Abstract
Free full text

Molecular, functional, and evolutionary analysis of sequences specific to Salmonella.
Abstract
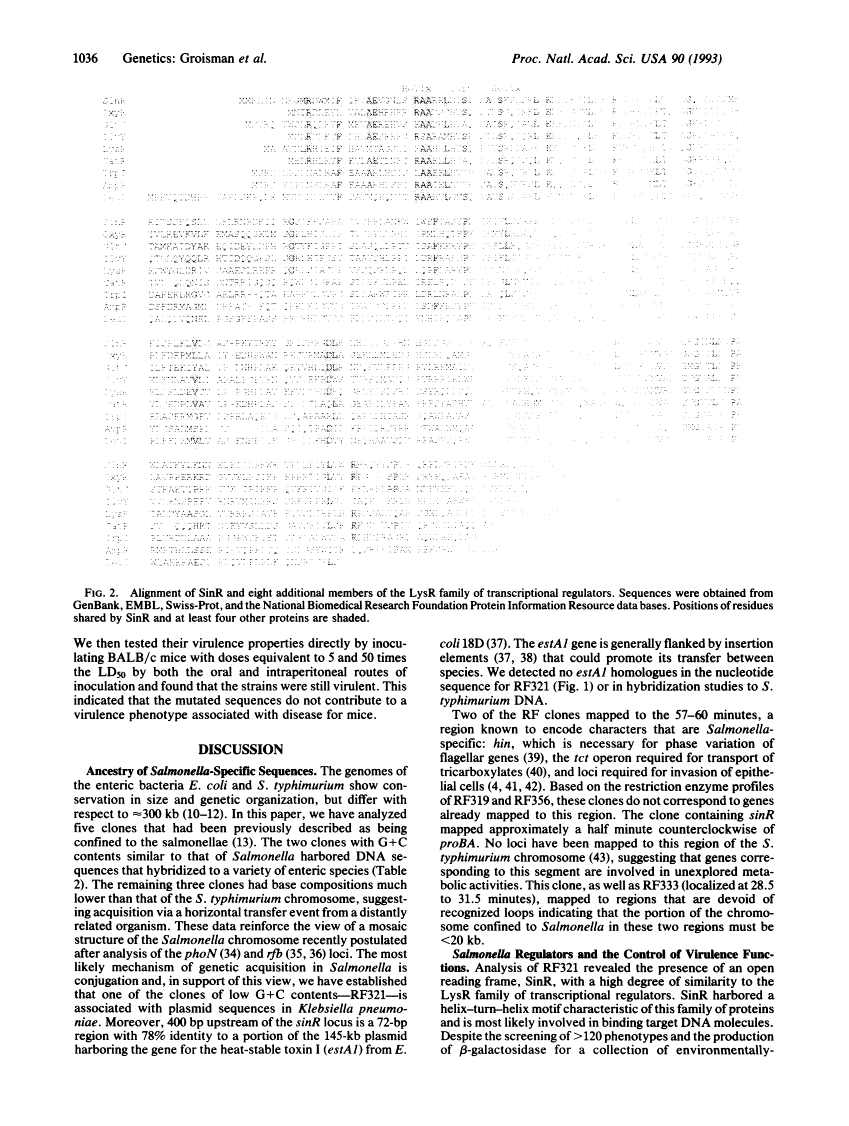
In that salmonellae have been implicated in an unprecedented array of diseases, sequences found to be specific to this species are often thought to be involved in the virulence attributes not seen in other enteric bacteria. To identify the molecular, genetic, and phenotypic characteristics that differentiate bacterial species, we analyzed five cloned DNA fragments that were originally described as being confined to Salmonella. Most of these segments mapped to unique positions on the Salmonella typhimurium chromosome indicative of independent evolutionary events, and three had G+C contents considerably lower than that of the Salmonella genome, suggesting that they arose through horizontal transfer. The nucleotide sequence was determined for one of the clones exhibiting an atypical base composition. This 4.9-kb fragment contained an open reading frame with structural similarity to the LysR family of transcriptional regulators. Strains harboring deletions in this region were tested for > 120 phenotypic characteristics including the effects on a collection of environmentally regulated lac gene fusions. In addition, all deletion strains behaved like the wild-type parent when tested for virulence in mice.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.5M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Finlay BB, Heffron F, Falkow S. Epithelial cell surfaces induce Salmonella proteins required for bacterial adherence and invasion. Science. 1989 Feb 17;243(4893):940–943. [Abstract] [Google Scholar]
- Galán JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6383–6387. [Europe PMC free article] [Abstract] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5189–5193. [Europe PMC free article] [Abstract] [Google Scholar]
- Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989 Feb 24;243(4894 Pt 1):1059–1062. [Abstract] [Google Scholar]
- Groisman EA, Heffron F, Solomon F. Molecular genetic analysis of the Escherichia coli phoP locus. J Bacteriol. 1992 Jan;174(2):486–491. [Europe PMC free article] [Abstract] [Google Scholar]
- Groisman EA, Saier MH., Jr Salmonella virulence: new clues to intramacrophage survival. Trends Biochem Sci. 1990 Jan;15(1):30–33. [Abstract] [Google Scholar]
- Galán JE, Curtiss R., 3rd Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991 Sep;59(9):2901–2908. [Europe PMC free article] [Abstract] [Google Scholar]
- Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. [Europe PMC free article] [Abstract] [Google Scholar]
- Tsen HY, Wang SJ, Roe BA, Green SS. DNA sequence of a Salmonella-specific DNA fragment and the use of oligonucleotide probes for Salmonella detection. Appl Microbiol Biotechnol. 1991 Jun;35(3):339–347. [Abstract] [Google Scholar]
- Jeter RM, Roth JR. Cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1987 Jul;169(7):3189–3198. [Europe PMC free article] [Abstract] [Google Scholar]
- Escalante-Semerena JC, Suh SJ, Roth JR. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990 Jan;172(1):273–280. [Europe PMC free article] [Abstract] [Google Scholar]
- Foster JW, Spector MP. Phosphate starvation regulon of Salmonella typhimurium. J Bacteriol. 1986 May;166(2):666–669. [Europe PMC free article] [Abstract] [Google Scholar]
- Aliabadi Z, Warren F, Mya S, Foster JW. Oxygen-regulated stimulons of Salmonella typhimurium identified by Mu d(Ap lac) operon fusions. J Bacteriol. 1986 Mar;165(3):780–786. [Europe PMC free article] [Abstract] [Google Scholar]
- Benson NR, Goldman BS. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J Bacteriol. 1992 Mar;174(5):1673–1681. [Europe PMC free article] [Abstract] [Google Scholar]
- Krane DE, Hartl DL, Ochman H. Rapid determination of nucleotide content and its application to the study of genome structure. Nucleic Acids Res. 1991 Oct 11;19(19):5181–5185. [Europe PMC free article] [Abstract] [Google Scholar]
- Groisman EA, Casadaban MJ. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. [Europe PMC free article] [Abstract] [Google Scholar]
- Lawrence JG, Ochman H, Hartl DL. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991 Aug;137(8):1911–1921. [Abstract] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. [Europe PMC free article] [Abstract] [Google Scholar]
- Muto A, Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. [Europe PMC free article] [Abstract] [Google Scholar]
- Ochman H, Whittam TS, Caugant DA, Selander RK. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. [Abstract] [Google Scholar]
- Storz G, Tartaglia LA, Ames BN. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990 Apr 13;248(4952):189–194. [Abstract] [Google Scholar]
- Rossen L, Shearman CA, Johnston AW, Downie JA. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA,B,C genes. EMBO J. 1985 Dec 16;4(13A):3369–3373. [Europe PMC free article] [Abstract] [Google Scholar]
- Bartowsky E, Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol Microbiol. 1991 Jul;5(7):1715–1725. [Abstract] [Google Scholar]
- Viale AM, Kobayashi H, Akazawa T, Henikoff S. rbcR [correction of rcbR], a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J Bacteriol. 1991 Aug;173(16):5224–5229. [Europe PMC free article] [Abstract] [Google Scholar]
- Groisman EA, Saier MH, Jr, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 1992 Apr;11(4):1309–1316. [Europe PMC free article] [Abstract] [Google Scholar]
- Wyk P, Reeves P. Identification and sequence of the gene for abequose synthase, which confers antigenic specificity on group B salmonellae: homology with galactose epimerase. J Bacteriol. 1989 Oct;171(10):5687–5693. [Europe PMC free article] [Abstract] [Google Scholar]
- Verma N, Reeves P. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989 Oct;171(10):5694–5701. [Europe PMC free article] [Abstract] [Google Scholar]
- Dallas WS. The heat-stable toxin I gene from Escherichia coli 18D. J Bacteriol. 1990 Sep;172(9):5490–5493. [Europe PMC free article] [Abstract] [Google Scholar]
- So M, Heffron F, McCarthy BJ. The E. coli gene encoding heat stable toxin is a bacterial transposon flanked by inverted repeats of IS1. Nature. 1979 Feb 8;277(5696):453–456. [Abstract] [Google Scholar]
- Simon M, Zieg J, Silverman M, Mandel G, Doolittle R. Phase variation: evolution of a controlling element. Science. 1980 Sep 19;209(4463):1370–1374. [Abstract] [Google Scholar]
- Widenhorn KA, Somers JM, Kay WW. Genetic regulation of the tricarboxylate transport operon (tctI) of Salmonella typhimurium. J Bacteriol. 1989 Aug;171(8):4436–4441. [Europe PMC free article] [Abstract] [Google Scholar]
- Elsinghorst EA, Baron LS, Kopecko DJ. Penetration of human intestinal epithelial cells by Salmonella: molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5173–5177. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee CA, Jones BD, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1847–1851. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanderson KE, Roth JR. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. [Europe PMC free article] [Abstract] [Google Scholar]
- Groisman EA, Chiao E, Lipps CJ, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7077–7081. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5054–5058. [Europe PMC free article] [Abstract] [Google Scholar]
- Curtiss R, 3rd, Kelly SM. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect Immun. 1987 Dec;55(12):3035–3043. [Europe PMC free article] [Abstract] [Google Scholar]
- Dorman CJ, Chatfield S, Higgins CF, Hayward C, Dougan G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect Immun. 1989 Jul;57(7):2136–2140. [Europe PMC free article] [Abstract] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975 Apr 11;188(4184):107–116. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.90.3.1033
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc45805?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
BanLec-eGFP Chimera as a Tool for Evaluation of Lectin Binding to High-Mannose Glycans on Microorganisms.
Biomolecules, 11(2):180, 28 Jan 2021
Cited by: 5 articles | PMID: 33525574 | PMCID: PMC7912117
In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD.
Sci Rep, 6:37858, 25 Nov 2016
Cited by: 15 articles | PMID: 27886269 | PMCID: PMC5122947
Mannose-Binding Lectin Inhibits the Motility of Pathogenic Salmonella by Affecting the Driving Forces of Motility and the Chemotactic Response.
PLoS One, 11(4):e0154165, 22 Apr 2016
Cited by: 5 articles | PMID: 27104738 | PMCID: PMC4841586
Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK.
BMC Genomics, 14:365, 31 May 2013
Cited by: 29 articles | PMID: 23725633 | PMCID: PMC3680342
Interplay between Fur and HNS in controlling virulence gene expression in Salmonella typhimurium.
Comput Biol Med, 42(11):1133-1140, 04 Oct 2012
Cited by: 10 articles | PMID: 23040276
Go to all (54) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The origin and evolution of species differences in Escherichia coli and Salmonella typhimurium.
EXS, 69:479-493, 01 Jan 1994
Cited by: 27 articles | PMID: 7994120
Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism.
Genetics, 130(3):429-443, 01 Mar 1992
Cited by: 60 articles | PMID: 1372579 | PMCID: PMC1204862
Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection.
Proc Natl Acad Sci U S A, 93(19):10434-10439, 01 Sep 1996
Cited by: 99 articles | PMID: 8816818 | PMCID: PMC38402
Molecular basis of the interaction of Salmonella with the intestinal mucosa.
Clin Microbiol Rev, 12(3):405-428, 01 Jul 1999
Cited by: 198 articles | PMID: 10398673 | PMCID: PMC100246
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: AI29554
NIGMS NIH HHS (1)
Grant ID: GM40995