Abstract
Free full text
Maintaining Tumour Heterogeneity in Patient-Derived Tumour Xenografts
Abstract
Pre-clinical models often fail to capture the diverse heterogeneity of human malignancies and as such lack clinical predictive power. Patient-derived tumour xenografts (PDXs) have emerged as a powerful technology: capable of retaining the molecular heterogeneity of their originating sample. However, heterogeneity within a tumour is governed by both cell-autonomous (e.g. genetic and epigenetic heterogeneity) and non-cell-autonomous (e.g. stromal heterogeneity) drivers. Whilst PDXs can largely recapitulate the polygenomic architecture of human tumours, they do not fully account for heterogeneity in the tumour microenvironment. Hence, these models have substantial utility in basic and translational research in cancer biology; but study of stromal or immune drivers of malignant progression may be limited. Similarly, PDX models offer the ability to conduct patient specific in vivo and ex vivo drug screens, but stromal contributions to treatment responses may be under-represented. This review discusses the sources and consequences of intratumour heterogeneity and how these are recapitulated in the PDX model. Limitations of the current generation of PDXs are discussed and strategies to improve several aspects of the model with respect to preserving heterogeneity are proposed.
Introduction
Despite remarkable advances in our understanding of the progression of human malignancies and the molecular events that underpin tumour survival, new therapies often fail to show significant efficacy in clinical trials. Projects such as The Cancer Genome Atlas and METABRIC have demonstrated the remarkable heterogeneity across tumours previously believed to be of the same subtype (1). It could be argued that clinical trials fail to sufficiently stratify patients based on relevant biomarkers of drug response: the response rate of an unscreened population to a molecularly targeted therapy typically lies between 10 and 20% (2). Patient stratification based on molecular determinants of drug efficacy and tumour heterogeneity allows for significantly greater responses - exemplified by the success of ALK kinase inhibitors in EML4-ALK positive non small cell lung cancers (3). However, even with patient stratification, clinical responses can be fleeting, often adding only 6-12 months before disease progression (2). Thus, understanding intertumour heterogeneity is the first step toward improved drug efficacy but cannot fully account for tumour relapse.
Intratumour heterogeneity is governed by both cell-autonomous (e.g. genomic and epigenomic heterogeneity) and non-cell-autonomous (e.g. stromal heterogeneity) factors. This heterogeneity has clinical implications in patient specific responses to therapy and the rapid emergence of resistance to targeted therapies (4). By capturing intra as well as intertumour heterogeneity, PDX models have a clear advantage over traditional models, supporting their use in oncological drug discovery and preclinical development. PDX models largely recapitulate cell-autonomous drivers of heterogeneity: exhibiting genomic clonal dynamics reminiscent of their originating tumour sample (5,6). Moreover, phenotypically distinct isogenic cellular clones have been shown to drive resistance to chemotherapy in colorectal PDX models (7). The tumour microenvironment has long been known to play an essential role in tumour progression and its role in drug response is becoming apparent (8,9). Although PDXs retain the 3D architecture found in human tumours, stromal and immune interactions may be altered by inter-species compatibility and cellular component deficiencies in host models.
The poor performance of so many investigational drugs suggests preclinical tumour models lack clinical predictive power. Indeed, one of the most often cited reasons for clinical failure is a lack of preclinical models that recapitulate the complexity of human cancers. It is with this in mind that many research and pharmaceutical groups alike have turned to PDX models (10,11). The establishment and predictive power of PDXs has been reviewed recently elsewhere (11). This review will focus on the limitations of current PDX models and how these can be addressed in the future, specifically in terms of maintaining the heterogeneous nature of human cancers (summarized in Table 1). PDXs are arguably the best models of tumour heterogeneity, and therefore perhaps the most powerful tools for investigating tumour biology. However, they may fail to fully account for many non-cell-autonomous drivers of heterogeneity (Figure 1A), and should be adapted if they are to reach their full potential as predictors of clinical efficacy in cancer drug development.
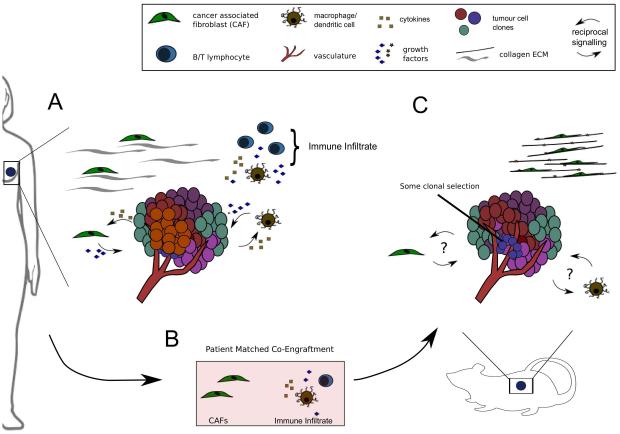
A shows a primary breast tumour including some sources of heterogeneity found in the native microenvironment. Reciprocal signalling pathways between tumour cells and tumour associated macrophages (TAMs) and cancer-associated fibroblasts (CAFs) are highlighted. Extracellular matrix (ECM) is shown as collagen fibres with associated fibroblasts and macrophage/dendritic cells with T/B Lymphocytes are shown as part of the immune infiltrate, though other cell types (NK cells, myeloid derived suppressors etc.) have been omitted for simplicity.
Panel B highlights patient matched fibroblasts and immune cells as possible candidates for co-engraftment in the next generation of PDX models. Current PDXs established in NSG mice lack an adaptive immune system and may have impaired innate immune cell infiltrates and cytokine signalling due to defective IL-2 receptor (33). Cancer associated fibroblasts are known to contribute to treatment response, although murine fibroblasts are present in PDX models, it is unclear how faithfully these recapitulate their human counterparts (26).
Panel C shows a PDX tumour in its native microenvironment. Questions over whether pro- and anti- tumour CAF/TAM signalling pathways are present to the same extent in PDX models as in the primary tumour are highlighted. Stromal and tissue architecture can have profound effects on transcriptional regulation but are often overlooked in the establishment of PDX models (25). To highlight potential differences in ECM organisation between the native microenvironment and that of the PDX, here the ECM is shown as highly organised collagen fibres with closely associated myofibroblasts.
Table 1
Table 1 shows sources of tumour heterogeneity, their consequences in translational and basic cancer biology and how they are currently represented in PDX models. Strategies to improve the model, by better representation of both cell-autonomous (genomic and epigenomic clones etc.) and non-cell-autonomous (stroma, immune infiltrate etc.) drivers of heterogeneity are proposed.
| Source of Heterogeneity | Implications for Basic and Translational Research | Representation in current PDTX models | Future Prospects | |
|---|---|---|---|---|
|
| ||||
| Cell-Autonomous |
 | Minor KRAS subclones predict resistance to EGFR targeted therapies in colorectal cancer.4 Overall clonal diversity correlates with drug resistance in ovarian and oesophageal cancers.16,17 | Genomic clones reconstructed in a panel of 15 breast cancer PDTX models revealed ongoing clonal dynamics. Polyclonal engraftment was possible, but clonal selection was clearly evident.5 | International collaborations such as the EuroPDX Consortium should facilitate sharing of expertise and eventually lead to increased engraftment with less pronounced clonal selection.10 |
 | Epigenetic ‘attractor states’ increase the phenotypic heterogeneity within the tumour and hence widen the pool of cellular clones able to contribute to treatment resistance.20 | Colorectal and breast PDTX models show five distinct cellular clonal phenotypes.6,7 ‘Type IV’ quiescent clones were responsible for resistance to chemotherapy in colorectal cancer.7 | PDTX models which better reflect the native tumour microenvironment should allow for more appropriate epigenetic clonal diversity. | |
|
| ||||
| Non Cell-Autonomous |
 | Pro- and anti- tumour properties are attributed to different populations of cancer associated fibroblasts (CAFs). Heterogeneous CAF or MSC populations could confer heterogeneity on the tumour bulk.26 | Human stromal components are replaced by murine equivalents on PDTX passage. It is unclear how closely mouse fibroblasts mimic their human counterparts in supporting tumour growth. Human fibroblast cell lines are heterogeneous in their ability to promote treatment resistance.26,27 | Patient matched stromal components should be sourced whenever possible. Although human fibroblasts can be expanded in vitro, cell sorting may reduce engraftment efficiency of tumour cells and this should be considered. |
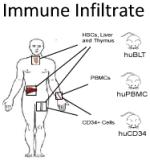 | The role of the immune infiltrate on tumour progression is highly complex. Checkpoint inhibitors and other immunotherapeutics are promising new treatment strategies in oncology. | Highly immunodeficient mice are used for PDTX implantation. The NSG strain is characterised by a lack of mature lymphocytes, the absence of functional NK cells, defective macrophages and defective dendritic cells.33 | HuPDTX immune models remain a substantial technical challenge. But the implications for study of tumour biology are profound. | |
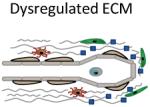 | Regulated ECM maintains tissue architecture and stem cell compartments. Loss of structure in cancer could contribute to oscillation between distinct transcriptional programs.21 | Matrigel is currently used to increase engraftment efficiency. Growth factors present in this murine basement membrane extract could support preferential engraftment of specific cell types. ECM is tissue specific, however in PDTX models, ectopic implantation is commonly used.23,25 | The ECM is tissue specific and orthotopic models should be considered where possible. Synthetic human alternatives to Matrigel should be investigated. | |
Clonal dynamics and tumour heterogeneity in PDX models
Through the course of tumour initiation and progression, cancerous cells undergo repeated mutational events that may or may not result in increased fitness relative to neighbouring cells. Dramatic increases in fitness are seen with the acquisition of key driver mutations early in a tumours evolution, for example the loss of TP53 may lead to clonal dominance. However, selection operates on phenotypes in response to stress inducing events, which may be stable or transient (12). A gain of fitness in one clone relative to another does not necessarily imply the loss of the latter in favour of the former. Rather, clonal populations within the tumour exist dynamically in space and time; competing, and perhaps cooperating, to further increase fitness of the tumour population as a whole (13) . Clonal dynamics derived from tumours’ inherent heterogeneity are thus extremely complex and can play key roles in tumour progression and development.
This conceptual framework of clonal evolution in cancer predicts several clinically observable features (14). Firstly, every mutation or copy number aberration (CNA) present in the bulk tumour need not be present in all cells; indeed, spatial variation exists in a tumour’s clonal composition. The existence of multiple subclones explains variable response rates to therapy, even within a single tumour mass, and the rapid emergence of drug resistance. For instance, the presence of a minor KRAS mutant clone can predict colorectal cancer patients who will develop resistance to epidermal growth factor receptor (EGFR) targeted therapy (4).
Our group has previously shown breast cancer consists of at least 10 distinct molecular subtypes with significant differences in disease outcome and treatment responses (1). Furthermore, we have helped delineate the diverse, variable clonal composition of triple negative breast cancers (TNBCs) (15). By allelic frequency measurements of 2,414 somatic mutations in 104 TNBCs, a complete spectrum of molecular and clonal compositions were characterised at diagnosis. Aside from the prognostic features of specific rare subclones (4), there is an association between clonal diversity and treatment resistance for at least some tumour types – notably ovarian (16) and oesophageal (17). Basal-like TNBCs have previously been linked with shorter disease free survival compared to non-basal-like TNBCs and tend to be associated with higher clonal diversity (15). Furthermore, integration of genomic and drug response data from breast cancer PDX models generated in our lab, show polygenomically engrafted tumours are more resistant to therapy than monogenomically engrafted tumours (Bruna et al., (2015) manuscript in preparation). Clearly more work is still to be done, but it seems likely that the clonal composition of tumours will have future utility in predicting disease outcome and informing treatment choice.
Multiple groups have attempted to define clonal dynamics based on either lentiviral barcoding (cellular clones) or mutational clustering (genomic clones) by population and single cell based computational approaches (5,18). For example, Eirew et al., reconstructed the genomic clonal dynamics of a panel of breast cancer PDX models using PyClone; a Bayesian clustering method for grouping somatic mutations (5). In each of the 15 cases examined, clonal diversity was reduced by xenotransplantation. This varied from extreme engraftment bias, selecting minor clones present in the sample of origin, to only moderate clonal selection. Remarkably, similar clonal dynamics were observed in parallel xenografts established from the same sample. In a separate study, Ding et al., found PDX models established from a basal like breast cancer were more representative of the patient’s metastatic lesion than the primary tumour (19). These observations suggest deterministic mechanisms underline the clonal selection found on engraftment. Eirew et al., further observed variable clonal dynamics between PDXs established from different molecular subtypes; underscoring the need for better representation of tumour molecular subtypes (1,5). In summary, PDX models can at least partially recapitulate the complex clonal dynamics of human malignancies and engraftment biases may represent non-stochastic selection events, which define a PDX model rather than limit its utility. The variable tumour- and subtype-dependent engraftment rates (low for some tumour types) and frequencies in the population mean that PDX programs representative of intertumour heterogeneity may require large multicentre collaborative efforts (such as the EuroPDX consortium (10)) and centralization of models.
Clonal evolution is a continuous process, and may be substantially altered by the selective pressures applied during chemo- and targeted therapy. Coupled with spatial heterogeneity within the tumour this may result in the need for multiple-site repeat biopsies to decipher the clonal composition and dynamics of a tumour and inform treatment choice. Clearly these procedures will be highly invasive and may not be technically possible; it is with this in mind that many researchers have turned to liquid biopsies (14,20). If our hypotheses are correct, modelling of clonal evolution in patient and matched PDX by computational approaches (such as PyClone (5) or CloneHD (18)) will allow unprecedented basic and translational research into clonal drivers of tumour progression and treatment response. Such studies will also be necessary to determine the longevity of PDX models of each cancer subtype, as it is unlikely that PDXs will remain patient-relevant and genomically stable in perpetuity.
It has been proposed that aberrant DNA methylation patterns in cancer can blur the lines between distinct phenotypic ‘attractor states’ (21). Thus, it may be important to consider cellular clones as drivers of malignant progression, independent of their genetic background. To track these isogenic cellular clones, lentiviral tagging has been utilised to mark individual cells, and their progeny, in breast and colorectal PDX models (6,7). In an elegant study by Kreso et al., it was found that minor ‘type IV’ subclones in colorectal PDXs were able to repopulate the tumour bulk after treatment with chemotherapy (7). These quiescent cell populations were genetically similar to their highly proliferative counterparts, and were later linked to the BMI1+ population thought to act as reserve stem cells of the intestinal and colonic crypts (22). As the cellular clones defined in this study were isogenic, their phenotype may have been driven by microenvironmental cues capable of modulating cellular transition between distinct gene expression patterns or epigenetic attractor states (23). Hence, the microenvironment’s composition could profoundly alter both a cells propensity to malignancy and the heterogeneity we hope to preserve in PDX models. Regardless, we should not underestimate the significance of even a partial translation of vastly heterogeneous human diseases into experimental model systems.
Limitations of the PDX model: tumour-extrinsic sources of heterogeneity
Aside from clonal dynamics driven by intrinsic differences in a cell’s genetic or epigenetic background, intratumour heterogeneity can be influenced by tumour-extrinsic factors in the non-cell-autonomous compartment (Figure 1A and Table 1) (23). Cellular interactions with the extracellular matrix (ECM) can alter gene expression programs, drive differentiation and profoundly alter cell behaviour. As cancers develop, tight regulation of the ECM is lost and tissue architecture begins to degrade (8). A recent study by Wang et al., provides direct evidence that ECM dependent signalling confers dynamic switching between TGFBR3 (transforming growth factor β receptor 3) and JUND (jun D protooncogene) related expression signatures (24). ECM driven oscillations between signalling pathways such as those described could have profound effects on propensity to malignancy. Furthermore, solid state ECM interactions are necessary for cells to maintain stem cell properties and regulated ECM helps maintain the stem cell niche (25). As cancer is often associated with a blurring of the boundaries between stem and differentiated cells, it is possible that a loss of structured ECM is essential for the stability of multiple sub-dominant cellular clones within a tumour (21). In PDX models, Matrigel™ is often used to increase the engraftment efficiency, however it is worth noting that this is a murine basement membrane extract and suitable synthetic human alternatives are available. The presence of growth factors in Matrigel may favour the engraftment of one cell type over another. Finally, as ECM structure is tissue specific (25), researchers should consider the use of orthotopic transplantations where possible.
The tumour microenvironment is further characterised by an influx of stromal cells. Infiltrating cancer associated fibroblasts (CAFs) can often confer resistance to cytotoxic and targeted therapies (9), however, recent studies confer on fibroblasts a degree of plasticity, with anti-tumour properties observed in some populations (26). Due to the high levels of CAF infiltrates seen in some tumour types, heterogeneity within their population would undoubtedly confer differential properties to the tumour bulk. We, and others, have found that human stromal cells are gradually replaced by murine equivalents upon engraftment in the mouse, suggesting that implanted human cells retain the ability to recruit murine accessory cells to their niche. However, it should be noted that some differences exist between ligand repertoires of human and murine fibroblasts (27). Clearly stromal architecture and activity is mimicked in the murine host, however it is currently unclear how this reflects human stroma with regards supporting tumour growth and development.
It is with this in mind that many have begun to investigate the co-engraftment of human mesenchymal stem cells (MSCs) or CAF cell lines in PDXs (Figure 1B and C). Here, care must be taken; the role of MSCs in tumour development is still controversial and may represent patient (or tissue) specific differences (28). If non-patient-matched sources of stromal cells are implanted, heterogeneity between tumours derived from different patients could be lost. Moreover, fibroblast cell lines vary considerably in their ability to confer resistance to cytotoxic therapies through hepatocyte growth factor (HGF)/c-Met signalling (9,27). Patient-derived fibroblasts can be isolated from tumour samples and expanded in vitro, thus, co-engraftment of matched stromal components should be considered wherever possible. This would significantly increase the advantages these models already have in retaining the complex heterogeneity found in patient samples.
Considering the crucial role of the immune system in tumour progression, perhaps the most obvious disadvantage of PDX models is the necessity for severely immunodeficient host animals. Tumour cells are broadly thought to be antigenic: point mutations in coding exons in a developed tumour results in a large repertoire of neoantigens. Targeting of these neoantigens can lead to significant CD8+ cytotoxic T-cell infiltration and tumour cell death. However, most tumours eventually progress and evade the immune system - often through the dominant inhibitory effects of suppressive pathways (so called ‘immune checkpoints’ such as CTLA-4/B7 and PD-1/PD-L1). This is supported by the prognostic value of the CD8+ to FOXP3+ (Cytotoxic to Regulatory T-cell) ratio in many solid tumours, and the recently reported clinical efficacy of a variety of checkpoint inhibitors (29,30).
The pro-inflammatory microenvironment established by CD8+ T-cells, M1 polarised tumour associated macrophages (TAMs), NK cells and others can lead to the recruitment of numerous immune suppressive components. TAMs and myeloid derived suppressor cells have been implicated in resistance to anti-angiogenic therapy. Additionally, macrophage and CD4+ T-cell recruitment following intensive chemotherapy in breast cancer patients is associated with significantly reduced recurrence free survival (30). Clearly, the co-engraftment of immune components into PDX models (Figure 1B) would facilitate both the study of novel therapies targeting tumour-immune interactions and allow for basic research into patient specific cross-talk between tumour progression and immune surveillance. Clone or patient specific differences in tolerization of dendritic cells, macrophage and neutrophil polarity and regulatory T-cell (Treg) infiltration could provide multiple novel insights into tumour biology.
The most robust reconstitution of the human immune system in immunodeficient mice is seen when bone marrow derived stem cells are co-engrafted along with liver and thymus fragments (BMT model) (31). However, the highly invasive multi-site biopsy required renders this model impractical for patient-matched humanised PDX models (huPDXs). CD34 marks a population of hematopoietic stem and progenitor cells (HSPCs) found in the blood and bone marrow. Mice implanted with CD34+ cells from umbilical cord blood show robust multilineage engraftment of human immune populations, though with reduced functionality relative to BMT mice. During chemotherapy, CD34+ cells leave the bone marrow and enter the circulation. If patients are given Granulocyte-Macrophage Colony Stimulating Factor (GM-CSF; Leukine™) to aid recovery from chemotherapy, numbers of CD34+ cells in the circulation have been known to exceed the bone marrow itself. Hence, CD34+ cells could potentially be harvested from patient’s blood to reconstitute a functional, patient-matched, immune system in mouse models (huCD34 model). As such an immune system would mature in the mouse and human immune cells would undergo central tolerance to mouse antigens during development. However, as they would not be exposed to patient antigens during this process, it is likely that they would mount a rapid non-self response to any subsequently engrafted human tissue. One possible solution comes from a recent study by Cosgun et al., showing immunodeficient adult mice carrying a mutation in the Kit receptor could support robust, uniform and sustained engraftment of CD34+ cells (32). If patient-matched CD34+ HSPCs were engrafted in adult Kit mutant mice with an established PDX tumour, it is possible that a functional immune system could develop in the presence of both human and mouse antigens.
The alternative, to engraft fully mature human immune cells, is used in the huPBMC model. Here, mononuclear cells from the peripheral blood (PBMCs) are isolated and implanted in immunodeficient mice. PBMCs comprise around 75% CD4/CD8+ T cells with the remainder primarily containing B and NK cells. As such, these models are well suited to short-term experiments where lymphocyte function is of primary concern. Aside from the lifetime of circulating cells, the major limitation of the huPBMC model is the rapid onset of graft versus host disease (GVHD) as engrafted cells mount an immune response against host murine tissue. The onset of GVHD can be delayed somewhat by the use of NOD scid gamma (NSG) strains lacking MHC I (33).
Undoubtedly the reconstitution of a patient-matched immune system in PDX models would be extremely valuable in the development of novel oncological drugs and in particular immunotherapeutics. The extent to which findings would be valid in the context of immune hyperactivation (against mouse in huPBMC or against tumour in huCD34) is unclear.
Conclusions and future prospects – toward a huPDX model
The PDX is arguably the most faithful model of cell-autonomous drivers of malignant progression. Sharing of expertise and resources through collaborative initiatives such as the EuroPDX consortium will lead to improved engraftment efficiencies and increase the breadth of tumour subtypes available in these model systems, contributing to a better representation of cancer heterogeneity in the lab (10).
Cooperation and competition between genetically or phenotypically distinct subclonal populations is thought to drive tumour growth, resistance to therapy and recurrence. While cell-autonomous sources of heterogeneity are clearly recapitulated in PDX models, current techniques fail to properly account for non-cell-autonomous factors. The microenvironment has long been known to play a significant role in tumour progression, but an incomplete understanding of stromal influences makes co-engraftment of non-patient-matched cell compartments a risky strategy. In order to maintain PDXs as models of diversity of human malignancies we must take care to engraft only patient-matched stromal components. CAFs have known roles in resistance to targeted therapy. It should, therefore, be a priority to ensure patient fibroblasts are maintained in the tumour xenograft.
Reconstituting a patient-matched immune system in PDX models is a significant challenge. Standard methods of engrafting either CD34+ HSPCs or mature circulating PBMCs will likely lead to inappropriate immune responses against human or murine tissues respectively. In order to study long-term tumour-immune interactions, a reconstituted immune system must simultaneously maintain tolerance to its human donor and acquire tolerance to its new host. A solution could come from engrafting both mature and naïve cell types. Maturing cells in the thymus and bone marrow would acquire central tolerance to murine tissues whilst mature cells in the periphery could act to suppress inappropriate responses against human antigens.
PDX models are capable of recapitulating the complexity of human malignancy remarkably well. These models have extraordinary utility in basic cancer research and beyond this have demonstrated clinical predictive power, allowing multiplexed screening of novel therapeutics in vivo. However, the aforementioned limitations of this model must be carefully considered when interpreting data. Although a significant amount of research is still needed, patient-matched huPDX models with co-engrafted stromal and immune components would offer unprecedented opportunity to study tumour biology and would be invaluable models in oncological drug development. In the future, huPDX models could allow researchers and clinicians to both predict and explain tumour response to novel targeted therapies.
Acknowledgements
Establishment of our Patient-Derived Tumour Xenograft models would not be possible without the dedicated work of the Cambridge Breast Unit, the Cambridge Institute Biological Resources Unit and our collaborators in EuroPDX consortium (http://www.EuroPDX.eu). JWC is grateful to OB Harris for critical review of the first draft of this manuscript. Additionally, we are particularly grateful to the patients who donated tissues to further our research and Cancer Research UK for supporting all authors.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-15-0727
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/75/15/2963.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/0008-5472.can-15-0727
Article citations
The roles of patient-derived xenograft models and artificial intelligence toward precision medicine.
MedComm (2020), 5(10):e745, 25 Sep 2024
Cited by: 0 articles | PMID: 39329017 | PMCID: PMC11424683
Review Free full text in Europe PMC
Syngeneic Mouse Models for Pre-Clinical Evaluation of CAR T Cells.
Cancers (Basel), 16(18):3186, 18 Sep 2024
Cited by: 0 articles | PMID: 39335157 | PMCID: PMC11430534
Review Free full text in Europe PMC
Modeling respiratory tract diseases for clinical translation employing conditionally reprogrammed cells.
Cell Insight, 3(6):100201, 18 Sep 2024
Cited by: 0 articles | PMID: 39391007 | PMCID: PMC11462205
Review Free full text in Europe PMC
Single-cell analysis of breast cancer metastasis reveals epithelial-mesenchymal plasticity signatures associated with poor outcomes.
J Clin Invest, 134(17):e164227, 03 Sep 2024
Cited by: 0 articles | PMID: 39225101 | PMCID: PMC11364385
Advancements and challenges in developing in vivo CAR T cell therapies for cancer treatment.
EBioMedicine, 106:105266, 01 Aug 2024
Cited by: 0 articles | PMID: 39094262 | PMCID: PMC11345408
Review Free full text in Europe PMC
Go to all (186) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Complex genetic and histopathological study of 15 patient-derived xenografts of aggressive lymphomas.
Lab Invest, 102(9):957-965, 29 Apr 2022
Cited by: 3 articles | PMID: 35488033 | PMCID: PMC9420679
Ewing Sarcoma PDX Models.
Methods Mol Biol, 2226:223-242, 01 Jan 2021
Cited by: 10 articles | PMID: 33326106
Patient-derived tumor immune microenvironments in patient-derived xenografts of lung cancer.
J Transl Med, 16(1):328, 26 Nov 2018
Cited by: 11 articles | PMID: 30477533 | PMCID: PMC6260563
Patient-derived xenografts (PDXs) as model systems for human cancer.
Curr Opin Biotechnol, 63:151-156, 18 Feb 2020
Cited by: 37 articles | PMID: 32070860
Review
Funding
Funders who supported this work.
Cancer Research UK (2)
Functional genomics of breast cancer
Professor Carlos Caldas, University of Cambridge
Grant ID: 16942
Functional genomics of breast cancer
Professor Carlos Caldas, University of Cambridge
Grant ID: A16942
National Institute for Health Research (NIHR) (1)
Grant ID: NF-SI-0611-10154





