Abstract
Free full text

Systematic VCP-UBXD Adaptor Network Proteomics Identifies a Role for UBXN10 in Regulating Ciliogenesis
Abstract
The AAA-ATPase VCP (also known as p97 or CDC48) uses ATP hydrolysis to “segregate” ubiquitinated proteins from their binding partners. VCP acts via UBX-domain containing adaptors that provide target specificity, but targets and functions of UBXD proteins remain poorly understood. Through systematic proteomic analysis of UBXD proteins in human cells, we reveal a network of over 195 interacting proteins, implicating VCP in diverse cellular pathways. We have explored one such complex between an unstudied adaptor UBXN10 and the intraflagellar transport B (IFT-B) complex, which regulates anterograde transport into cilia. UBXN10 localizes to cilia in a VCP-dependent manner and both VCP and UBXN10 are required for ciliogenesis. Pharmacological inhibition of VCP destabilized the IFT-B complex and increased trafficking rates. Depletion of UBXN10 in zebrafish embryos causes defects in left-right asymmetry, which depends on functional cilia. This study provides a resource for exploring the landscape of UBXD proteins in biology and identifies an unexpected requirement for VCP-UBXN10 in ciliogenesis.
The VCP AAA-ATPase, an evolutionarily conserved ATP-driven “segregase”, is a central regulator of protein quality control and ubiquitin-mediated signaling1. VCP functions in diverse processes such as ER associated degradation, macro and selective autophagy and DNA damage responses (DDR)2,3. ATP hydrolysis by VCP remodels protein complexes, unfolds ubiquitylated substrates for degradation, and extracts proteins embedded in membranes or chromatin. The prevailing model suggests that specific adaptor proteins target VCP to cellular structures and substrates where VCP activity is required3.
The largest family of adaptors contains the ubiquitin X domain (UBXD), which adopts a ubiquitin fold and associates with the N-terminus of VCP (Fig. 1a)4. A subset of UBXD proteins also contain ubiquitin associated domains (UBA), to recognize polyubiquitin chains on substrates5. Other cofactors utilize shorter motifs to associate with VCP6. Some studies have suggested the involvement of distinct adaptors in specific processes, however UFD1L-NPLOC4 association with VCP is required in some cases for adaptor binding7.
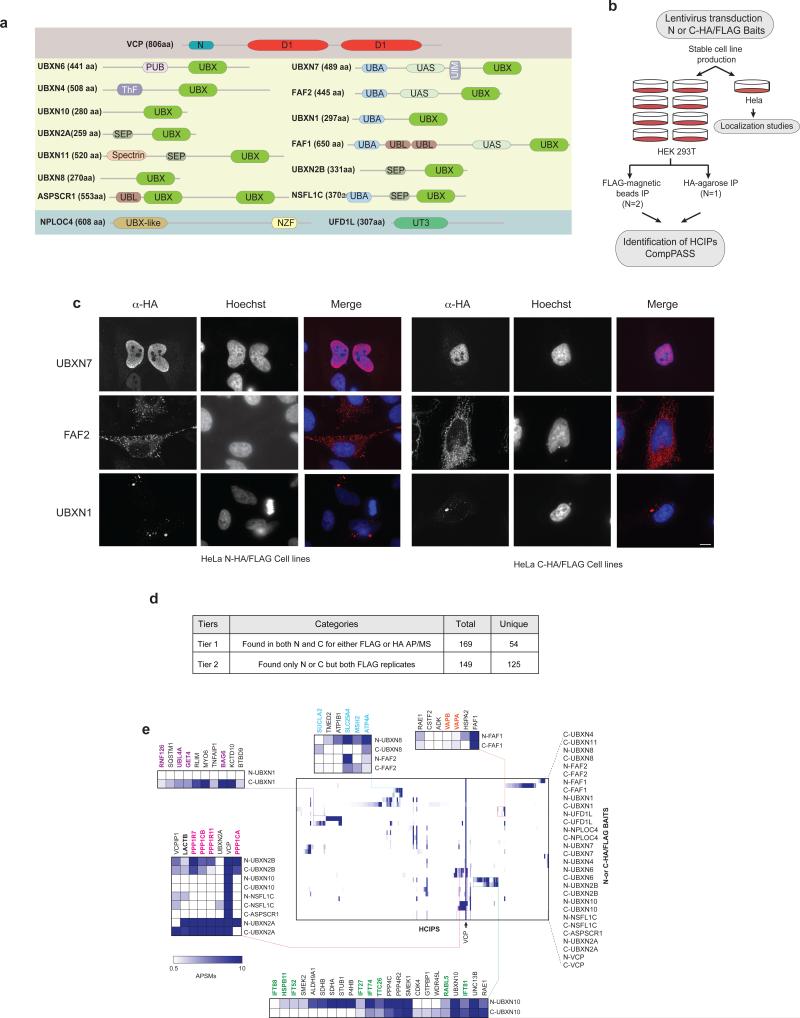
(a) Domain organization of VCP and UBXD proteins used as baits in the AP-MS studies. (D1, D2: ATPase domains, PUB: Peptide N-glycosidase/Ubiquitin associated, ThF: Thioredoxin like Fold, UBA: Ubiquitin associated, UIM: ubiquitin interaction motif, UBL: Ubiquitin –Like, SEP: Shp, EYC, p47, NZF: Npl4 Zinc Finger, UT3: Ufd1 Truncation 3).
(b) Pipeline for interaction proteomics and localization experiments.
(c) Localization of selected baits in HeLa cells visualized by α-HA staining. Scale bar, 10 μm.
(d) Organization of interactions observed in 6 AP-MS experiments into categories (Tiers) based on the criteria indicated under “categories”.
(e) Heat map of HCIPs (represented by APSM) identified for all baits. HCIPs were filtered using a NWD ≥ 0.98, Z ≥ 4, APSM ≥ 2 between biological duplicates. Hierarchical clustering was carried out based on interaction profiles. Selected complexes or clusters have been highlighted in detail and relevant members are color-coded.
Our understanding of how VCP is targeted to substrates is limited and only a subset of adaptors have been studied in detail. Even adaptors such as UBXN7 whose role in Cullin RING Ligase (CRL) function has been reported, the full repertoire of targets and biological pathways remain unknown. VCP is mutated in human neurodegenerative disorders including Inclusion Body Myopathy, Paget's disease of the bone and Frontotemporal Dementia (IBMPFD) and Amyotrophic Lateral Sclerosis (ALS)8,9. A significant fraction of disease-specific mutations cluster to the N-terminus where adaptors bind, suggesting that alterations in the repertoire of adaptors associated with VCP could contribute to disease10,11. Thus, a more complete understanding of adaptors and their targets is needed to fully understand VCP function.
Here, we employed a previously described interaction proteomics platform12,13 to analyze the VCP-UBXD interaction landscape, including several adaptors that are hitherto unstudied. The VCP-adaptor network is linked with a vast array of biological processes and we explore a interaction between the unstudied adaptor UBXN10 and the intraflagellar transport B (IFT-B) complex involved in cilia biogenesis.
Cilia are microtubule-based structures that are required for proper signaling in virtually every cell of the human body14. Defects in cilia formation or function cause a host of multi-system disorders referred to as ciliopathies. Cilia are assembled and disassembled by distinct IFT complexes that regulate anterograde (IFT-B) and retrograde (IFT-A) trafficking of ciliary components15,16 in a Bardet–Biedl syndrome (BBSome)17 complex-dependent manner. While hundreds of candidate ciliary proteins have been identified, their functions remain largely uncharacterized18–20. Through biochemical studies coupled with genetic and pharmacological disruption of VCP-UBXN10 function, we reveal a role for this complex in promoting ciliogenesis in mammalian cells in culture and in zebrafish. The VCP-UBXD network elaborated here provides a resource for further elucidation of the targeting mechanisms and biological functions of VCP.
RESULTS
Mapping the VCP-UBXD adaptor protein network
We used an unbiased comparative proteomic approach to develop a physical map of the VCP-UBXD network in 293T cells stably expressing one of thirteen FLAG-HAUBXD proteins, UFD1L, NPLOC4, or VCP via lentiviral transduction (Fig. 1a,b). Because UBX adaptors have protein interaction domains near their N and C-termini (Fig. 1a), we created both N- and C-terminally tagged proteins, with the exception of N-tagged UBXN11 and ASPSCR1, which failed to generate stable cell lines (Supplementary Table 1). Immunoblotting with available antibodies demonstrated expression levels of the tagged protein ranging from 0.6-4.5-fold compared with the endogenous protein (Supplementary Fig. 1a). We also created HeLa cells expressing N or C-terminally tagged VCP or its adaptors and determined their sub-cellular localization (Fig. 1b,c, Supplementary Fig. 2), allowing correlation with the known localization of interaction partners. These 30 cell lines were subjected to two independent α-FLAG magnetic bead affinity purification-mass spectrometry (AP-MS) experiments and one α-HA agarose AP-MS experiment (Fig. 1b, Supplementary Fig. 1b). To filter the >400 proteins identified for each bait for high confidence interacting proteins (HCIPs), we employed the CompPASS platform, which uses a database containing AP-MS derived APSM (assembled peptide spectral match) data for >100 unrelated bait proteins to identify non-specific contaminants. CompPASS determines HCIPs based on a normalized weighted D score (NWD-Score) measuring the frequency with which interactors appear in the database, the abundance (APSM-based), and the reproducibility of detection in technical replicates12,13,21. An APSM-based Z-score was also employed for identifying proteins that are often seen in AP-MS experiments but are selectively enriched with particular baits12,13,21. We designated interactors as HCIPs if they passed three criteria: 1) NWD ≥ 0.98, 2) Z ≥ 4, and 3) APSM ≥ 2. Proteins scoring below this threshold may still represent bona fide interactors, but could be weakly associated or sub-stoichiometric. In the majority of cases, bait abundance and the overall number of HCIPs was similar between N- and C-terminally tagged proteins (Supplementary Fig. 1b).
Overview and topology of the VCP-adaptor network
We organized HCIPs into tiers based on their reproducible identification in both α-FLAG and α-HA datasets (Fig. 1d, Supplementary Fig. 1d). Overall, we identified 169 HCIPs (54 unique proteins) from both N- and C-terminal baits for either α-FLAG or α-HA purified-complexes, which we designate as Tier 1 HCIPs (Fig. 1d, Supplementary Fig. 1c, Supplementary Table 2). We identified 149 HCIPs (125 unique proteins) in replicate analyses in either N- or C-terminally tagged baits from the α-FLAG dataset, which we refer to as Tier 2 (Fig. 1d, Supplementary Fig. 1c, Supplementary Table 2). Finally, 680 interactions (481 unique proteins) were found only once across all baits (Tier 3); these HCIPs are of lower confidence than Tier 1, 2 HCIPs (Supplementary Fig. 1c, Supplementary Table 2). The majority of Tier 1 and 2 HCIPs had adjusted p values ≤ 0.05 based on a hypergeometric test that evaluated the likelihood of nonrandom association among bait and prey observations across all LC-MS analyses in each dataset (Supplementary Table 4). As described below, we observed extensive validation for selected HCIPs using reciprocal interaction approaches.
Hierarchical clustering revealed that while the majority of HCIPs were unique to individual UBXD proteins, a subset were found in common between baits; for example, UBXN2A and UBXN2B associated with a phosphatase complex (Fig. 1e). A number of HCIPs, were common to many UBXD proteins, including the known VCP-associated deubiquitinating enzyme VCPIP, the BAG6 complex (UBXN1, FAF2), and CUL3 adaptors TNFAIP1, KCTD10, KCTD13 (UBXN1, FAF1, UBXN7). It was evident that some interactions are dependent upon the position of the epitope tag; UBXN1-HA-FLAG associated with the BAG6 complex but HA-FLAG-UBXN1 did not (Fig. 1e).
Of the 89 interactions reported in BioGRID in high-throughput AP-MS experiments with the adaptors analyzed here, 23 were identified as Tier 1, 2 HCIPs, (Fig. 2a, Supplementary Table 3). Of 69 interactions identified by immunoprecipitation-immunoblotting (IP-IB), 17 were present in Tier 1, 2 and finally for yeast two-hybrid data, 4 of 56 reported interactions were identified in Tier 1, 2 (Fig. 2a). A number of BioGRID interactions were also identified in our dataset but did not pass our stringent filtering criteria (65 interactions in APMS, 38 interactions in IP-IB and 17 interactions in yeast two hybrid) (Fig. 2a). Furthermore, 29 of 39 interactions reported in a previous study examining 5 UBA-UBXD adaptors by AP-MS5 were identified in our study (9 in Tier 1 and 2 and 20 below our filtering criteria, Fig. 2b). Moreover, our study identified 83 Tier 1, 106 Tier 2 that are not reported in BioGRID, and thereby represents the largest collection of candidate UBXD-interacting proteins reported to date.
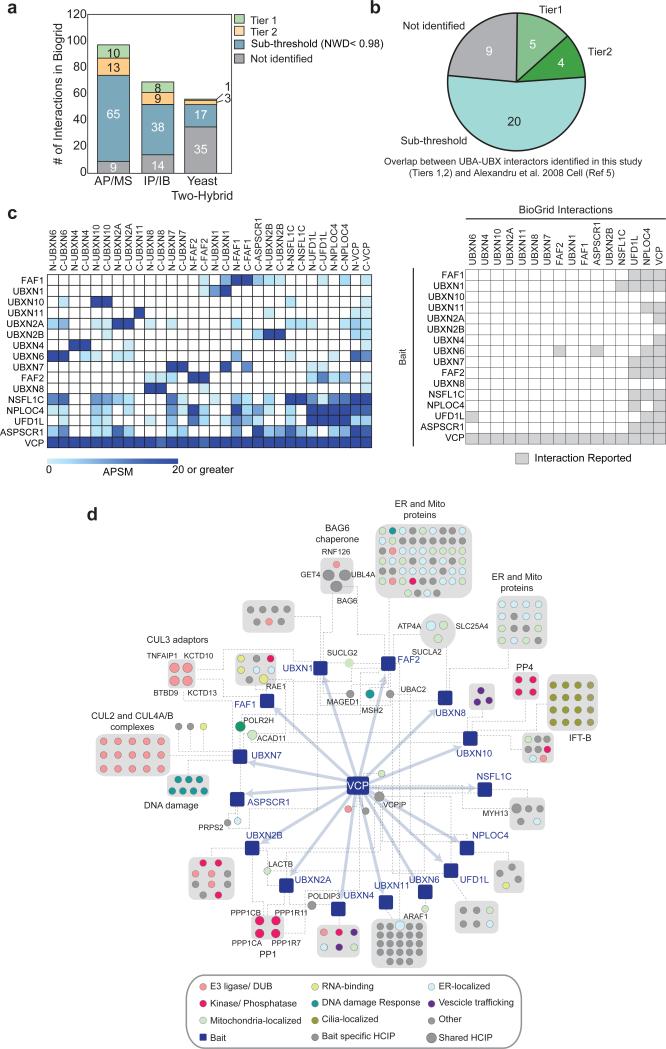
(a) Comparison of interacting proteins for 12 UBXD, UFD1L, and NPLOC4 proteins identified here with those reported in BioGRID. (AP-MS and IP-IB data in BioGRID available for 13 adaptors, yeast two-hybrid data available for 6 adaptors). See Supplementary Table 3.
(b) Comparison of interacting proteins for UBA-UBXD proteins found previously (Reference 6).
(c) Interactions observed among UBXD, UFD1L, and NPLOC4 proteins in the data reported here (left panel) and reported previously in BioGRID (right panel).
(d) Integrated interaction map of the VCP-UBXD network. HCIPs were filtered using NWD ≥ 0.98, Z ≥ 4, APSM ≥ 2 between α-FLAG biological duplicates and additionally verified in α-HA AP-MS. For clarity gene names have been omitted, except when HCIPs are shared between UBXD proteins. Interactions between UBXD adaptors have been omitted for clarity.
Placement of UBXD proteins within cellular modules
We identified interactions among UBXD proteins that were not previously reported, thereby extending potential crosstalk between UBXD proteins possibly assembled on the same VCP hexamer. Of the 14 UBXD proteins in BioGRID, 8 have been reported to interact with UFD1L and/or NPLOC4. In addition, UBXN6 has been reported to associate with FAF2 and ASPSCR1, and NSFL1C has been reported to interact with UBXN11 (Fig. 2c). In our dataset, UFD1L and/or NPLOC4 associated with 11 of 14 UBXD proteins (Fig. 2c). UBXN2A and UBXN6 associated with 6 and 5 distinct UBXD proteins, respectively (Fig. 2c). ASPSCR1 has been proposed to function in the assembly of the VCP hexamer22 and associated with 6 UBXD proteins (Fig. 2c). While we cannot rule out the possibility that the interactions we observe may represent adaptor exchange post-lysis, it suggests that VCP-adaptor modules may be more complex than currently appreciated.
We organized the VCP-adaptor network globally and individually, focusing on Tier 1, 2 HCIPs (Fig. 2d, 3a-g, Supplementary Fig. 3, Supplementary Table 2). Gene Ontology analysis of Tier 1, 2 HCIPs displays enrichment for numerous biological processes, including ubiquitin signaling, protein trafficking, membrane transport, DDR, and metabolism (Fig. 2d, Supplemental Fig. 1d). HCIPs frequently localize in the same compartment as the bait. For example, nuclear UBXN7 (Fig. 1c) associated with transcription-coupled nucleotide repair machinery, other DDR proteins, and CRL components (Fig. 2d, ,3b3b)6,23. VCP-UBXN2B mediates homotypic membrane fusion and promotes ER and Golgi biogenesis during interphase24–26. Phosphorylation of UBXN2B has been proposed to regulate this process27, however the in vivo kinase is presently unknown. We identified two kinases – GSK3β and CDC42BPB – in association with UBXN2B (Fig. 3e).
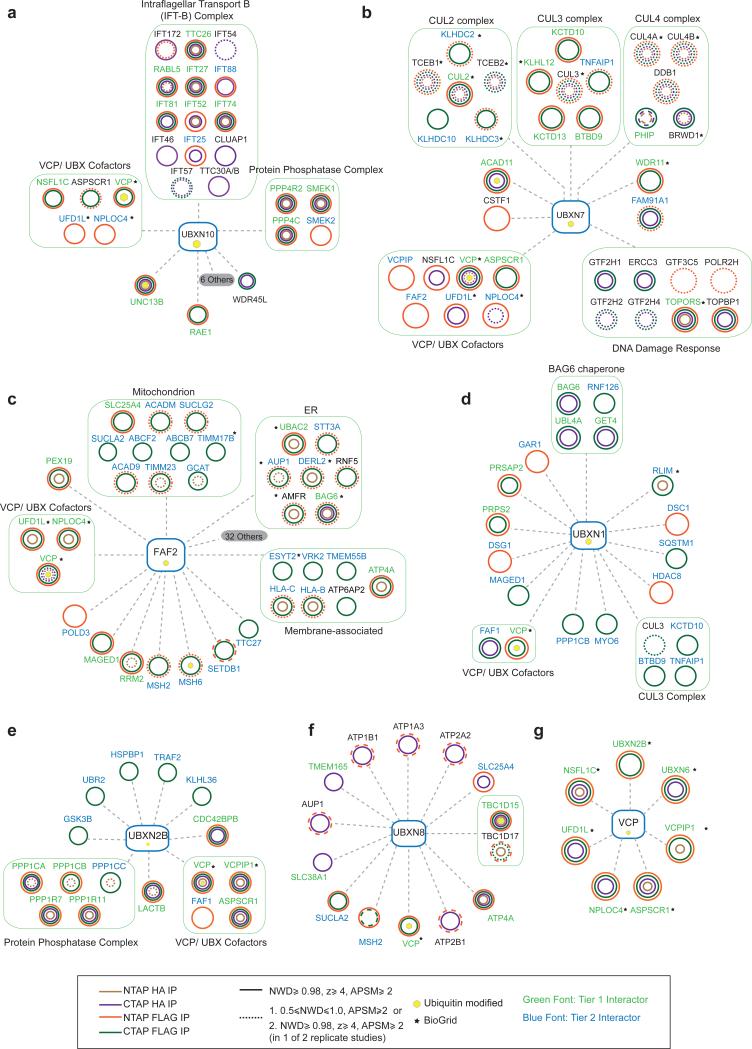
(a-g) Consolidated interaction maps of HCIPs identified across N and C-tag α-FLAG and α-HA APMS studies. (a) UBXN10, (b) UBXN7, (c) FAF2, (d) UBXN1, (e) UBXN2B, (f) UBXN8 and (g) VCP. Legend describes criteria used for inclusion.
Several UBXD-associated proteins associate with membrane components. We found that the membrane-tethered FAF2 (also called UBXD8) that functions in ERAD and lipid droplet homeostasis21,28 associated with AMFR, DERLIN2, AUP1, BAG6 and UBAC2 (Fig. 2d, ,3c)3c) as expected. We unexpectedly observed partial co-localization of FAF2 with mitochondria (Fig. 1c and Supplementary Fig. 1e) and observe several mitochondrial proteins in association with FAF2, although we cannot rule out post-lysis association (Fig. 3c). Further studies are required to determine if FAF2 has a role in mitochondrial function. UBXN8, also implicated in ERAD29, shared interactors with FAF2 (Fig. 1e, ,2d,2d, 3c,f), suggesting possible overlapping functions and also interacted with TBC1D15 and TBC1D17, which were recently reported to function in mitophagy30 a process that requires VCP31.
We performed reciprocal interaction experiments to validate the network. AP-MS experiments using 7 HCIPs as baits, identified the corresponding endogenous UBXD adaptor in 6 of 7 baits (Fig. 4a). We carried out transient-transfection and immunoprecipitation of 19 HCIPs, and validated 16 (Fig. 4b-o). These results are consistent with previous studies indicating ~70% of HCIPs identified using CompPASS can be validated by reciprocal approaches12,13. Thus, our study suggests that the VCP UBXD network is linked to numerous biological pathways, and that certain UBXD proteins may function in concert by associating within the same VCP hexamer.
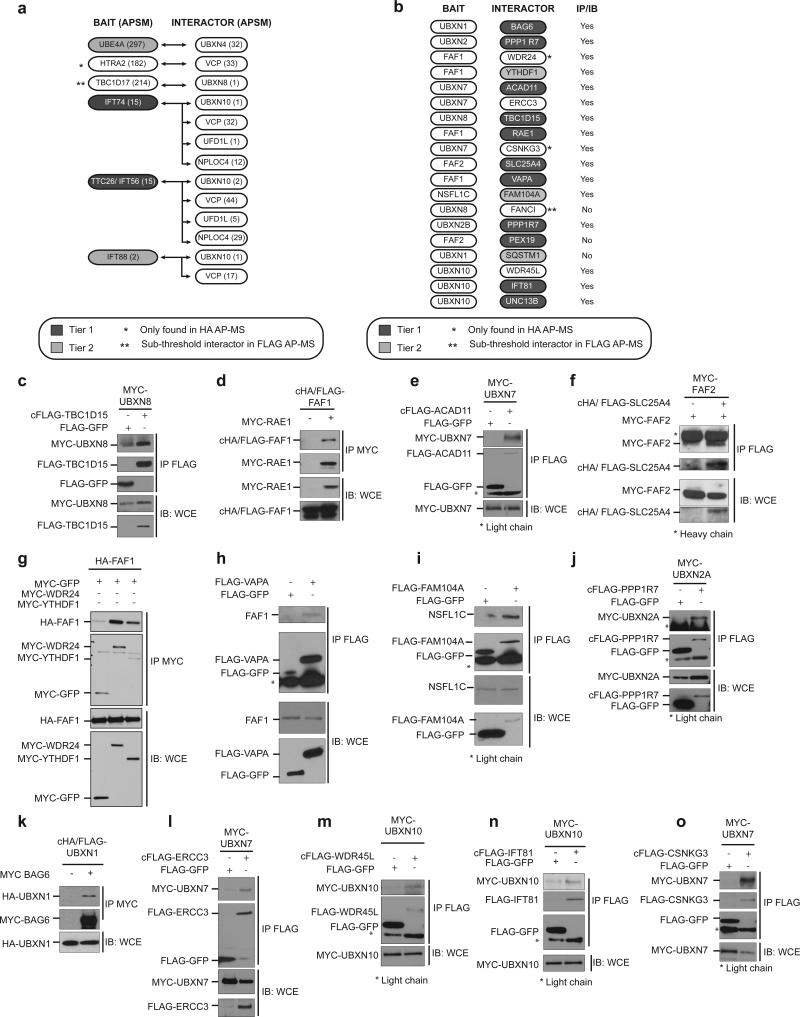
(a) Summary of interactor validation by AP-MS. Table represents APSMs for baits and HCIPs for reciprocal validation studies in HEK-293T cells. FANCD2 AP-MS did not identify UBXN8 and is not shown.
(b) Summary of interactor validation by immunoprecipitation and immunoblotting. HEK-293T cells were transiently transfected with epitope tagged constructs. Interactors were immunoprecipitated and probed for associated UBXD adaptor.
(c-o) Validation of interactors summarized in (b). Uncropped images of blots are shown is Supplementary Fig. 8.
UBXN10 interacts with the IFT-B trafficking complex
UBXN10 is a previously unstudied “UBX-domain only” adaptor (Fig. 1a). Proteomic analysis of N and C-terminally tagged UBXN10 identified 8 members of IFT-B as HCIPs, and 8 members that were below the CompPASS threshold (Fig. 2d, ,3a,3a, ,5a).5a). We also identified a PP4-SMEK phosphatase complex, vesicle sorting proteins (UNC13 A, B and WDR45L) and UFD1L-NPLOC4. We did not detect any IFT-A subunits indicating the interaction is specific to IFT-B (Fig. 3a, Table S2). Reciprocal AP-MS with 3 HCIPs (IFT88, IFT74 and TTC26) identified VCP, UBXN10 and UFD1L-NPLOC4 (Fig. 4a, ,5a),5a), and we validated UBXN10 interaction with IFT81 and WDR45L by co-immunoprecipitation (Fig. 4m,n). No IFT-B subunits were detected in other UBXD AP-MS experiments (Supplementary Table 1), indicating specificity for UBXN10.
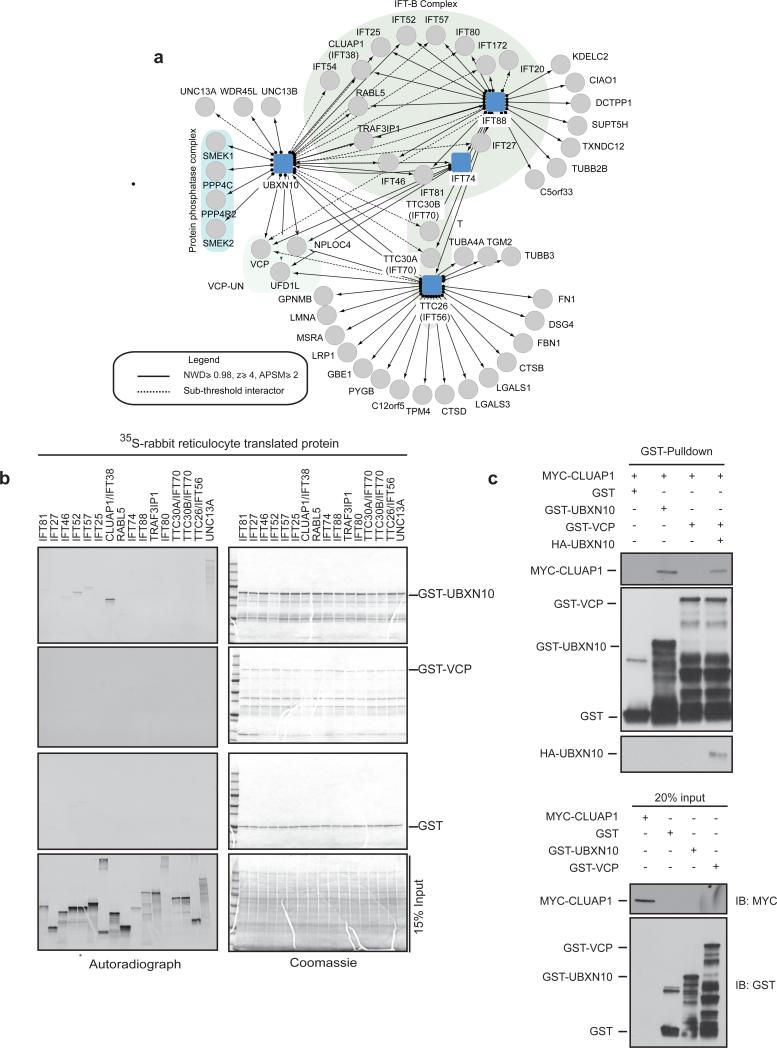
(a) Interaction map of UBXN10 and selected IFT-B subunits (IFT74, IFT88 and TTC26/IFT56) used as baits in AP-MS studies.
(b) UBXN10 binds specifically to the CLUAP1/IFT38 subunit of IFT-B. 35S-labelled-IFT-B subunits were translated in rabbit reticulocyte lysates and incubated with GST alone or GST-UBXN10 or GST-VCP purified from E.Coli. GST pull-downs were resolved by SDS-PAGE, stained with Coomassie and subjected to autoradiography. * : 35S-labelled- IFT52 was inadvertently loaded twice in the input gel alone. (c) UBXN10 links VCP to CLUAP1. The indicated GST-tagged proteins were incubated with in vitro translated MYC-CLUAP1. GST-VCP was incubated with in vitro translated HA-UBXN10 prior to incubation with MYC-CLUAP1. GST pulldowns were resolved on SDS-PAGE and immunoblotted for the indicated proteins. Uncropped images of blots are shown is Supplementary Fig. 8.
To validate the UBXN10-IFT-B interaction, we co-translated 15 MYC-tagged IFT-B subunits with HA-UBXN10 in an in vitro HeLa extract translation system, and tested interactions by co-immunoprecipitation (Supplementary Fig. 4a). In this system, extract-derived IFT-B subunits assemble with the translated subunit as determined by AP-MS and co-immunoprecipitation (Supplementary Fig. 4b-d). We found that 14 of 15 IFT-B subunits, associated with HA-UBXN10 (Supplementary Fig. 4a). UBXN10 also associated with UNC13A in this assay (Supplementary Fig. 4a).
To identify direct binding partners for UBXN10, we translated 15 radio-labeled IFTB subunits in rabbit reticulocyte lysates, and performed GST-pulldowns with E. coli-purified GST-VCP or GST-UBXN10. UBXN10 associated most efficiently with CLUAP1 (Fig. 5b, Supplementary Fig. 4e). Weaker interactions with IFT46, 52 and 57 were also detected, but these subunits also bound GST alone at low levels, suggesting a non-specific interaction (Supplementary Fig. 4e). We did not observe a direct interaction between IFT-B and GST-VCP above that observed with GST alone (Fig. 5b, Supplementary Fig. 4e). However, addition of HA-UBXN10 enabled the association of CLUAP1 with VCP (Fig. 5c). Thus, UBXN10 likely facilitates recruitment of VCP to IFT-B via CLUAP1.
UBXN10 localizes to cilia in a VCP-dependent manner
Given the association of UBXN10 with IFT-B and the finding that UBXN10 mRNA is enriched in cilia-rich tissues32, we examined whether UBXN10 localized to the primary cilium. We stably expressed GFP-UBXN10 in hTERT-RPE1, IMCD or LLC-PK1 cells and observed localization within cilia either along the length of the cilium, as discrete puncta within the cilium, or at the ciliary tip as reported for IFT-B subunit IFT88 (Fig. 6a,c, Supplementary Fig. 5a). Live cell imaging in LLC-PK1 cells, demonstrated GFP-UBXN10 trafficking in a bi-directional manner within cilia at rates similar to IFT88 (Fig. 6b, Supplementary Video 1). Furthermore UBXN10-mCHERRY and EYFP-IFT-B co-trafficked in the same complex, as determined by total internal reflection fluorescence (TIRF) microscopy, strongly suggesting that UBXN10 migrates within the cilium with IFT-B (Fig. 6c, Supplementary Video 2). We were unable to visualize VCP or UFD1L-NPLOC4 within cilia due to their high abundance in the cytoplasm (Supplementary Fig. 5b). However, VCP has been identified in previous proteomic studies of purified cilia19,33.
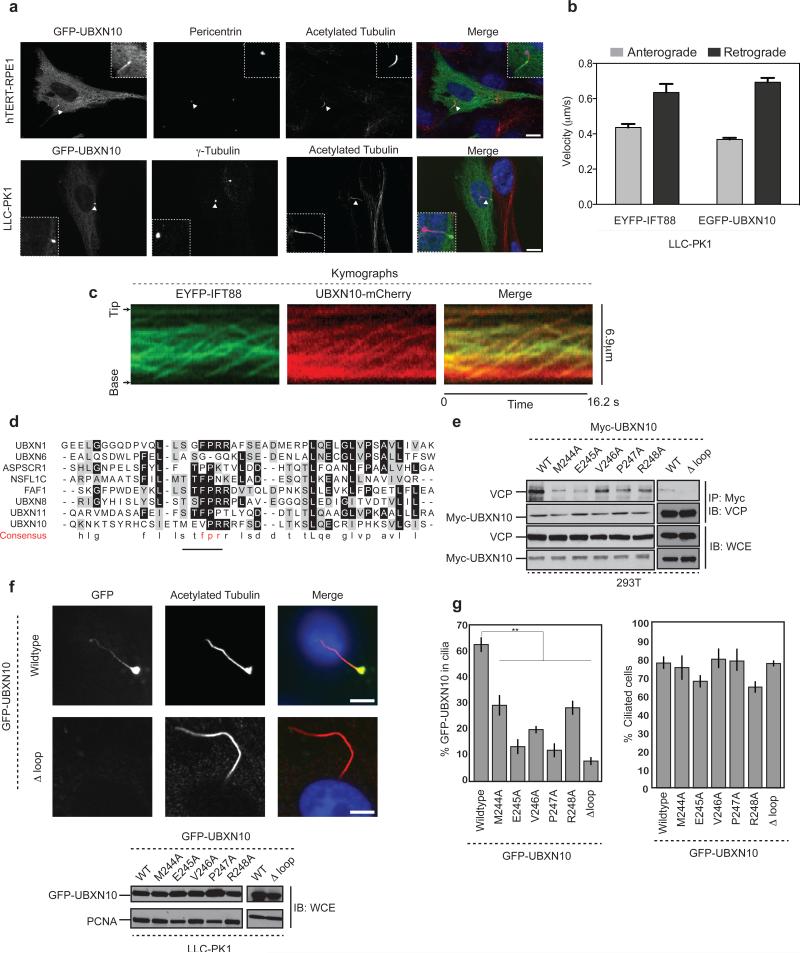
(a) GFP-UBXN10 localizes to cilia in multiple cell types. hTERT-RPE1 or LLC-PK1 cells stably expressing GFP-UBXN10 were starved for 48 h to induce cilia formation, fixed and stained with antibodies to acetylated tubulin (cilia) and pericentrin (centrosome). Images represent maximal intensity projections of multiple 0.2 μM z-stacks. Scale bar,10 μm.
(b) Comparison of EYFP-IFT88 and EGFP-UBXN10 anterograde and retrograde velocities in LLC-PK1 cells. n = 57, 38, 79 and 47 cells pooled from 2 independent experiments for anterograde and retrograde velocity measurements for EYFP-IFT88 and EGFP-UBXN10 respectively. Error bars represent mean ± s.d.
(c) Kymographs derived from TIRF microscopy of IMCD EYFP-IFT88 and UBXN10-mCHERRY cells demonstrating co-migration of UBXN10 with IFT-B within cilia.
(d) CLUSTAL-based sequence alignment of the loop region in the UBX domain that interacts with VCP. The FPR motif that interacts with VCP is replaced with TMEVPR in UBXN10.
(e) Mutations in the UBX domain impair UBXN10 interaction with VCP. MYC-UBXN10 wild-type or the indicated mutants were transiently transfected into HEK-293T cells. MYC immunoprecipitates were resolved by SDS PAGE and probed for endogenous VCP.
(f) Confocal images of LLC-PK1 stable cell lines expressing GFP-UBXN10 wildtype or the Δ–loop mutant. Scale bar is 5 μm. Bottom panel: Levels of GFP-UBXN10 protein in LLC-PK1 cells.
(g) Quantification of GFP-UBXN10 signal within cilia (left) and the total number of ciliated cells for each mutant (right). n = 222 (Wildtype), 275 (M244A), 228 (E245A), 255 (V246A), 230 (P247A), 310 (R248A) 245 (Δ Loop) pooled from 3 independent experiments. Error bars represent mean ± s.e.m. Statistical significance was calculated using unpaired two-tailed Students t-test. p values compared to wildtype for all mutants ≤ 0.001 **: p≤ 0.001. Uncropped images of blots are shown is Supplementary Fig. 8.
We asked if UBXN10 interaction with VCP is required for its ciliary localization. Structural modeling of UBXN10, based on a FAF1-VCP complex34, identified a Met-Glu-Val-Pro-Arg (MEVPR) motif as a possible UBXN10-VCP interaction interface (Fig. 6d, Supplementary Fig. 5c). Mutation of each residue or deletion of the loop (Δloop) reduced binding to VCP in vivo (Fig. 6e) and decreased localization of GFP-UBXN10 to cilia, despite equal levels of expression (Fig. 6f,g). These data indicate that UBXN10 requires VCP interaction to localize within cilia.
VCP-UBXN10 is required for ciliogenesis
Loss of IFT-B subunits leads to the loss or shortening of cilia whereas loss of IFT-A results in malformed cilia that accumulate IFT proteins35,36. Therefore, we examined the involvement of VCP and UBXN10 in cilia formation and maintenance. Depletion of either VCP or UBXN10 in hTERT-RPE1 cells with multiple siRNAs resulted in a reduction in the number of cilia (ascertained by acetylated tubulin staining) (Fig. 7a). Similar results were observed in LLC-PK1 and hTERT-RPE1 cells expressing GFP-UBXN10 (Supplementary Fig. 6a,b). Cell cycle analysis upon depletion of VCP or UBXN10 did not identify any overt delay in G0/G1 accumulation (Fig. 7b), indicating that the phenotypes observed are not a result of secondary cell cycle effects previously observed upon VCP depletion. Co-depletion of UFD1L-NPLOC4 by siRNA led to a 15-18% reduction in ciliogenesis (Fig. 7c).
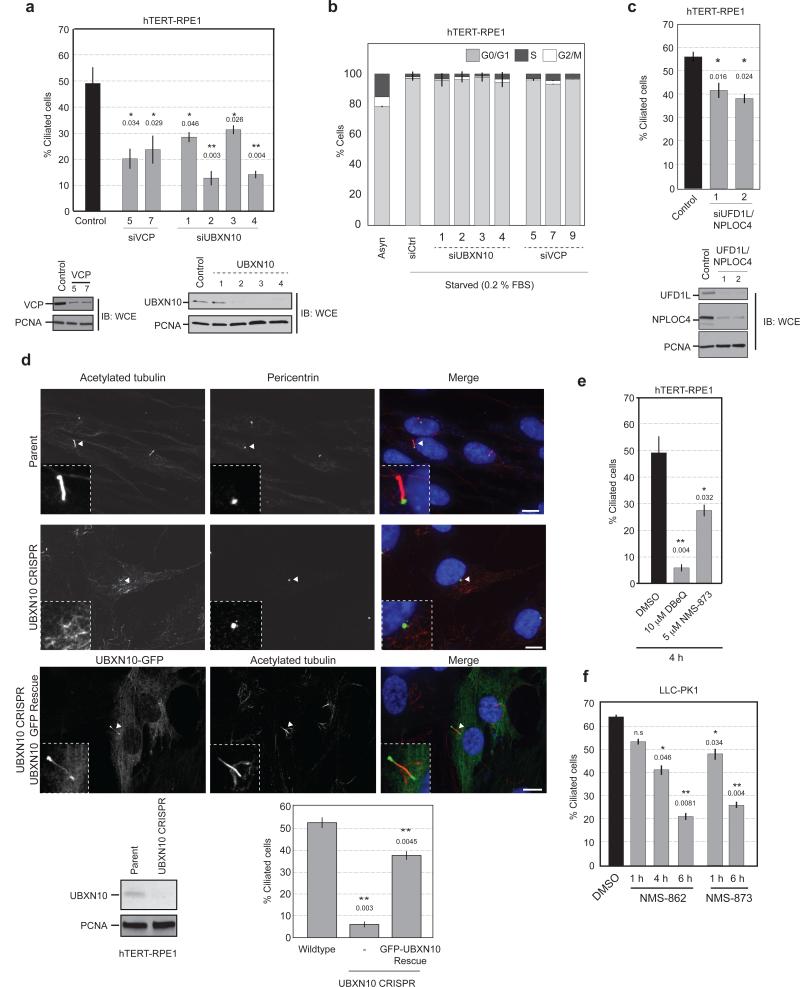
(a) hTERT-RPE1 cells were transfected with siRNAs targeting VCP or UBXN10. Cells were fixed and stained with acetylated tubulin and the number of cilia were quantified. n = 268 (siControl), 220 (siVCP-5), 208 (siVCP-7), 129 (siUBXD3-1), 136 (siUBXD3-2), 142 (siUBXD3-3), 157 (siUBXD3-4) pooled from 2 independent experiments. Error bars represent mean ± s.e.m. Statistical significance calculated using unpaired, two-tailed Students t-test, p values compared to siControl are shown **/*: p≤ 0.01, 0.05 respectively.
(b) Cell cycle analysis of hTERT-RPE1 cells depleted of VCP or UBXN10 analyzed by flow cytometry. n=30,000 live gated cells from 3 biological replicates. Error bars represent mean ± s.d.
(c) hTERT-RPE1 cells were depleted of UFD1L, NPLOC4 withANOVA 2 separate siRNA pairs. The percentage of ciliated cells was determined. n = 165 (siControl), 110 (siUFD1L/NPLOC4-1), 112 (siUFD1L/NPLOC4-2) pooled from 2 independent experiments. Error bars represent mean ± s.e.m. Statistical significance calculated using unpaired, two-tailed Students t-test, p values compared to siControl are shown. *: p≤ 0.05.
(d) UBXN10 knockout hTERT-RPE1 cells generated via CRISPR-CAS9 have impaired ciliogenesis. Re-expression of GFP-UBXN10 reinstates ciliogenesis. The number of ciliated cells were quantified as before. n = 263 (Parent), 218 (UBXN10 CRISPR), 204 (UBXN10-GFP Rescue) pooled from 3 independent experiments. Error bars represent mean ± s.e.m. Statistical significance was calculated using unpaired two-tailed Students t-test. p values compared to Parent are shown. **: p≤ 0.01. Scale bar, 10 μm.
(e,f) hTERT-RPE1 (e) or LLC-PK1 were starved for 48 h. Cells were treated with the indicated VCP inhibitors, and the percentage of ciliated cells was determined as before. n (hTERT-RPE1) = 121 (DMSO), 145 (DBeQ), 134 (NMS-873). n(LLC-PK1) = 154 (DMSO), 132 (NMS-862, 1h), 149 (NMS-862, 4h), 121 (NMS-862, 6h), 138 (NMS-873, 1h), 161 (NMS-873, 6h) pooled from 2 independent experiments. Error bars represent mean ± s.e.m. Statistical significance was calculated using unpaired, two-tailed Students t-test, p values compared to DMSO are shown. n.s, not significant. **/*: p≤ 0.01, 0.05. Uncropped images of blots are shown is Supplementary Fig. 8.
To definitively implicate UBXN10 in ciliogenesis, we used CRISPR-Cas9 to disrupt the UBXN10 gene in hTERT-RPE1 cells and observed a significant decrease in the number of cilia (Fig. 7d, Supplementary Fig. 6c). This defect was rescued by expression of GFP-UBXN10, that correctly localized to the cilium (Fig. 7d, Supplementary Fig. 6c).
We also considered the possibility that VCP-UBXN10 was required for the maintenance of pre-formed cilia. Because this scenario could not be addressed via prolonged siRNA depletion or deletion studies, we acutely inhibited VCP activity in cells using specific small molecule inhibitors37,38. hTERT-RPE1 or LLC-PK1 cells were acutely treated with DMSO, the ATP-competitive inhibitor DBeQ, or the allosteric inhibitors NMS-862 and NMS-873 (Supplementary Fig. 6d) and the percentage of ciliated cells was determined. Within 1 hour of treatment, loss of cilia could be discerned in both cell types (Fig. 7e,f). At time points longer than 6 hours it was difficult to find cilia for quantification (Fig. 7e,f). These results demonstrate a requirement of VCP and UBXN10 in the formation and maintenance of cilia.
The stability and activity of IFT-B is dependent on VCP catalytic activity
IFT complexes move with well-defined velocities in a cell-type dependent manner. To test whether VCP function is required for IFT-B trafficking, we combined small molecule VCP inhibitors with time-lapse imaging of EYFP-IFT88 and CSAP-Cherry (a centriolar marker used for ciliary orientation) in IMCD cells. Treatment of cells with VCP inhibitors for 4-6 hours led to a dramatic reduction in the length and number of cilia (Fig. 8a, Supplementary Fig. 7b). Therefore, ciliated cells were treated acutely with a high concentration of inhibitors to achieve physiologically relevant levels in a rapid time frame (<5 min) and cells immediately imaged to measure IFT velocity (Supplementary Fig. 6d). Acute inhibition caused a significant increase in anterograde and retrograde velocities of EYFP-IFT88 (Fig. 8b, Supplementary Fig. 7a). These results are consistent with studies in Chlamydomonas suggesting disassembly of cilia increases IFT-B velocities possibly due to the lack of cargo on IFT-B39. At longer times with lower concentrations of DBeQ, the anterograde and retrograde velocities were moderately elevated but not to the extent seen with acute treatment. However, under these conditions, there was also a discernable loss of cilia (Fig. 8a,b and Supplementary Fig. 7a). We asked if VCP inhibition impacted UBXN10 trafficking within cilia. TIRF microscopy in IMCD EYFP-IFT88-UBXN10-mCHERRY cells acutely treated with NMS-873 demonstrated a decrease in the number of cilia with UBXN10 trafficking compared to control cells, suggesting that UBXN10 may be uncoupled from IFT-B (Fig. 8c, Supplementary Fig. 7c). We also examined whether VCP inhibition altered IFT-A and BBSsome trafficking. We acutely treated IMCD GFP-IFT4340 cells with NMS-873 and observed a ~20% decrease in both anterograde and retrograde velocities of IFT-A (Figure 8d). Furthermore, the BBSome subunit BBS5 was depleted from cilia over time in response to VCP inhibition (Figure 8e,f). These studies suggest that VCP inhibition may alter the steady-state rates of protein transport within cilia.
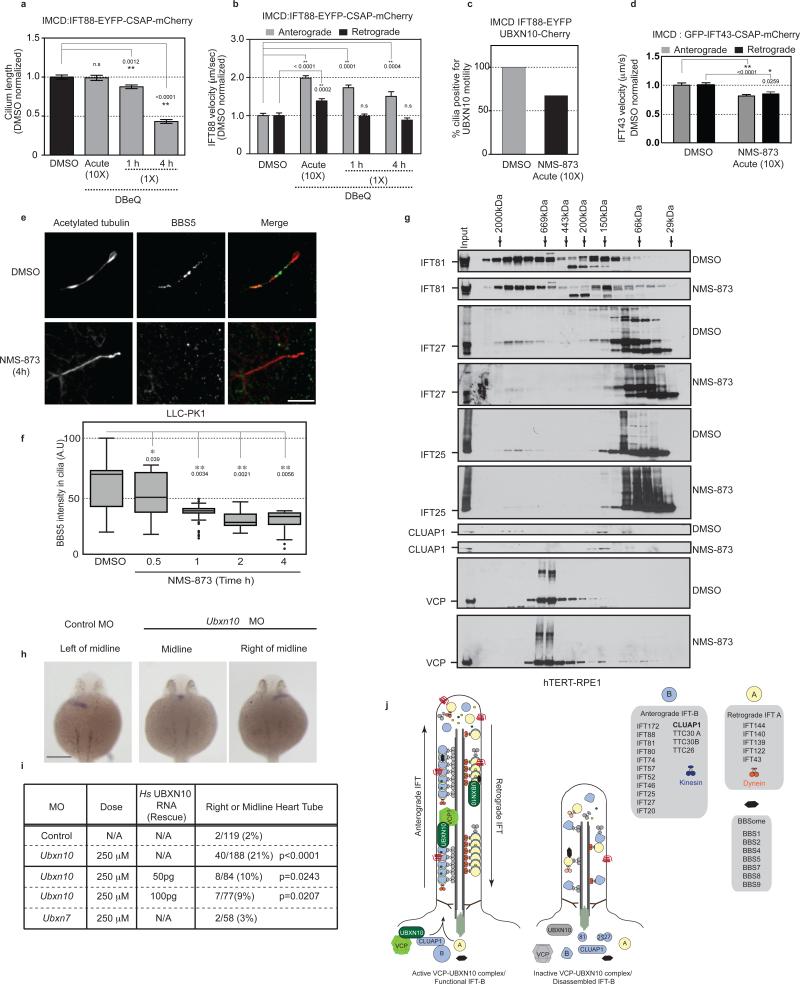
(a) Inhibition of VCP causes cilia shortening. Cells were treated with 5 μM (1X) or 50 μM (10X) DBeQ. n = 66 cells pooled from 2 independent experiments. Error bars represent mean ± s.e.m. Statistical significance calculated using ANOVA, p values shown. n.s, not significant **: p≤ 0.01.
(b) Acute treatment with DBeQ increases anterograde and retrograde IFT-88 velocities. n = 56 cells pooled from 2 independent experiments. Error bars represent mean ± s.e.m. Statistical significance calculated using ANOVA, p values shown. n.s, not significant. **: p≤ 0.01.
(c) VCP inhibition uncouples UBXN10 from IFT-B. Cells were treated with DMSO or 50 μM NMS-873 for < 5 min. Cilia that displayed motile EYFP-IFT88 particles were measured. The fraction of such cilia with observable UBXD3-Cherry motility was determined. n = 75 (DMSO) and 82 (NMS-873) cells.
(d) Acute treatment with NMS-873 decreases anterograde and retrograde IFT-43 velocities. n = 40 cells pooled from 2 independent experiments. Error bars represent mean ± s.e.m. Statistical significance calculated using ANOVA, p values shown. **/*: p≤ 0.01, 0.05.
(e,f) VCP inhibition causes loss of ciliary BBS5. Cells were treated with 5 μM NMS-873 and the intensity of BBS5 within cilia was quantified. n (left to right) = 75, 68, 69, 33, 42 cells pooled from 2 independent experiments. Maxima, centre, minima and quartiles (Q1,2,3) left to right : (100, 60, 22, 43, 70, 73), (77, 53, 20 37, 51, 71), (49, 37, 18, 37, 39, 41), (47, 31, 20, 27, 29, 36), (39, 31, 8, 28, 34, 37). Error bars represent mean ± s.d. Statistical significance calculated using ANOVA, p values shown. **/*: p≤ 0.01, 0.05. Scale bar, 5 μm.
(g) VCP inhibition leads to destabilization of IFT-B complex. Cells were treated with 5 μM NMS-873 for 4 h. Lysates were fractionated by gel filtration and probed for the indicated proteins. IFT25-27 specific bands determined by siRNA knockdown (Supplementary Fig. 7c).
(h,i) Depletion of zebrafish Ubxn10 causes defects in left/right asymmetry. Whole-mount in situ staining with the pan-cardiomyocyte marker cmlc2 demonstrating position of heart tube. Scale bar, 200 μm (h). Summary of 3 independent MO experiments. p values calculated using Fishers exact test (i).
(j) Model. Uncropped images of blots are shown is Supplementary Fig. 8.
Assembly and regulation of IFT complexes in vivo is poorly understood. We examined whether VCP inhibition affected the composition of IFT-B complexes. hTERT-RPE1 cells were treated with either DMSO or NMS-873 and lysates were fractionated by gel filtration. Immunoblotting for IFT-B subunits revealed multiple complexes, IFT81 and IFT57 are in two complexes at ~1-2 MDa and 150 KDa (Fig. 8g, Supplementary Fig. 7d). In contrast, heterodimeric IFT25 and IFT2741 were abundant at 66 KDa but were also detected in the 1-2 MDa complex. VCP inhibition resulted in a reduction of IFT81, IFT25 and IFT27 in the 1-2 MDa size fractions and an increase in smaller molecular weight fractions (Fig. 8g). Recent studies suggest that IFT25 and 27 exist in two pools, one that associates with the IFT-B complex and a heterodimeric form that regulates access of the BBSome to cilia42,43, which is recapitulated by our gel filtration results. CLUAP1 was present in a 1-2 MDa complex and a ~150 kDa complex (also occupied by UBXN10), and VCP inhibition resulted in a reduction in the 1-2 MDa form of CLUAP1 and the 150 kDa form of UBXN10 (Fig. 8g, Supplementary Fig. 7d). We noted that NMS-873 treatment resulted in a decrease in UBXN10 protein levels (Supplementary Fig. 7d), therefore we asked if VCP inhibition affected UBXD adaptor levels in cells. Treatment of LLC-PK1 cells with NMS-873 led to a reduction in UBXN10 as well as UBXN7 and FAF2 at 4-6 hours post inhibitor treatment (Supplementary Fig. 7f). However, we observed a rapid loss of ciliary GFP-UBXN10 within 1 hour of inhibitor treatment (when protein levels are unaltered relative to control) (Supplementary Fig. 7g). This suggests that VCP inhibition results in the loss of UBXN10 from cilia and that the decline in the adaptor levels maybe a secondary effect of prolonged inhibition. At present, we cannot determine if this effect is due to defects in entry or exit of UBXN10 from cilia. Taken together, these results suggest that VCP inhibition results in selective loss of a subset of proteins from the IFT-B complex.
Loss of UBXN10 in zebrafish results in loss of left-right asymmetry
In zebrafish, defects in ciliary function can be discerned by defects in left-right assymetry44 (Fig. 8h). To examine the involvement of UBXN10 in cilia-related functions in vivo, we depleted UBXN10 in one-cell zebrafish embryos using morpholinos (MO) and determined heart tube position by in-situ hybridization. In control embryos, the heart tube was positioned on the left of the body axis as expected. In contrast, 21% of the Ubxn10 MO embryos displayed abnormal heart positioning to the midline or to the right (Fig. 8h,i). Co-injection of human UBXN10 mRNA partially rescued the defect (9%) (Fig. 8i). We observed no significant defect in cardiac positioning using a Ubxn7 MO indicating specificity for Ubxn10 (Fig. 8i). These studies support our hypothesis that the VCPUBXN10 complex has conserved roles in ciliogenesis.
DISCUSSION
This work provides a comprehensive analysis of the VCP-UBXD protein interaction network, identifying over 195 HCIPs, and greatly expands the potential breadth of cellular VCP function by: (1) identifying stable interacting partners for UBXDs with no known function, (2) implicating an adaptor and associated complexes in a known VCP pathway thereby informing further mechanistic studies, and (3) identifying candidate targets for certain well-studied VCP adaptors. Further work is required to elucidate functional roles for modules identified here.
While a role for ubiquitin-dependent signaling has long been proposed in cilia-related processes45,46, only recently have E3s and candidate substrates been identified47–50. Our identification of a role for VCP-UBXN10 in cilia formation and maintenance at the cellular and organismal level was unexpected but suggests a role for VCP's segregase activity in either trafficking of IFT-B itself in cilia or in trafficking of IFT-B cargo. As UBXN10 has no known ubiquitin interaction domains, further studies are necessary to understand the role, if any, of ubiquitin in VCP-UBXN10 function within cilia.
We find that UBXN10 recruits VCP to IFT-B via direct interaction with CLUAP1 and that UBXN10 traffics within cilia along with IFT-B in a bi-directional manner. While previous proteomic studies detect VCP in purified cilia26,41, we have been unable to directly visualize VCP within cilia. However, since an interaction between VCP and UBXN10 is required for UBXN10 trafficking to the cilia, we favor the possibility that the VCP-UBXN10 complex traffics within cilia. Further studies are required to test whether VCP enters cilia and whether this requires UBXN10. VCP-UBXN10 depletion or pharmacological inhibition of VCP causes the shortening and eventual loss of cilia and our biochemical studies suggest that this maybe due in part to defects in the assembly of IFT25-IFT27 (and perhaps other subunits) within IFT-B. Furthermore, acute treatment of ciliated cells with VCP inhibitors led to increased anterograde and retrograde IFT-B velocities, whereas IFT-A velocities were decreased, indicating a mis-coordination of IFT particle trafficking. We also observe loss of BBS5 in cilia upon VCP inhibition. In C.elegans the BBSome is thought to link IFT-A and B complexes within cilia24, as genetic deletion of BBSome subunits results in loss of coordinated IFT-A and B trafficking.
Our results suggest that interfering with VCP activity leads to the acute loss of UBXN10 from primary cilia. Subsequently, we detect the disruption of IFT complexes biochemically. We speculate that loss of specific subunits from IFT-B may disable critical interactions with IFT-A, the BBSome complex, cargo and/or motors, leading to cilia shortening and eventual collapse (Figure 8j). Whether this regulation of IFT-B occurs within cilia or in the cell body is presently unclear and will be the focus of future studies. Our proteomic study identifies biologically relevant functions for VCP adaptors and provides a rich resource for the further exploration of VCP function in numerous cellular processes.
Online Methods
Cell Culture and generation of cell lines
HEK293T and HeLa were from ATCC. The following cell lines were originally from ATCC but were gifts from the specified groups. hTERT-RPE1 - from Prasad Jallepalli (MSKCC) - verified for human by MS/MS. Mouse Inner Medullary Collecting Duct (IMCD) - from David M. Cohen (Oregon Health & Science University) - verified for mouse by MS/MS. Pig kidney epithelial (LLC-PK1) cells - from Joseph Bonventre (Brigham and Women's Hospital) - not species verified. All cell lines were negative for mycoplasma based on PCR. No cell lines used in this paper were listed in the ICLAC database of commonly misidentified cell lines
Cell lines stably expressing EYFP-IFT88 were generated using a blasticidin-resistant retroviral vector as previously described51. The fluorescently tagged centriole and spindle–associated protein (CSAP)52 was utilized as a ciliary orientation marker. The mCherry tagged CSAP was cloned into a hygromycin-resistant retroviral pBABE vector and introduced into IMCD3 and LLC-PK1 as described previously51. Xenopus laevis IFT43 was subcloned into blasticidin resistant pHAGE N-GFP vector. UBXN10 was cloned into lentiviral vectors to be expressed as a N-EGFP or C-mCHERRY tagged protein. Stable cell lines were selected with blasticidin.
All cell lines were cultured in Dulbecco's modified Eagle's medium, supplemented with 10 % fetal bovine serum (FBS), GlutaMAX, 100 units mL−1, penicillin, 0.1 mg ml−1 streptomycin, and 0.1 mM nonessential amino acids. Cells were maintained in a humidified, 5.0% CO2 atmosphere at 37 °C. hTERT-RPE1, IMCD3 (clone m368-2) and LLC-PK1 (clone p368-7) cells stably expressing EYFP-IFT88 and mCherry-CSAP were transferred from a culture flask after detachment using Trypsin-EGTA into 18 mm diameter glass coverslips (No. 1.5, Electron Microscopy Sciences, Hatfield, PA). The cells were grown to high confluency to promote the highest frequency of ciliation.
Generation of CRISPR cell lines
The CRISPR-Cas9 gene editing system was used to generate UBXN10 knockout cell lines in hTERT-RPE1 cells. The guide sequence 5’ ATAAGGCCCAAGTCCGCCAA 3’ was cloned into the pX330 vector carrying the hSpCas9 and transiently transfected into hTERT-RPE1 cells along with a pHAGE-HA/FLAG-GFP carrying a blasticidin resistance cassette using Lipofectamine 3000 (Invitrogen). 36 h post-transfection the cells were pulsed briefly with 5 μg ml−1 blasticidin for a further 24-36 h. The surviving cells were allowed to recover in drug-free media for 24 h and the serially diluted into 96 well plates for clonal selection. CRISPR targeted clones were expanded and UBXN10 locus was PCR amplified and Sanger sequenced. We identified a frame-shift at Pro12 that introduces a premature stop codon at the endogenous UBXN10 locus.
Interaction Proteomics and Mass Spectrometry Analysis
Interaction proteomics was performed as described previously12,13. Briefly, 293T or HeLa cells were transduced with a lentiviral vector expressing either N- or C-HA/FLAG tagged VCP or its UBX-domain containing adaptors and stable cell lines selected with puromycin. HEK-293T cells from 8 × 15 cm dishes were harvested for duplicate α-FLAG AP/MS studies. The biological duplicates were processed in parallel as follows. Cells were lysed in 3 ml of 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5 % Nonidet P40, 1 mM DTT and protease and phosphatase inhibitors. Clarified lysates were filtered through 0.45 μm spin filters (Millipore Ultrafree-CL) and immunoprecipitated with 60 μl anti-FLAG magnetic beads per replicate (Sigma). Complexes were washed 4× with lysis buffer, and 3× with PBS and eluted with FLAG peptide at room temperature. Elutions were reductively carboxymethylated, and TCA precipitated. TCA-precipitated proteins were trypsinized, purified with Empore C18 extraction media (3 M), and analyzed via LC-MS/MS with a LTQ-Velos linear ion trap mass spectrometer (Thermo) with an 18 cm3 125 μm (ID) C18 column and a 50 min 8%–26% acetonitrile gradient. Complexes were analyzed twice by LC-MS to generate technical duplicates. AP-MS experiments using HA resin (Sigma) were also performed on a single IP but with technical duplicates. Spectra were searched with Sequest against a target-decoy human tryptic UNIPROT-based peptide database, and these results were loaded into the Comparative Proteomics Analysis Software Suite (CompPASS)12,13, to identify high confidence candidate interacting proteins (HCIPs). Individual experiments were analyzed using a stats table derived from analogous AP-MS data for 41 (for FLAG AP/MS) or 168 (for HA AP/MS) unrelated proteins to determine normalized weighted D-scores (NWD-score) and Z-scores based on spectral counts.
To identify bait-associated proteins, proteins were filtered at a 3% false discovery rate for those with a NWD-score ≥ 0.98, Z-score ≥ 4, and average assembled peptide spectral matches (APSMs) ≥ 2 in both biological duplicates (for FLAG AP/MS). An identical cut-off was used for the single HA AP/MS for each tagged bait. Baits were removed from all HCIP lists prior to HCIP calculation. To evaluate the statistical significance of each bait-prey interaction, a hypergeometric test was applied comparing the null hypothesis that detection of bait and prey was independent across LC-MS analyses, versus the alternative hypothesis that bait and prey were detected preferentially together (Supplementary Table 4). For purposes of this analysis, AP-MS data were separated according to the location of the affinity tag (N- versus C- terminus) and according to whether HA or FLAG purification was performed. P-values were subsequently corrected to account for multiple hypothesis testing using the method of Benjamini and Hochberg53. HCIPs designated in this study generally had adjusted p values ≤ 0.05 (Supplementary Table 4). The only exceptions were for interactors that associated with VCP itself and for UBXD-UBXD associations. Because the very abundant VCP protein (~1% of the total cellular protein) was ubiquitously detected across FLAG AP-MS analyses, detection-based statistical analyses such as the hypergeometric test described here will be insensitive to associations with its interactors. Some UBXD-UBXD associations similarly fail to meet significance cutoffs in this statistical analysis due to high frequency of detection across these LC-MS analyses. This list of HCIPs from the α-FLAG dataset was cross-referenced with the interactors in the α-HA dataset and interaction maps were generated via Cytoscape. In situations where known members of an associating complex were identified in the IP but did not pass put cut-offs, these were included in Fig. 3 and Supplementary Figure 3. APSMs for the HCIPs identified in the FLAG datasets were bait normalized and plotted in a heatmap form using Multi-experiment Viewer (MeV).
Validation Studies
A subset of interactors were used to reciprocally validate the interaction with either VCP or the UBXD adaptor. Interactors were cloned into lentiviral vectors containing a N-HA/FLAG tag. Stable cell lines were generated for 7 interactors and analyzed by AP-MS as described above. IFT-B stable cell lines were generated in hTERT-RPE1 cells. Nineteen interactors were cloned into epitope tagged vectors and transiently transfected with its cognate tagged UBXD into 293T cells. 48 h post-transfection, cells were lysed and clarified lysates were used to immunoprecipitate the tagged HCIP and immunoblotted for the UBXD protein.
Antibodies, siRNAs and Reagents
The rabbit VCP (Cat # A300-588A, 1:1000 dilution), rabbit UBXN7 (Cat # A303-865A, 1:1000 dilution), rabbit FAF1 antibody (Cat # A302-810A, 1:1000 dilution) were from Bethyl. Rabbit FAF2 (Cat # S0495, 1:1000 dilution) was from Epitomics. Rabbit UBXN1 (Cat # AB10041, 1:1000 dilution) was from Millipore. Rabbit UBXN10 antibody was from Sigma (Cat # HPA028564, 1:500 dilution), mouse UFD1L antibody was from BD Biosciences (Cat # 611642, 1:1000 dilution), rabbit NPLOC4 (Cat # HPA021650, 1:1000 dilution) and mouse Acetylated tubulin (Cat # T7451 clone 6-11B-1, 1:10,000 dilution) antibodies were from Sigma, Mouse HA antibody was from Covance (Cat # MMS-101P, clone 16B12, 1:1000 dilution), mouse PCNA (Cat # SC-56, clone PC10, 1:1000 dilution), mouse MYC (Cat # SC-40, Clone 9E10, 1:1000), Tomm20 (Cat # SC-11415, 1:500 dilution) and rabbit γ-tubulin (Cat # SC-10732, 1:500 dilution) antibodies were from Santa Cruz. Rabbit Pericentrin antibody was from Abcam (Cat # AB4448, 1:500 dilution). IFT81 (Cat # 11744-1-AP, 1:1000 dilution), IFT57 (Cat # 11083-1-AP, 1:1000 dilution), IFT25 (Cat # 15732-1-AP, 1:1000 dilution), IFT27 (Cat # 15017-1-AP, 1:1000 dilution) and BBS5 (Cat # 14569-1-AP, 1:100 dilution) rabbit antibodies were from Proteintech. Rabbit Cluap1 was from Genetex (Cat # GTX106897, 1:500 dilution). (DBeQ was purchased from Biovision, NMS-862 was from Ryan Scientific and NMS-873 was from Glixx Labs. Sequences of siRNAs from Dharmacon (Thermo). Sequences (5’-3’) for siRNAs: UBXN10-1 (CAGCCAAACUCACUAAAAUA), UBXN10-2 (GGUGUGCGCUCAUCAUAUA), UBXN10-3 (UACCGACACUGCAGCAUUG), UBXN10-4 (GCUGCUUGCUGUUAGAUCA), IFT25 (SMARTpool siRNA, Cat. # M-010908-01-0005), IFT27 (SMARTpool siRNA, Cat. # M-009565-00-0005), UFD1L-1 (AAUCAAGCCUGGAGAUAUU), UFD1L-2 (GACCAAACCCGACAAGGGCA), NPLOC4-1 (AAUAAUGGCUUCUCGGUUU), NPLOC4-2 (GGACACCUAUUUCCUAAGU). VCP siRNAs were from Invitrogen, VCP-5 (ACUUCCUGAUGAGAAGUCCCGUGUU), VCP-7 (CCCAAGAUGGAUGAAUUGCAGUUGU).
In vitro Translation Reactions
For HeLa in vitro translation reactions the indicated proteins (MYC-IFT-B subunits and HA-UBXN10) were cloned into pT7CFE Gateway compatible vectors and in vitro translated in HeLa lysate using the 1-Step couple in vitro expression system (Thermo). Samples were incubated at 30 °C for 5 h and centrifuged briefly to remove any aggregated material. 20 % of the reaction was removed for input loading and the remaining reaction was diluted and used for immunoprecipitation reactions. Samples were incubated with MYC agarose overnight, washed in lysis buffer and resolved on a gel. Membranes were probed with HA antibody.
GST, GST-UBXN10 or GST-VCP were expressed from the pDEST15 N-GST Gateway compatible vector. Rosetta cells transformed with the indicated constructs were induced to express protein with 1 mM IPTG overnight at 30 °C. Cell pellets were lysed in 50 mM Tris Cl [pH 7.8], 150 mM KCl, 0.1% NP-40, 1 mM DTT and protease inhibitors. Lysates were sonicated briefly and clarified by centrifugation. Proteins were purified using GSH agarose (Novagen) and left coupled to beads in storage buffer containing 50 % glycerol. GST-VCP was incubated with in vitro-translated HA-UBXN10 for 1 h on ice.
IFT-B subunits in pT7CFE vectors were translated in rabbit reticulocyte lysate (Promega) in the presence of 5 μCi of 35S (Perkin Elmer) for 1 hour are 30 °C. 15 % of the reaction was saved as input and the remaining reaction was diluted and incubated with GST tagged proteins for 2 h at 4 °C. The beads were extensively washed and the samples then resolved by SDS PAGE. Interactions were visualized by autoradiography. The experiment was performed 2 independent times.
Immunofluorescence and Microscopy
HeLa, hTERT-RPE1, IMCD or LLC-PK1 cells were grown on cover slips and fixed with 4 % paraformaldehyde at room temperature for 15 min. Cells were washed in PBS, permeabilized in 0.2 % Triton X-100 for 5 min, washed again and then blocked in 1 % BSA in PBS for 1 h. The coverslips were incubated with the indicated antibodies for 1 h in a humidified chamber, washed and incubated for a further hour with appropriate Alexa-Fluor conjugated secondary antibodies (Invitrogen) for 1 h in the dark. Cells were washed with PBS, nuclei were stained with Hoechst and mounted on slides. All images were collected with a Yokogawa CSU-X1 spinning disk confocal on a Nikon Ti-E inverted microscope equipped with 100× Plan Apo NA 1.4 objective lens. The indicated fluorophores were excited with either a 491 nm, 561 nm or 642 nm laser line via an AOTF controlled solid-state laser launch. Emission spectra were collected with a quad band pass polychroic mirror and Chroma emissions filters ET525/50m (488nm), ET620/60m (561) and ET700/75 (642). Images were acquired with a Hamamatsu ORCA-AG cooled CCD camera controlled with MetaMorph 7 software. 10-15 z-series optical sections were collected with a step size of 0.2 microns using a Prior Proscan focus motor. Z-series were deconvolved using AutoQuant blind deconvolution software, and displayed as maximum z-projections. Gamma and brightness were adjusted on displayed images (identically for compared image sets using MetaMorph 7 software). Ciliated cells were counted based on acetylated tubulin staining using Pericentrin as an orientation marker. To determine BBS5 localization to cilia, the intensity of BBS5 within cilia (based on the acetylated tubulin staining) was quantified and normalized to the length of each cilium measured in MetaMorph. For immunoflourescene based quantification studies, fields of cells were chosen based on the Hoechst (nuclei) channel to prevent biased image acquisition. On average 150-350 cells were quantified for each sample in two separate experiments. All experiments were performed at least 2 independent times.
IFT velocity measurements and TIRF
Velocity measurements of IFT88, IFT43 and UBXN10 were performed as described previously51. The cells were rinsed with PBS and subjected to the transport measurements by inversion of the monolayer onto a larger size (35 mm) coverslip and mounted on the stage of a Nikon Eclipse Ti inverted microscope equipped with a Nikon Plan Fluor 100×A 1.4 NA objective lens and a CoolSnap-HQ (Photometrics, Tucson, AZ). The Nikon FITC and HcRED1 cubes were used to efficiently reflect 488 nm and 575 nm wavelengths correspondingly and pass the emission wavelengths into the CCD camera detection channel. The acquisition settings were kept constant for all samples so that valid comparisons could be made between measurements from different data sets. Acquisition parameters were set within the linear range of the CCD camera detection. The acquired fluorescence image time series were subjected to the kymograph analysis using the ImageJ software package (http://rsbweb.nih.gov/ij). The standard deviations of the recovered mean values were obtained from the analysis of the trajectories of multiple particles.
The ciliary particle transport in cells stably co-expressing EYFP-IFT88 and UBXN10-mCherry was sampled via total internal reflection fluorescence (TIRF) microscopy. Similarly to EPI-fluorescence imaging, coverslips were mounted on the stage of a Nikon Eclipse Ti inverted microscope equipped with a Nikon motorized TIRF illuminator and a Nikon CFI Apo TIRF 100X oil 1.49 NA objective lens. The fluorophores were excited using 491nm and 561nm 50mW solid state lasers coupled together using a laser merge module with acousto-optic tunable filter (Spectral Applied Research, Richmond Hill, ON). TIRF image time series were acquired with a Hamamatsu ORCA-D2 CCD camera controlled with MetaMorph 7 software. For particle velocity measurement, one hundred images were continuously (49.44 ms read-out time per frame) collected using an exposure time of 100 ms and 2×2 binning. The acquisition parameters were set within the linear range of the CCD camera detection. The acquired fluorescence image time series were subjected to the kymograph analysis of the ImageJ software package (http://rsbweb.nih.gov/ij). The standard deviations of the recovered mean values were obtained from the analysis of the trajectories of multiple particles.
For drug treatments, cells on coverslips were treated with either 50 μM DBeQ or NMS-862 or NMS-873, inverted immediately onto 35mm coverslip and immediately imaged. These measurements lasted for 5 min. For longer incubations, cells were treated with either 5 μM (NMS-862 or NMS-873) or 10 μM (DBeQ) for 1, 4 or 6 h. These experiments were performed 2 independent times.
Morpholino-based Studies in Danio rerio
Zebrafish were maintained according to established IACUC protocols. Wild type (strain AB) zebrafish embryos were used. Embryos were used from 0-26 hpf, at which time the sex of the animal is not known. Zebrafish male and female adults used in matings were < 1 year old. Embryos were injected at the 1-cell stage with morpholinos (MOs) (GeneTools) designed to block either the ATG start site or an exon-intron splice site of the target gene (Ubxn10 MO, ATG start site, 5’ ACGTGCATCCTTTCTCCAGAGGACA 3’, Ubxn7 MO, splice site 5′ TTTTGGATTCTCCACCCGAAGCCAT 3’). MOs were injected at concentrations of 250-330 μM. Hs GST-UBXN10 RNA (50-100 pg) was injected into embryos along with the morpholino for rescue studies. Embryos were fixed in 4 % PFA at 26 h post-fertilization (hpf) for in situ hybridization using published methods (http://zfin.org/ZFIN/Methods/ThisseProtocol.html). Gene expression was visualized using the pan-cardiomyocyte marker cmlc2. Zebrafish embryos from a single clutch were allocated to experimental groups (i.e., control and morpholino-injected groups) at the 1-cell stage prior to MO injection, at which time they are indistinguishable from one another and the genetic makeup of each embryo is unknown (zebrafish are not isogenic). The number of morphant embryos displaying abnormal gene expression (cardiac jogging laterality defects) was compared to control embryos, and statistical significance was determined by Fisher's exact test. These experiments were performed 3 independent times.
Gel Filtration Analysis
hTERT-RPE1 cells were grown in 15 cm dishes and starved in 0.2 % FBS for 48 h. Cells were treated with either DMSO or 5 μM NMS-873 (in DMSO) for 4 h. Cells were harvested and lysed in 1 ml of lysis buffer 50 mM HEPES pH 7.4, 150 mM KCl, 1mM EGTA, 1 mM MgCl2, 10 % glycerol, and 0.05 % NP-40. Cells were centrifuged for 1 h at 13,000 rpm. Samples were loaded on a Superose 6 size 10/300 GL column (GE Health Sciences) and 0.5 ml fractions were collected. Total protein from each fraction was TCA precipitated and washed with acetone 3 times. Samples were dried and resuspended in SDS sample buffer. Equal volumes of fractions were resolved by SDS PAGE and samples were resolved on SDS PAGE and immunoblotted with the indicated antibodies.
Statistical Analyses
No statistical method was used to predetermine sample size. Sample sizes for all relevant studies were chosen based on previously published studies our groups. The experiments were not randomized. For immunoflourescene-based quantification studies, fields of cells were chosen based on the Hoechst (nuclei) channel to prevent biased image acquisition. The investigators were not blinded to allocation during experiments and outcome assessment. When gene expression data was gathered from experimental groups following in situ hybridization for cmlc2, the researcher knowingly scored the control group first to determine what normal gene expression looks like
Supplementary Material
1
2
3
4
5
6
7
8
9
ACKNOWLEDGMENTS
This work was supported by NIH grants R37-NS083524 and RO1-AG011085 to J.W.H. We would like to thank the Nikon Imaging Center (Harvard Medical School) for microscopy assistance. We would like to thank John Wallingford (UT Austin) for the Xenopus laevis IFT-43 construct and Maxene Nachury and Andrew Nager (Stanford) for helpful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
J.W.H. and M.R. conceived the project. M.R. prepared cell lines, performed immunofluorescence in cells and zebrafish, cilia length measurements, performed and analyzed AP-MS experiments (with the assistance of J.R.L), and performed biochemical studies. M.S. performed cilia trafficking experiments under the direction of J.S. M.G. performed zebrafish experiments under the direction of W.G. E.L.H performed the statistical analysis to determine p values for the MS dataset, M.R. and J.W.H. wrote the paper with input from all authors.
Datasets and Accession Codes
The UBXN10 UBX domain was modeled onto the crystal structure of the FAF1 UBX domain in complex with the VCP N-terminus (PDB accession code 3QWZ) using Phyre2.0 (http://www.sbg.bio.ic.ac.uk/phyre2/html).
All MS data has been deposited on PeptideAtlas and can be accessed via: http://www.peptideatlas.org/PASS/PASS00701.
COMPETING FINANCIAL INTERESTS
J.W.H. is a consultant for Biogen and Millennium: The Takeda Oncology Company.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb3238
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4610257
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
AAA+ ATPase chaperone p97/VCP<sup>FAF2</sup> governs basal pexophagy.
Nat Commun, 15(1):9347, 29 Oct 2024
Cited by: 0 articles | PMID: 39472561 | PMCID: PMC11522385
VCP/p97-associated proteins are binders and debranching enzymes of K48-K63-branched ubiquitin chains.
Nat Struct Mol Biol, 08 Jul 2024
Cited by: 0 articles | PMID: 38977901
Emerging insights into CP110 removal during early steps of ciliogenesis.
J Cell Sci, 137(4):jcs261579, 28 Feb 2024
Cited by: 0 articles | PMID: 38415788
Review
Exploring the role of ubiquitin regulatory X domain family proteins in cancers: bioinformatics insights, mechanisms, and implications for therapy.
J Transl Med, 22(1):157, 15 Feb 2024
Cited by: 0 articles | PMID: 38365777 | PMCID: PMC10870615
Review Free full text in Europe PMC
VCF1 is a p97/VCP cofactor promoting recognition of ubiquitylated p97-UFD1-NPL4 substrates.
Nat Commun, 15(1):2459, 19 Mar 2024
Cited by: 1 article | PMID: 38503733 | PMCID: PMC10950897
Go to all (57) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
HPA - The Human Protein Atlas
- (1 citation) HPA - HPA028564
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Corrigendum: Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis.
Nat Cell Biol, 18(3):347, 01 Mar 2016
Cited by: 1 article | PMID: 26911913
Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations.
Nat Cell Biol, 13(9):1116-1123, 07 Aug 2011
Cited by: 155 articles | PMID: 21822278 | PMCID: PMC3246400
UBXD1 is a VCP-interacting protein that is involved in ER-associated degradation.
Biochem Biophys Res Commun, 382(2):303-308, 09 Mar 2009
Cited by: 25 articles | PMID: 19275885
Insights into adaptor binding to the AAA protein p97.
Biochem Soc Trans, 36(pt 1):62-67, 01 Feb 2008
Cited by: 94 articles | PMID: 18208387
Review
Funding
Funders who supported this work.
NIA NIH HHS (2)
Grant ID: R01-AG011085
Grant ID: R01 AG011085
NINDS NIH HHS (2)
Grant ID: R37-NS083524
Grant ID: R37 NS083524





