited by: 6 articles | PMID: 33229516 | PMCID: PMC7733803
Structural basis for differential neutralization of ebolaviruses.
Viruses, 4(4):447-470, 05 Apr 2012
Cited by: 51 articles | PMID: 22590681 | PMCID: PMC3347318
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCATS NIH HHS (2)
Grant ID: UL1 TR000445
Grant ID: UL1 TR000445-06
NCI NIH HHS (2)
Grant ID: P30 CA068485
Grant ID: P30 CA68485
NCRR NIH HHS (2)
Grant ID: UL1 RR024975-01
Grant ID: UL1 RR024975
NHLBI NIH HHS (1)
Grant ID: T32 HL069765
NIAID NIH HHS (9)
Grant ID: T32 AI007244
Grant ID: 1U19AI109711
Grant ID: U19 AI109711
Grant ID: R01 AI089498
Grant ID: U19AI109762
Grant ID: R01AI089498
Grant ID: U01 AI082156
Grant ID: U01AI082156
Grant ID: U19 AI109762
NIDDK NIH HHS (2)
Grant ID: P30 DK058404
Grant ID: DK058404
NIGMS NIH HHS (2)
Grant ID: P41 GM103310
Grant ID: GM103310





