Abstract
Free full text

Engineered bacterial outer membrane vesicles with enhanced functionality
Abstract
We have engineered bacterial outer membrane vesicles (OMVs) with dramatically enhanced functionality by fusing several heterologous proteins to the vesicle-associated toxin ClyA of Escherichia coli. Similar to native unfused ClyA, chimeric ClyA fusion proteins were found localized in bacterial OMVs and retained activity of the fusion partners, demonstrating for the first time that ClyA can be used to co-localize fully functional heterologous proteins directly in bacterial OMVs. For instance, fusions of ClyA to the enzymes β-lactamase and organophosphorus hydrolase resulted in synthetic OMVs that were capable of hydrolyzing β-lactam antibiotics and paraoxon, respectively. Similarly, expression of an anti-digoxin single-chain Fv antibody fragment fused to the C-terminus of ClyA resulted in designer “immuno-MVs” that could bind tightly and specifically to the antibody’s cognate antigen. Finally, OMVs displaying green fluorescent protein fused to the C-terminus of ClyA were highly fluorescent and, as a result of this new functionality, could be easily tracked during vesicle interaction with human epithelial cells. We expect that the relative plasticity exhibited by ClyA as a fusion partner should prove useful for: (i) further mechanistic studies to identify the vesiculation machinery that regulates OMV secretion and to map the intracellular routing of ClyA-containing OMVs during invasion of host cells; and (ii) biotechnology applications such as surface display of proteins and delivery of biologics.
Introduction
Protein translocation is a highly conserved process that is essential to all life. Extracellular secretion of virulence factors is a strategy utilized by invading bacteria to establish a colonization niche, communicate with host cells, and modulate host defense and response. With few exceptions, bacterial protein secretion systems are characterized by the membrane translocation of a single protein or else small protein complexes.1; 2; 3; 4; 5 Recently, however, the production and release of outer membrane vesicles (OMVs) has been demonstrated as a novel secretion mechanism for the transmission of a diverse group of proteins and lipids to mammalian cells.6 OMVs are small proteoliposomes with an average diameter of 50–200 nm that are constitutively released from the outer membrane of pathogenic and non-pathogenic species of Gram-negative bacteria during growth.7 Biochemical analysis has demonstrated that OMVs are comprised of outer membrane proteins, lipopolysaccharide, phospholipids and soluble periplasmic proteins,8; 9 the latter of which become trapped in the vesicle lumen during release from the cell surface. OMVs are largely devoid of inner membrane and cytoplasm components although several studies indicate that chromosomal, phage and plasmid DNA can infiltrate OMVs as a means of OMV-mediated transfer of genetic information between bacteria.10; 11; 12; 13
An intriguing yet poorly understood phenomena pertaining to OMVs is the observation that certain membrane and/or soluble periplasmic proteins are enriched in vesicles while others are preferentially excluded. The majority of these enriched proteins happen to be virulence factors including, for example, Escherichia coli cytolysin A (ClyA),14 enterotoxigenic E. coli heat labile enterotoxin (LT),8 and Actinobacillus actinomycetemcomitans leukotoxin,15 whereas proteins that are excluded from OMVs include numerous unidentified outer membrane (OM) proteins15 as well as E. coli DsbA.14 The preferential exclusion of proteins raises the interesting possibility that a yet-to-be determined sorting mechanism exists in the bacterial periplasm for discriminatory loading of a highly specific subset of proteins into OMVs.14; 16 Moreover, the observation that certain virulence factors are enriched in vesicles suggests that OMVs may play a key role in bacterial pathogenesis by mediating transmission of active virulence factors and other bacterial envelope components to host cells. Indeed, numerous vesicle-associated virulence factors (e.g., adhesins, immunomodulatory compounds, proteases and toxins) have been shown to induce cytotoxicity, confer vesicle binding to and invasion of host cells, and modulate the host immune response.8; 17; 18; 19; 20
To date, one of the best studied vesicle-associated virulence factors is the 34-kDa cytotoxin ClyA (also called HlyE or SheA) found in pathogenic and non-pathogenic E. coli strains 14; 21 and also in Salmonella enterica serovars Typhi and Paratyphi A.22 Structural studies indicate that the water-soluble form of ClyA is a bundle of four major α-helices, with a small surface-exposed hydrophobic beta-hairpin at the “head” end of the structure, and the N- and C-termini at the “tail” end23 while lipid-associated ClyA forms an oligomeric pore complex comprised of either 8 or 13 ClyA subunits.24; 25 Expression of the clyA gene is silenced in non-pathogenic E. coli K-12 laboratory strains by the nucleoid protein H-NS26 but is derepressed in H-NS-deficient E. coli, thereby inducing cytotoxicity towards cultured mammalian cells.27 More recent evidence indicates that ClyA is exported from E. coli in OMVs and retains a cytolytically active, oligomeric conformation in the vesicles.14 However, the route by which ClyA manages to cross the bacterial IM and assemble in OMVs remains a mystery, as it carries no canonical signal peptide21 and is not N-terminally processed.28 Also undetermined is the role that ClyA plays in vesicle-mediated interactions with mammalian cells. Thus, in the present study, we sought to engineer synthetic membrane vesicles (s-MVs) with non-native functions that could be used for a wide range of applications including, for instance, the analysis of the complete ClyA translocation process. Specifically, we have programmed s-MVs with enhanced functionality by creating chimeras between heterologous proteins such as green fluorescent protein (GFP) or β-lactamase (Bla) and ClyA. Using these engineered vesicles, we have determined that ClyA is capable of co-localizing a variety of structurally diverse fusion partners to the surface of E. coli and their released vesicles, but only when the periplasmic disulfide bond-forming machinery was present. Importantly, these cell- and OMV-associated proteins retained their biological activity, suggesting that the functionality of natural OMVs can be easily expanded via the expression of ClyA chimeras.
Results
GFP co-localizes in outer membrane vesicles when fused to ClyA
Previous studies demonstrated that genetic fusions between E. coli ClyA and reporter proteins such as Bla and GFP were efficiently translocated across the cytoplasmic membrane29; 30 and that localization was independent of the position (N- or C-terminus) of ClyA in the fusion protein.29 Separately, Wai and coworkers demonstrated that ClyA was exported from laboratory strains of E. coli cells via OMVs composed of outer membrane and periplasm.31 These same authors reported that ClyA was significantly enriched in OMVs relative to other lumenal and membrane-bound OMV proteins.14 Based on these results, we hypothesized that proteins fused to the N- or C-terminus of ClyA would be efficiently co-localized in OMVs and would retain their native function following vesicle localization. To test this, we first generated fusion constructs between GFP and the N- or C-terminus of ClyA. Expression of these fusion proteins in the OMV hyper-producing strain JC803132 followed by purification of vesicles from cell-free culture supernatants yielded uniform s-MVs (Fig. 1a) with an average diameter (Fig. 1b) and zeta-potential (data not shown) that were nearly indistinguishable from naked OMVs produced from plasmid-free JC8031 cells. This result was consistent with earlier findings that vesicle density and size were unaltered due to the incorporation of a heterologous vesicle protein.33 A significant level of ClyA-GFP or GFP-ClyA was localized in vesicles whereas unfused GFP expressed alone was not detected in the s-MV preparations (Fig. 1c). Consistent with earlier studies of ClyA localization,14; 31 subcellular fractionation of E. coli cells revealed that unfused ClyA accumulated in the cytoplasm, periplasm and OMV fractions (Fig. 1d). Likewise, the addition of GFP as an N- or C-terminal passenger protein resulted in a similar pattern of localization (Fig. 1d), although the amount of ClyA fusions in the insoluble fraction clearly increased compared to unfused ClyA (data not shown). Both fusion proteins were fluorescent in the cytoplasm, periplasm, and OMV fractions (Fig. 1e and f) and the whole cell fluorescence associated with cells expressing ClyA-GFP or GFP-ClyA was nearly as bright as cells expressing GFP alone (data not shown). Taken together, the data clearly indicate that GFP was compatible with ClyA translocation as chimeras between these two proteins co-localized in OMVs without significant losses in fluorescence activity.
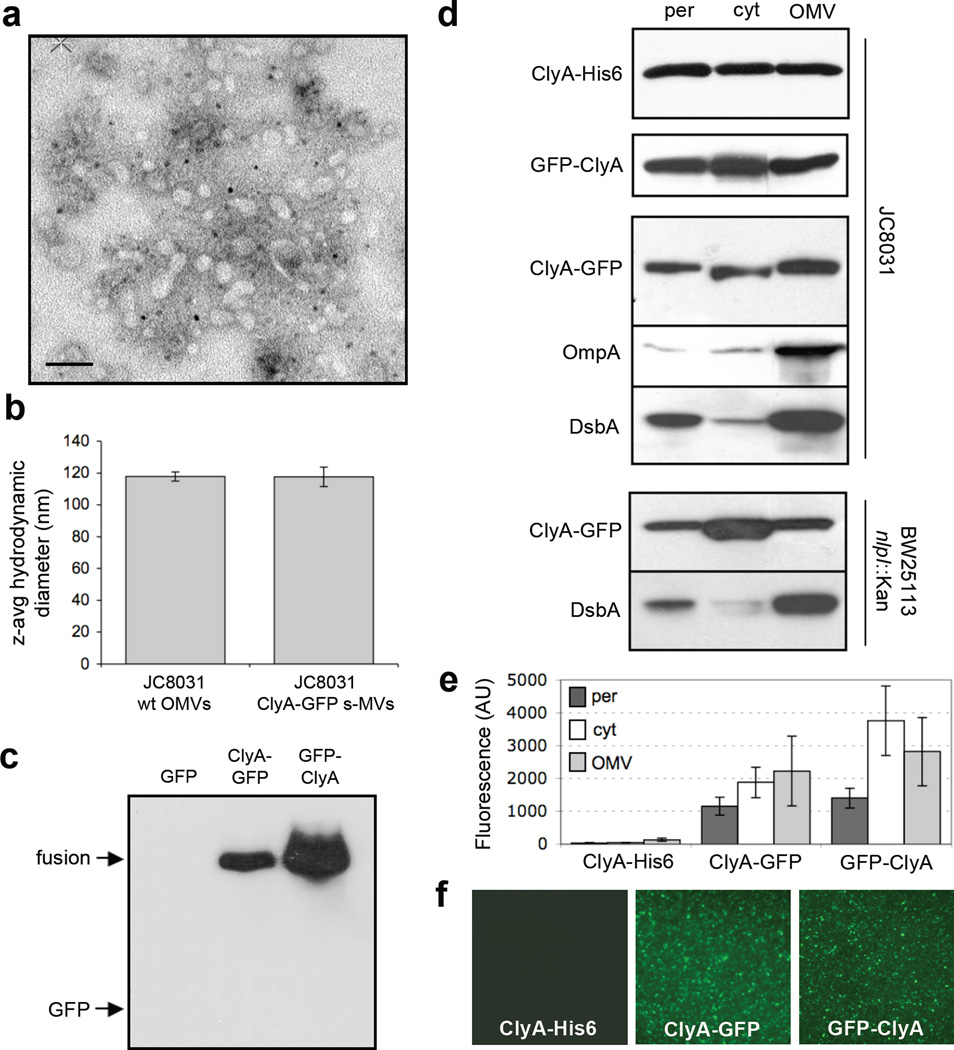
Subcellular localization of ClyA and ClyA fusions. (a) Electron micrograph of vesicles derived from JC8031 cells expressing ClyA-GFP. Bar is equal to 100 nm. (b) Z-average particle size of 1 ml vesicle suspensions containing ~30 µg/ml total protein obtained from plasmid-free or ClyA-GFP-expressing JC8031 cells. Error bars represent the standard deviation of 3 replicates. (c) Western blot of vesicle fractions isolated from E. coli strain JC8031 expressing GFP, ClyA-GFP and GFP-ClyA. Blot was probed with anti-GFP serum. (d) Western blot and (e) GFP fluorescence of periplasmic (per), cytoplasmic (cyt) and vesicle (OMV) fractions generated from JC8031 or BW25113 nlpI::Kan cells expressing ClyA-His6, ClyA-GFP and GFP-ClyA. ClyA-His6 blot was first probed with anti-polyhistidine. ClyA-GFP and GFP-ClyA blots were probed with anti-GFP. Following stripping of membranes, blots were reprobed with anti-OmpA serum or anti-DsbA serum as indicated. All fractions were generated from an equivalent number of cells. (f) Fluorescence microscopy of vesicles generated from JC8031 cells expressing ClyA-His6, ClyA-GFP and GFP-ClyA.
The quality of the fractionation procedure was confirmed by the observation that endogenously expressed outer membrane protein OmpA was always found predominantly in the OMV fraction (shown for cells expressing ClyA-GFP, Fig. 1d), consistent with earlier studies,14 while GroEL was found exclusively in the cytoplasmic fraction (data not shown) and DsbA in the periplasmic fraction (Fig. 1d). In our hands, DsbA also accumulated to high levels in the OMV fraction (Fig. 1d). While it is common for periplasmic proteins to become entrapped in OMVs,8; 9; 14; 33 the presence of DsbA was unexpected based on the findings of Wai and coworkers who reported that this protein was excluded from their OMV fractions.14 One explanation for this discrepancy might have been our use of the tolRA mutant strain JC8031, which despite its tendency to secrete copious amounts of vesicles is also characterized by a leaky outer membrane.32; 34 However, similar patterns of ClyA-GFP and DsbA localization were observed following subcellular fractionation of ΔnlpI mutant cells (Fig. 1d) that is known to produce relatively large quantities of vesicles but does not exhibit membrane instability.34 Thus, the explanation that we currently favor, based on earlier observations,8 is that our use of a different host strain resulted in altered vesicle protein profiles.
To determine whether ClyA-GFP in the pelleted supernatant was associated with intact vesicles and not with released outer membrane fragments, we next tested whether the fusion protein co-migrated with both periplasmic and outer membrane material. To this end, pelleted supernatant from cells expressing ClyA-GFP was separated by density gradient centrifugation. Western blotting and densitometry analysis of the resulting fractions revealed a gradient profile for ClyA-GFP that peaked in fractions 6–8 (Fig. 2a and b), reminiscent of the gradient profile of OMV-associated α-hemolysin.35 As expected, the maximal GFP activity was detected in the same fractions that contained the ClyA-GFP-enriched OMVs (Fig. 2b and c) although weaker fluorescence could be detected in denser fractions (Fig. 2c). The outer membrane protein OmpA was similarly enriched in fractions 6–8 containing the majority of the ClyA-GFP, but strong bands also appeared in fractions 9 and 10 (Fig. 2a). Interestingly, DsbA was more evenly distributed between fractions 5–10 (Fig. 2a), indicating co-migration with vesicles that included a large portion of ClyA-GFP (fractions 6–8) as well as with vesicles that contained lesser amounts of ClyA-GFP (fractions 5, 9 and 10).
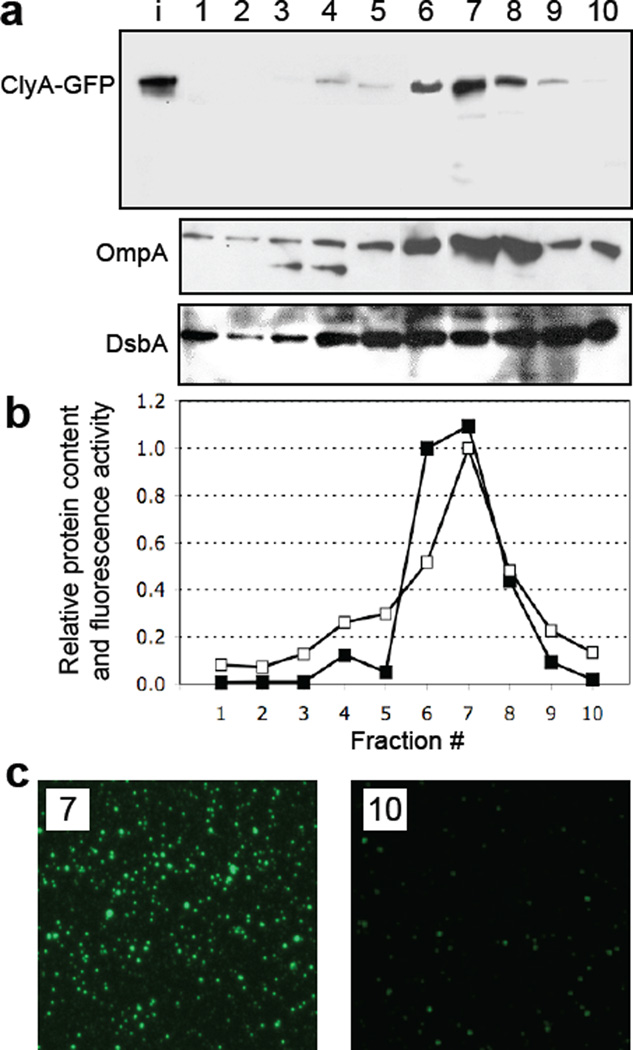
Density gradient fractionation of vesicles. (a) Electrophoretic analysis of the density gradient fractions from top (lane 1, lowest density) to bottom (lane 10, highest density) from JC8031 cells expressing ClyA-GFP. The bands corresponding to ClyA-GFP (top), OmpA (middle) and DsbA (bottom) were obtained with anti-GFP, anti-OmpA and anti-DsbA serum, respectively. Lane i represents input vesicles from the purified cell-free supernatant. (b) Quantification of the ClyA-GFP levels (filled squared) in each fraction as determined by densitometry using ImageJ software. Band intensity values were normalized to the maximum intensity corresponding to the ClyA-GFP measured in fraction 7. Also plotted is the GFP activity (open squares) measured in each fraction and normalized to the maximum activity, which also corresponded to fraction 7. (c) Fluorescence microscopy of vesicles that migrated in gradient fractions 7 and 10.
ClyA anchors correctly folded GFP to the outer surface of E. coli and to the surface of s-MVs
To determine the topology of the ClyA-GFP and GFP-ClyA chimeras, we probed the surface accessibility of GFP on whole cells and on vesicles. Previous studies showed that a fraction of the secreted ClyA remains located on the bacterial cell surface.14 Likewise, we observed that both ClyA-GFP and GFP-ClyA were localized to the cell surface as evidenced by the accessibility of the GFP moiety to cross-reacting antibodies. Specifically, positive immunofluorescent- and immunogold-labeling using anti-GFP antibodies was detected for JC8031 cells expressing ClyA fused with GFP but not in the cases of unfused ClyA or unfused GFP (Fig. 3, shown for unfused GFP and ClyA-GFP).
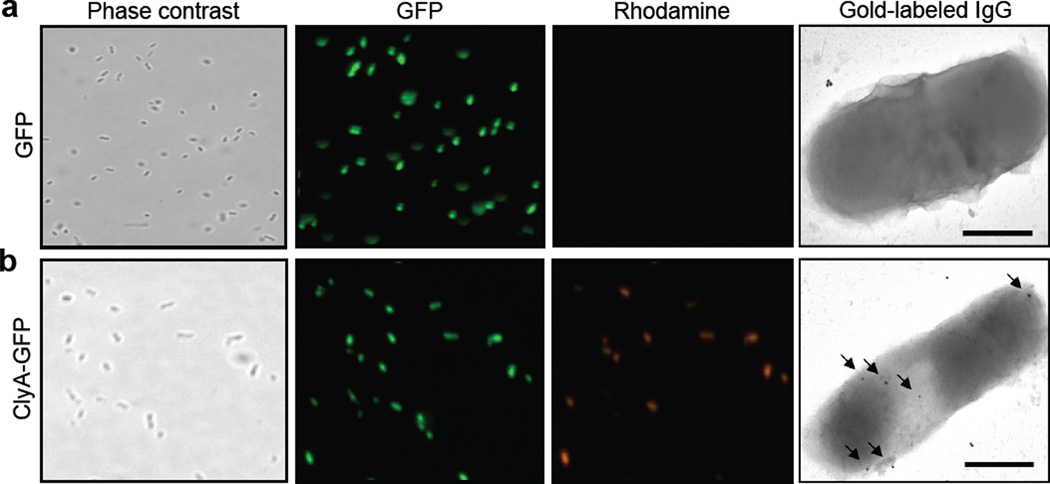
Microscopic analysis of ClyA expression. JC8031 cells grown at 37°C in LB were induced to express (a) GFP and (b) ClyA-GFP. For immunofluorescence microscopy, cells were treated with mouse monoclonal anti-GFP and subsequently with rhodamine-conjugated anti-mouse IgG. Panels show phase contrast microscopy and fluorescence microscopy using green and red emission filters as indicated. For immunoelectron microscopy, cells were treated with mouse monoclonal anti-GFP and subsequently with gold-conjugated anti-mouse IgG. Arrows indicate the 25 nm gold particles. The bars are equal to 500 nm.
To determine if the GFP associated with vesicles was similarly accessible to cross-reacting antibodies, immunofluorescent labeling of s-MVs was performed but only a weak immunofluorescent signal above background could be seen in this analysis (data not shown). This prompted us to explore a more sensitive and quantitative surface plasmon resonance (SPR)-based strategy for detecting vesicle-associated GFP. For this, biotinylated anti-E. coli antibodies (test channel) and bovine serum albumin (BSA, reference channel) were coupled to an SPR sensor chip through streptavidin binding. To verify that the SPR surface could capture our s-MVs, we introduced solutions containing varying amounts of intact s-MVs from fraction 7 above, where the initial concentration of s-MVs in this fraction was 13.5 ± 1.34 µg/µl. Following introduction of ClyA-GFP-containing s-MVs to the SPR, we immediately noticed that the test channel coated with anti-E. coli antibodies but not the BSA-coated reference channel was highly fluorescent (Fig. 4a), even after several PBS wash steps, indicating specific capture of our s-MVs on the SPR surface. SPR binding revealed concentration-dependent shifts in the SPR wavelength over a range of s-MV concentrations (0.02–0.70 µg/µl) (Fig. 4b and c). Importantly, there was no measurable change in SPR wavelength upon treatment of surface-captured s-MVs with PBS (data not shown), indicating that the fluorescent vesicles were stably and tightly bound to the immobilized anti-E. coli antibody. Finally, to determine whether our SPR strategy was suitable for detecting OMV-associated antigens, we introduced anti-GFP monoclonal antibodies into the test channel that contained surface-captured s-MVs displaying active GFP. Specific binding between anti-GFP antibodies and vesicle-associated GFP was confirmed by a marked increase in the SPR wavelength in the test channel (Fig. 4d, black line). Controls were performed where these s-MVs were treated with non-specific anti-His6× monoclonal antibodies or where s-MVs displaying unfused ClyA were captured on the SPR surface and treated with anti-GFP antibodies; both cases resulted in no detectable increase in the SPR wavelength (shown for anti-His6× treatment of Cly-GFP s-MVs in Fig. 4d, gray line).
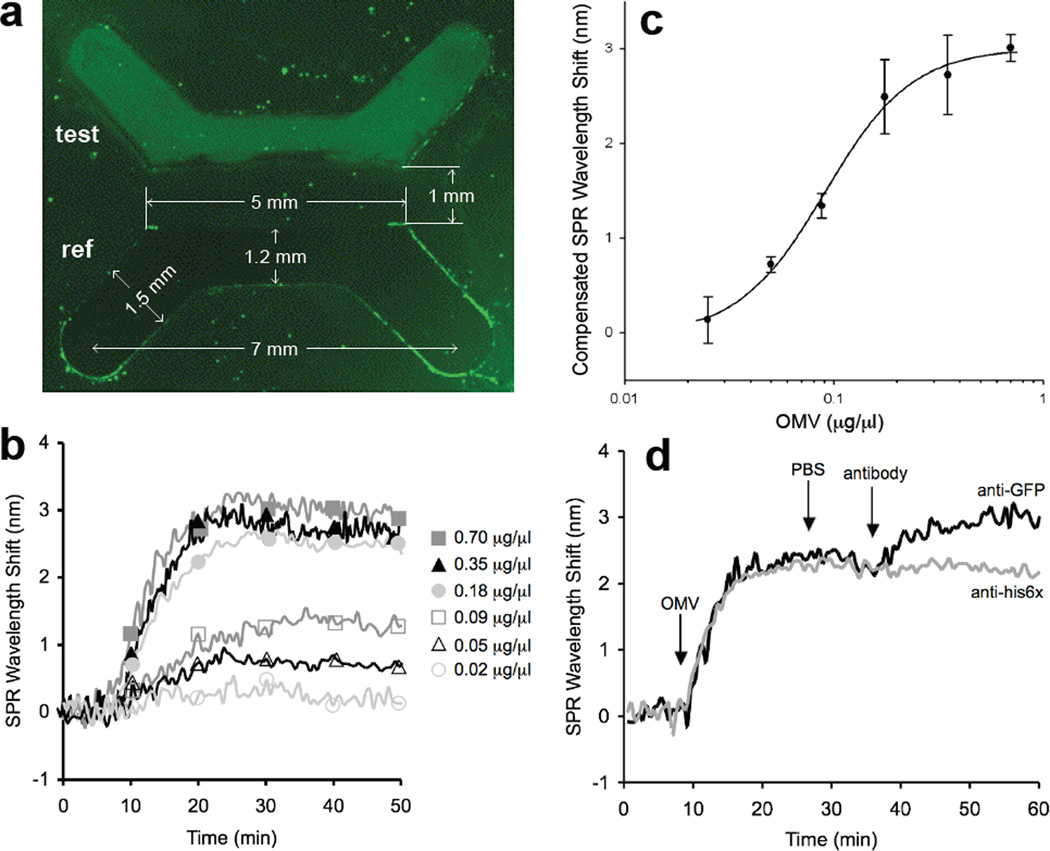
Detection of vesicles and vesicle-associated antigens via immuno-SPR. (a) Fluorescence microscopy analysis of the binding of fluorescent s-MVs (at a concentration of 0.35 µg/µl) to anti-E. coli antibody in the test channel and to the BSA-treated surface in the reference channel following 20 min of s-MV binding and 20 min PBS rinse. The measurements indicated represent the size of the PDMS master. (b) Overlaid SPR sensorgrams showing concentration-dependent binding of OMVs to immobilized anti-E. coli antibodies. For each binding experiment, 200 µl of OMV-containing samples (diluted to the concentrations indicated) was introduced to the test or reference channel for 20 min, followed by a 20 min PBS rinse. The SPR signal was recorded as wavelength shift (nm) versus time and plotted as a “sensorgram”. All binding experiments were performed at 25°C ± 1°C with a flowrate of 10 µl/min. Each vesicle sample was assayed in triplicate and the standard error was determined to be less than 5%. (c) Vesicle standard curve generated using the SPR immunosensor. The steady-state SPR signal change was calculated by subtracting the average SPR signal during the PBS wash step following OMV binding from the average SPR signal collected during the initial PBS wash step prior to OMV addition. The equation y = 0.92ln(x) + 3.63 with an R2 value of 0.95 describes the fit of a straight line through the logarithm of the data and was determined using SigmaPlot. The results are the average of the values calculated for the SPR signal change from three independent binding measurements with error bars showing ± standard error. It is noteworthy that the lower detection limit for this system was determined to be 0.01 µg/µl (10% of the compensated SPR wavelength shift in the standard curve) and that for vesicle concentrations ≥0.18 µg/µl, SPR wavelength shifts >2.5 nm were recorded, which is ~10× greater than the baseline signal. (d) Representative sensorgrams for antibody binding to s-MV surface-displayed GFP in test and reference channels. Channels were prepared identically so that fluorescent s-MVs were captured in both channels. The change in SPR signal over time was measured following addition of 1 µg/µl anti-GFP (black line) or 1 µg/µl anti-his6× (gray line) monoclonal antibody to surface-captured s-MVs. Antibody binding proceeded for 20 min followed by a 10 min PBS rinse. Each antibody was assayed in triplicate and the standard error was determined to be less than 5%.
We next performed Proteinase K (PK) susceptibility assays on ClyA-GFP and GFP-ClyA vesicles to determine whether the immuno-accessible GFP was protected by the vesicle structure. When s-MVs derived from JC8031 cells expressing ClyA-GFP or GFP-ClyA were incubated with PK in the absence of membrane-disrupting detergent, vesicle-associated fluorescence was completely abolished (Fig. 5a, shown for ClyA-GFP) suggesting that the majority of the functional GFP was surface exposed and not protected by the vesicle structure. Consistent with this, Western blot analysis confirmed that nearly all of the s-MV-associated ClyA-GFP was degraded to a lower molecular weight anti-GFP or anti-ClyA cross-reacting species upon treatment with PK (Fig. 5b, lanes 1–3 and 7–9). Interestingly, we observed a significant amount of PK-resistant material following identical treatment of GFP-ClyA s-MVs (Fig. 5b, lanes 4–6 and 10–12) that persisted even after incubation with 2–5× higher PK concentrations for 2× longer durations (data not shown). Since these PK-treated s-MVs were non-fluorescent but contained a considerable portion of PK-resistant GFP-ClyA, we conclude that only a fraction of the fusion localizes with functional GFP tethered outside the OMV while the remainder adopts an inactive conformation that is protected by the vesicle structure. Possible reasons for this include a localization defect caused by the relatively high level of expression for GFP-ClyA compared to ClyA-GFP and/or the apparent instability of the fusion as evidenced by the multiple anti-ClyA cross-reacting bands seen in the absence of PK (Fig. 5b, lane 10). For both chimeras, complete proteolytic digestion of GFP by PK occurred when membranes were disrupted by the addition of 1% SDS (Fig. 5b). Control experiments using purified, soluble ClyA-GFP showed that the protein was PK-sensitive both in the presence or absence of the detergent (data not shown).
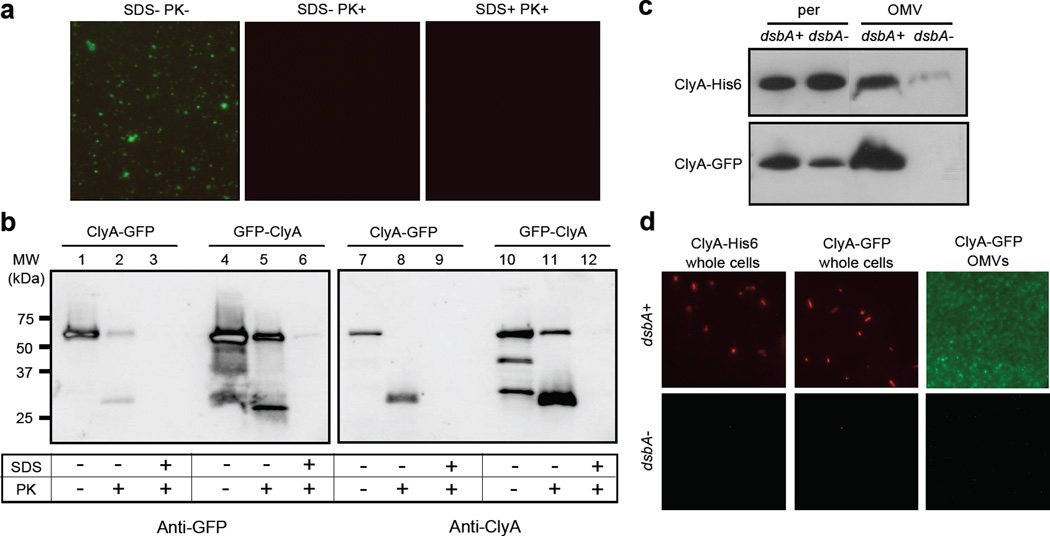
Biochemical and genetic analysis of ClyA localization. Proteinase K susceptibility of OMV-tethered GFP as determined by: (a) fluorescence microscopy of vesicles generated from JC8031 cells expressing ClyA-GFP treated with Proteinase K and SDS as indicated; and (b) Western blot of vesicles generated from JC8031 cells expressing ClyA-GFP or GFP-ClyA. Blots were probed with mouse anti-GFP (left panels) or anti-ClyA serum (right panels). Molecular weight (MW) ladder is marked at left. An equivalent number of vesicles was used in all cases. (c) Western blot analysis of periplasmic and OMV fractions from JC8031 (dsbA+) and JC8031 dsbA::Kan cells (dsbA-) expressing either ClyA-GFP or ClyA-His6 as indicated. (d) Immunofluorescence (left and center panels) of wt JC8031 (dsbA+, top) and JC8031 dsbA::Kan cells (dsbA-, bottom) expressing pClyA-GFP or pClyA-His6 as indicated and fluorescence (right panel) of vesicles derived from the same cells as indicated. For immunofluorescence, cells were treated with either mouse monoclonal anti-GFP or anti-polyhistidine antibodies and subsequently with rhodamine-conjugated anti-mouse IgG.
Periplasmic disulfide bond-forming machinery is required for localization of ClyA and ClyA fusions in OMVs
In previous work, ClyA in the periplasm was shown to adopt a monomeric conformation owing to an intramolecular disulfide bond formed between the cysteine residues at positions 87 and 285 in the polypeptide.36 The presence of the disulfide bond was sufficient to prevent oligomerization of ClyA and inactivate its native haemolytic activity. In agreement with this finding, Wai et al. reported that DsbA, the enzyme responsible for catalyzing the formation of disulfide bonds in periplasmic proteins, was absent from OMVs containing ClyA and that the absence of DsbA was necessary for ClyA to oligomerize into its haemolytic conformation.14 Contrary to the findings of Wai et al., we found that DsbA was co-localized in vesicles containing the ClyA fusions (see Fig. 1 above). Thus, we hypothesized that oxidized, monomeric ClyA might be favorable for efficient cell surface and vesicle localization of our fusions. To test our hypothesis, we compared expression of ClyA-GFP in strain JC8031 and an isogenic dsbA::Kan mutant derived from JC8031. Western blot analysis revealed that while both strains accumulated similar amounts of ClyA-GFP in the periplasm, only in cells with DsbA present was localization of ClyA-GFP observed (Fig. 5c). This was corroborated by a complete lack of fluorescence seen for OMVs derived from JC8031 dsbA::Kan cells expressing ClyA-GFP (Fig. 5d). Immunofluorescent staining of these same cells revealed that localization of ClyA-GFP to the bacterial cell surface was also dependent upon DsbA (Fig. 5d). To our surprise, DsbA-dependent vesicle localization was also observed for unfused ClyA suggesting that the periplasmic redox state is a critical factor in regulating the efficiency of protein localization into vesicles under the conditions tested here. Interestingly, we observed very little, to no, apparent cytotoxicity for naked OMVs from plasmid-free JC8031 cells, consistent with earlier studies,14 and also for s-MVs containing ClyA-His6, ClyA-GFP or GFP-ClyA (data not shown) suggesting that ClyA in our vesicles was not in its haemolytically-active, oligomeric conformation.
Engineered ClyA-GFP s-MVs are useful for visualizing vesicle interactions with eukaryotic cells
Previous studies have shown that vesicles derived from pathogenic E. coli or non-pathogenic E. coli strains can associate with eukaryotic cells.20; 33 Thus, we next investigated whether s-MVs functionalized with ClyA-GFP were suitable for tracking vesicle association with eukaryotic cells. Previous attempts at this focused on loading GFP into the lumen of vesicles following its transport into the periplasm by the twin-arginine translocation (Tat) pathway.33 However, GFP-containing OMVs were only weakly fluorescent and could not be tracked in host cells by microscopy, likely due to the low yield of GFP transport to the periplasm via the Tat system. To determine if s-MVs engineered with ClyA-GFP were bright enough for tracking studies, we performed vesicle-host cell co-incubation assays. Punctate green staining was observed following a 30 min incubation of HeLa cells with ~150 µg vesicles purified ClyA-GFP s-MVs and the intensity of this staining increased as the incubation time between HeLa cells and ClyA-GFP s-MVs increased (Fig. 6a). These results imply that vesicles persisted on the cell surface or had fused directly to the target cell membrane. To corroborate this, HeLa cells that had been incubated with purified ClyA-GFP-containing s-MVs were stained with a fluorescent version of the cell surface marker wheat germ agglutinin (WGA) and then washed with PBS. Confocal microscopy revealed that ClyA-GFP OMVs co-localized with WGA on the exterior of the cell (data not shown). Next, we explored the fate of ClyA-GFP vesicles by examining whether the appearance of punctate fluorescence was temperature dependent, a hallmark of cellular internalization.20; 37 HeLa cells incubated with vesicles containing ClyA-GFP at 4°C exhibited very low levels of cell-associated fluorescence (compare Fig. 6b). However, when HeLa cells were incubated with ClyA-GFP s-MVs at 4°C for 3 h and then shifted to 37°C for 3 hr, strong cell fluorescence was observed (Fig. 6b), leaving open the possibility that some s-MVs may be internalized0 at 37°C. A key factor in endocytosis is ganglioside M1 (GM1), which is a eukaryotic cell surface receptor for enterotoxins such as LT and cholera toxin (CT) and is required for endocytosis of LT-containing OMVs derived from non-pathogenic E. coli.20 Therefore, we tested whether the observed fluorescence of HeLa cells incubated with ClyA-GFP s-MVs was GM1-dependent. Indeed, fluorescence associated with HeLa cells was significantly decreased following incubation with purified ClyA-GFP s-MVs that had been pretreated with GM1 (Fig. 6c). Moreover, incubation with GM1-treated vesicles resulted in a small number of large fluorescent clusters and much less punctate green fluorescence than was observed for HeLa cells incubated with untreated ClyA-GFP OMVs (Fig. 6c). Thus, it appears that GM1 cell surface receptors may play an important role in mediating interactions between HeLa cells and engineered vesicles. Finally, to assay the cytotoxic effect of vesicles on target cells, we analyzed how cultured HeLa cells were affected by equivalent amounts of different vesicle preparations. In general, vesicles containing ClyA-His6 or ClyA-GFP exhibited virtually no detectable cytotoxicity (Fig. 6d), consistent with a monomoric, DsbA+ conformation14 for both ClyA and ClyA-GFP in our vesicles.
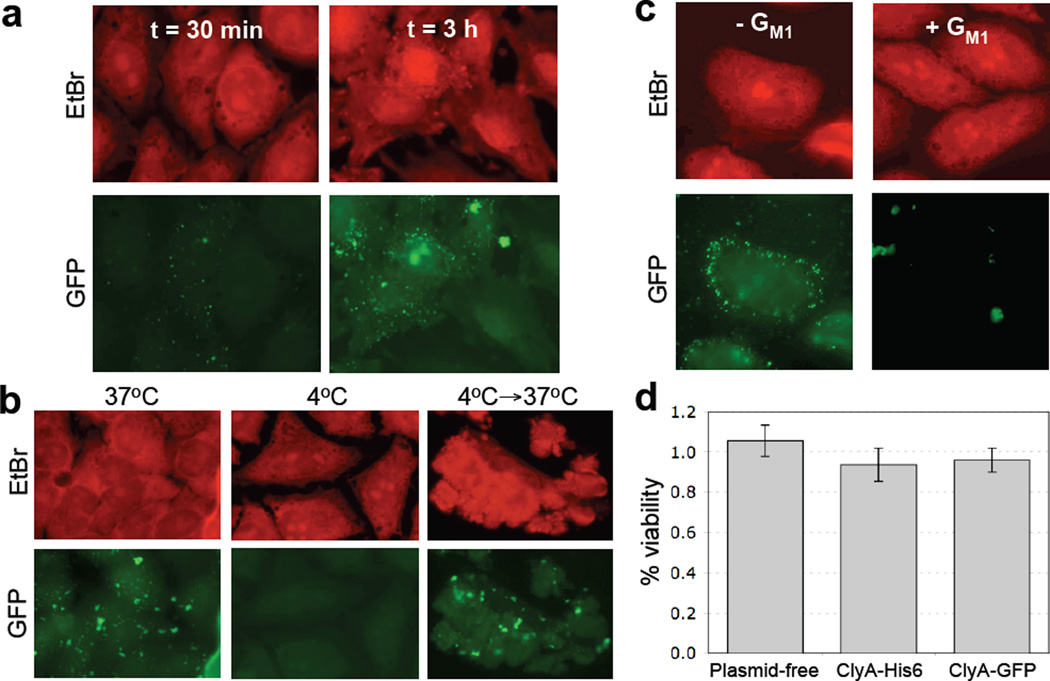
Interaction of ClyA-GFP vesicles with HeLa cells. (a) OMVs containing ClyA-GFP were incubated with HeLa cells at 37°C for 30 min or 3 h as indicated. Fixed cells were stained with 0.5 mg/mL ethidium bromide (EtBr, upper panels) and visualized by fluorescence microscopy. (b) Temperature dependence of OMV-HeLa cell interactions was examined by incubation of HeLa cells with GFP-ClyA OMVs at the temperatures indicated. An equivalent number of OMVs (~150 µg) was used in all cases. (c) Untreated (− GM1) or pretreated (+ GM1) OMVs from JC8031 cells expressing ClyA-GFP were incubated with HeLa cells at 37°C for 3 h. Fixed cells were stained with 0.5 mg/mL ethidium bromide (EtBr, upper panels) and visualized by fluorescence microscopy. An equivalent number of OMVs (~150 µg) was used in all cases. (d) Cytotoxicity of vesicles as measured using the MTS assay with HeLa cell cultures. % viability is reported as the viability of vesicle-treated HeLa cells normalized to the viability following treatment with PBS. HeLa cells were treated with vesicle solutions derived from plasmid-free JC8031 cells and JC8031 cells expressing ClyA-His6, ClyA-GFP (dark gray). An equivalent number of OMVs (~150 µg) was used in all cases. Each sample was assayed in triplicate with error bars showing ± standard error.
Heterologous proteins co-localized in s-MVs via ClyA retain their activity
To determine whether proteins other than GFP could be fused to ClyA while still retaining their function, we generated a series of N- and C-terminal fusions between ClyA and the following enzymes: β-lactamase (Bla), organophosphorus hydrolase (OPH) and β-galactosidase (LacZ). Similar to what was seen for GFP, ClyA-Bla resulted in localization of Bla to the surface of JC8031 cells and vesicles as determined using a nitrocefin hydrolysis assay (Table 1). Since Bla expressed in the cytoplasm was not localized to the cell surface or vesicles and since nitrocefin is relatively impermeable to the outer membrane,38; 39 these data provide strong evidence that the Bla moiety was externally localized on cells and vesicles. Similar results were obtained using the Bla substrate penicillin-G (data not shown), which also penetrates the outer membrane very poorly.40 Interestingly, as was seen above for the chimeras between ClyA and GFP, fusion of Bla to the N-terminus of ClyA resulted in a significantly lower level of Bla activity. Consistent with these results, fusion of ClyA to the N-terminus of the OPH enzyme conferred a significant level of OPH activity to the surface of cells and vesicles, whereas OPH-ClyA resulted in no measurable surface-associated OPH activity (Table 1). Since the OPH substrate paraoxon used in these experiments is membrane impermeable,41 we conclude that ClyA-OPH is oriented with OPH externally bound to both cells and vesicles. Also, since OPH activity depends upon homodimer formation,42 ClyA apparently tethers OPH in a conformation that allows for dimerization with a neighboring ClyA-OPH molecule. Finally, to determine if the ability to display multimeric enzymes is a general feature of ClyA-mediated surface exposure, we constructed ClyA fusions to the homotetrameric LacZ enzyme from E. coli.43 While expression of ClyA-LacZ resulted in strong cytoplasmic LacZ activity, there was no measurable LacZ activity on the surface of cells or their derived s-MVs (Table 1). In fact, there was no LacZ activity in the periplasm of cells expressing ClyA-LacZ (data not shown), consistent with the observation that the normally cytoplasmic LacZ protein contains sequences that hinder transport44 usually leading to a misfolded, and thus inactive, protein.
Table 1
ClyA-mediated display of enzymatically active proteins
| Construct | Cell surface activity | OMV activity |
|---|---|---|
| β-lactamase (Bla) | [ΔA486/min] | [ΔA486/min] |
| pBAD18-Kan | 0.03 | 0.02 |
| pΔss-Bla | 0.47 | 0.98 |
| pClyA-Bla | 8.53 | 7.05 |
| pBla-ClyA | 1.43 | 1.65 |
| Organophosphorus hydrolase (OPH) | [U*1000/OD600] | [U*1000/g total protein] |
| pBAD24 | 0.09 | 0.04 |
| pOPH | 0.36 | 1.63 |
| pClyA-OPH | 11.62 | 81.63 |
| pOPH-ClyA | 0.22 | 8.19 |
| β-galactosidase (LacZ) | [ΔA420/min] | [ΔA420/min] |
| pBAD24 | 0.02 | 0.02 |
| pLacZ | 0.02 (7.21)† | 0.03 |
| pClyA-LacZ | 0.04 (8.33)† | 0.03 |
All values represent the average of 3 replicate experiments where the standard error was <5%.
Single-chain antibody fragments (scFv) have been successfully used to create artificial immunoliposomes for targeting of these vesicles and their payloads to specific cell types.45 Along similar lines, we sought to create bacterial “immuno-MVs” by displaying scFv fragments on E. coli-derived vesicles. For these experiments, we made use of an scFv derived from the 26–10 monoclonal antibody that binds with high affinity (KD = 0.9 ± 0.2 × 10–9 M−1) to the cardiac glycoside digoxin (scFv.Dig).46; 47 Using a fluorescent conjugate of digoxin (Dig-BODIPY), we observed that expression of ClyA-scFv.Dig, but not scFv.Dig alone, resulted in cells and vesicles that were able to bind the fluorescent probe (Fig. 7a). Since Dig-BODIPY cannot permeate the outer membrane under standard conditions, the detection of Dig-BODIPY binding activity using intact cells indicates that scFvs were functionally displayed on the outer cell and vesicle surface. For comparison, cells expressing scFv.Dig fused to the well-characterized Lpp-OmpA hybrid OM anchor47 displayed uniform but notably weaker cell surface fluorescence and no detectable fluorescence on OMVs (Fig. 7a) despite the fact that wildtype OmpA localizes in OMVs (see also Fig. 1d above).
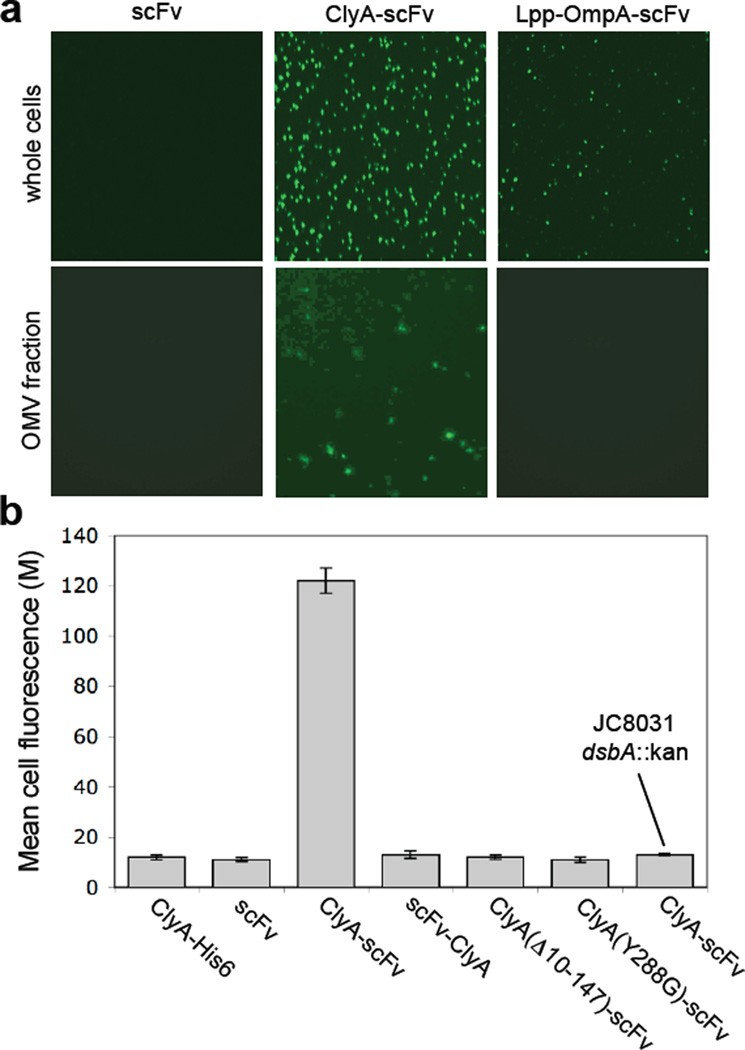
Creation of immuno-MVs via ClyA-scFv chimeras. (a) Fluorescence microscopy of whole cells and vesicles generated from JC8031 cells expressing scFv.Dig, ClyA-scFv.Dig or Lpp-OmpA-scFv.Dig as indicated. For these studies, cells were grown and induced at room temperature followed by fluorescent labeling of cells or their derived vesicles with 1 µM Dig-BODIPY for 1 h at room temperature. (b) Genetic analysis of scFv.Dig localization was performed using flow cytometric analysis of strains and plasmids as indicated. Cells were grown and induced at room temperature followed by labeling with 1 µM Dig-BODIPY for 1 h at room temperature. Fluorescence is reported as the mean fluorescence for each cell population and was assayed in triplicate with error bars showing ± standard error.
Lastly, we sought to determine if the capture of Dig-BODIPY by ClyA-scFv.Dig could be used as a genetic screen for ClyA localization. Consistent with the fluorescence microscopy results above, labeling of JC8031 cells expressing ClyA-scFv.Dig with Dig-BODIPY resulted in highly fluorescent cells as revealed by flow cytometry (Fig. 7b). However, when ClyA-scFv.Dig was expressed in JC8031 dsbA::Kan cells, fluorescence was completely abolished. Likewise, when scFv.Dig was expressed alone, as an N-terminal fusion to ClyA (scFv.Dig-ClyA), or as a C-terminal fusion to ClyA variants carrying mutations that were previously reported to disrupt translocation (e.g., deletion of 10–147 of the last C-terminal amino acids31 or substitution of the Tyr288 residue with Gly29), no measurable cell fluorescence was detected (Fig. 7b).
Discussion
This work describes the development and characterization of engineered synthetic membrane vesicles (s-MVs) created by genetic fusion of a recombinant polypeptide with the E. coli cytotoxin ClyA. In general, it was observed that most recombinant polypeptide fusions co-localized with ClyA to the bacterial cell surface and into OMVs. Specifically, we demonstrated that direct fusion of Bla, OPH, GFP and anti-digoxin scFv to the C-terminus of ClyA resulted in functional display of each protein on the surface of E. coli cells and their derived OMVs, giving rise to s-MVs with significantly expanded, non-native functionality (e.g., fluorescence, antigen binding). Interestingly, fusion of each of these proteins to the N-terminus of ClyA yielded unpredictable results. For instance, scFv.Dig-ClyA exhibited no detectable activity on the surface of cells or OMVs while GFP-ClyA resulted in the display of active protein. In the latter case, even though a portion of the fusions was active, a significant amount of non-fluorescent GFP-ClyA accumulated in OMVs. We also found that fusion of the enzymes Bla and OPH resulted in little to no activity on cells and OMVs when each enzyme was fused to the N-terminus of ClyA. These results are consistent with earlier studies where a 2-fold increase in secretion to the extracellular medium was observed for ClyA-Bla versus Bla-ClyA.29 At present, it is not known why N-terminal fusions to ClyA were inconsisten whereas C-terminal fusions invariably yielded well-displayed proteins that retain their biological function. Our results might be explained by inspection of the medium-resolution structure of membrane-bound ClyA showing that the C-terminus of ClyA is embedded deeper within membranes (i.e., closer to the outer surface) than the N-terminal portion, which occurs close to the periplasmic side of the outer membrane.24; 25 According to this model, fusions to the C-terminus of ClyA are closer to the outer surface and more likely to extend into the extracellular environment, especially with the addition of a flexible 5-residue Gly linker that was included in all of our fusions.
Based on its relative plasticity as a fusion partner, we expect ClyA to serve as a useful tethering module for: (1) dissecting the complete ClyA translocation pathway from bacteria to target host cells; and (2) biotechnological applications that rely on cell or OMV surface display, such as affinity maturation of antibody fragments and vaccine adjuvant development.46; 47; 49; 50 For instance, with respect to genetic analysis, expression of ClyA-GFP and ClyA-scFv.Dig both proved capable of reporting the localization of ClyA to the bacterial cell surface and into OMVs and revealed an essential role for DsbA in the translocation process. A simple model for ClyA assembly in vesicles proposed by Uhlin and coworkers, and supported by our findings, is that ClyA oligomerization and membrane insertion is governed by the redox state of ClyA.14 Here, however, we show that a more reducing environment in the periplasm, such as occurs when DsbA is absent, favors the localization of ClyA and ClyA fusions into vesicles. While the involvement of DsbA appears to be via direct oxidation of ClyA,14 we cannot rule out the possibility that DsbA is responsible for the oxidation of a periplasmic or membrane component(s) that mediate the localization of ClyA and/or the formation of OMVs.
With respect to biotechnology, the ability of ClyA to anchor and functionally display a variety of different prokaryotic and eukaryotic proteins on the surface of bacterial cells and vesicles should be useful for numerous applications. First, ClyA exhibits properties that make it an ideal carrier protein for bacterial cell surface display of peptides and proteins and compares favorably with the well-established Lpp-OmpA surface anchor as evidenced by scFv binding of fluorescently tagged digoxin. Second, by judicious selection and display of specific scFv fragments on OMVs, one might be able to redirect these engineered “immuno-MVs” to specific cell types, or otherwise engineer the host-vesicle interaction, in order to achieve a desired therapeutic or immunological response. Along these lines, we provide encouraging data that engineered vesicles retain their characteristic interactions with mammalian cells20; 33 and are almost completely non-cytotoxic. It is also noteworthy that expression of all ClyA fusions in this study had no measurable effect on the growth rate of bacterial cells relative to cells expressing unfused ClyA or empty vector controls, thus appreciable yields of cells or their derived OMVs should be attainable for any OMV-based application.
Materials and Methods
Bacterial Strains, Plasmids and Growth Conditions
The bacterial strains and plasmids used in this study are described in Table 2. Strain JCA were made by introducing the dsbA::Kan allele into JC8031 cells by P1 vir transduction using DHA as the donor. Plasmid pClyA was constructed by ligating the PCR-amplified clyA gene into pBAD18-Cm between SacI and XbaI sites. Insertion of DNA encoding either the gfpmut2 gene51; 52 or a 6× polyhistidine sequence between XbaI and HindIII sites resulted in plasmids pGFP-ClyA and pClyA-His6, respectively. Plasmid pGFP-ClyA was constructed by first cloning the gfpmut2 gene between SacI and XmaI sites of pBAD18-Cm followed by insertion of the clyA gene between XmaI and XbaI sites. Plasmid pGFP was constructed by ligating the PCR-amplified gfpmut2 gene between SacI and HindIII sites of pBAD18-Cm. For ClyA-X fusions in pBAD24, each of the PCR-amplified partner genes (except for bla) was inserted between XmaI and SphI sites followed by clyA ligation between NcoI and XmaI sites. X-ClyA fusions were similarly constructed in pBAD24 with the ligations of fusion partner between NcoI and XmaI, and clyA between XmaI and SphI. Control plasmids without clyA were constructed by inserting the fusion partner, X, between NcoI and SphI of pBAD24. For Bla fusions with ClyA, a similar strategy as described above was used for inserting clyA and bla into plasmid pBAD18-Kan between SacI and XmaI and SphI sites. The gene encoding the Lpp-OmpA-scFv.Dig chimera in pB18D was amplified and ligated into pBAD24 between NcoI and SphI, resulting in pB24D. To generate pClyA(Δ156–303)-scFv.Dig, pClyA-scFv.Dig was digested with HpaI and XmaI and then self-ligated via blunt-end ligation after removal of overhanging basepairs. To generate pClyA(Δ293–303)-scFv.Dig, DNA encoding the first 292 amino acids of ClyA was PCR-amplified and inserted in place of wt clyA in pClyA-scFv.Dig. Plasmid pClyA(Y288G)-scFv.Dig was generated with pClyA-scFv.Dig as template for site-directed mutagenesis using a Stratagene QuickChange® site-directed mutagenesis kit. Cells were grown in LB broth with appropriate antibiotics: ampicillin, 100 µg/ml; chloramphenicol, 25 µg/ml; and kanamycin, 50 µg/ml. Cell growth was maintained at 37°C unless otherwise noted. Protein synthesis was induced for 6 h by adding 0.2% arabinose when cells reached an OD600 ≈ 0.5.
Table 2
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype/Description | Source |
|---|---|---|
| 1292 | supE hsdS met gal lacY tonA | 32 |
| JC8031 | 1292 ΔtolRA | 32 |
| BW25113 | lacIq
rrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 64 |
| BW25113 nlpI::Kan | BW25113 nlpI::Kan created via the method of Datsenko and Wanner | 65 |
| DHB4 | MC1000 phoR Δ (phoA) PvuII Δ (malF)3 F′[lacIqZYA pro] | Laboratory stock |
| DHA | DHB4 dsbA::Kan | 66 |
| JCA | JC8031 dsbA::Kan | This study |
| pBAD18-Cm | araBAD promoter; pBR322 ori Cmr | 67 |
| pBAD18-Kan | araBAD promoter; pBR322 ori Cmr | 67 |
| pBAD24 | araBAD promoter; pBR322 ori Ampr | 67 |
| pClyA-His6 | E. coli clyA carrying a C-terminal 6× polyhistidine tag cloned in pBAD18-Cm | This study |
| pGFP | gfp-mut2 gene cloned in pBAD18-Cm | This study |
| pClyA-GFP | clyA gene fused to 5’ end of gfp-mut2 in pBAD18-Cm | This study |
| pGFP-ClyA | clyA gene fused to 3’ end of gfp-mut2 in pBAD18-Cm | This study |
| pΔss-Bla | Mature region of bla gene cloned in pBAD18-Kan | |
| pClyA-Bla | clyA gene fused to 5’ end of bla in pBAD18-Kan | This study |
| pBla-ClyA | clyA gene fused to 3’ end of bla in pBAD18-Kan | This study |
| pKEG01 | Flavobacterium sp. opd gene in pET78UF | 68 |
| pOPH | opd gene cloned in pBAD24 | This study |
| pClyA-OPH | clyA gene fused to 5’ end of opd in pBAD24 | This study |
| pOPH-ClyA | clyA gene fused to 3’ end of opd in pBAD24 | This study |
| pLacZ | lacZ gene cloned in pBAD24 | This study |
| pClyA-LacZ | clyA gene fused to 5’ end of lacZ in pBAD24 | This study |
| pB18D | anti-digoxin scFv fused to the 3’ end of lpp-ompA cloned in pBAD18 | 58 |
| pB24D | anti-digoxin scFv fused to the 3’ end of lpp-ompA cloned in pBAD24 | This study |
| pscFv.Dig | anti-digoxin scFv cloned in pBAD24 | This study |
| pClyA-scFv.Dig | clyA gene fused to 5’ end of anti-digoxin scFv in pBAD24 | This study |
| pscFv.Dig-ClyA | clyA gene fused to 3’ end of anti-digoxin scFv in pBAD24 | This study |
| pClyA(Δ293–303)-scFv.Dig | clyA with 10 C-terminal residues removed | This study |
| pClyA(Δ156–303)-scFv.Dig | clyA with 147 C-terminal residues removed | This study |
| pClyA(Y288G)-scFv.Dig | clyA with Tyr288 residue mutated to Gly | This study |
Cell culture
Human epithelial cervical carcinoma (HeLa) cells were obtained from the American Type Culture Collection (ATCC # CCL-2) and grown in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) supplemented with 10% NuSerum, and 1% penicillin/streptomycin. Cells were maintained at 37°C in a humidified atmosphere of 95% air, 5% CO2. For fluorescence microscopy experiments, cells were grown on 12-mm circular glass coverslips for two days prior to experimentation.
Subcellular fractionation
Cytoplasmic and periplasmic fractions from cells expressing fusion proteins were generated by the cold osmotic shock procedure53 and the pellet remaining after removal of the soluble fraction was collected as the insoluble fraction.
Isolation of bacterial vesicles
Vesicles were isolated from late log-phase bacterial cultures grown aerobically at 37°C in LB broth (unless otherwise indicated) essentially as described previously.14 Briefly, bacterial cells were removed by centrifugation at 5,000×g for 15 min at 4°C and the cell-free supernatants were filtered through a 0.2 µm-pore-size vacuum filter. Vesicles were collected from the filtered supernatant by ultracentrifugation at 141,000×g for 2 h at 4 °C in a 28 Ti rotor (Beckman Instruments, Inc.) and the pellet containing OMVs was carefully removed and suspended in PBS (pH 7.0). Vesicle preparations were plated on LB agar to confirm complete removal of bacterial cells. Vesicles preparations were kept at −20°C.
Outer membrane vesicles fractionation
Separation of pelleted outer membrane vesicle samples was performed as described.8 Briefly, vesicles were isolated as described above but suspended in 50 mM HEPES (pH 6.8), adjusted to 45% Optiprep (Sigma) in 0.15 ml and transferred to the bottom of 12-ml ultracentrifugation tubes. Different Optiprep/HEPES layers were sequentially added as follows: 0.9 ml 35%, 0.9 ml 30%, 0.66 ml 25%, 0.66 ml 20%, 0.33 ml 15% and 0.33 ml 10%. Gradients were centrifuged (180,000×g, 180 min, 4°C). A total of 10 fractions of equal volumes were sequentially removed and analysed by SDS-PAGE.
Vesicle characterization
The amount of vesicles in purified cell-free supernatant was determined by measuring the total protein concentration or the dry mass of vesicles according to published protocols.54 Particle size distribution and zeta potential of vesicle samples containing approximately 30 µg/mL total protein in 1 mL of PBS were measured in a Nanosizer Nano ZS instrument (Malvern Instruments) using standard protocols. Malvern Dispersion Technology Software was used for data acquisition and analysis, applying the general purpose algorithm for calculating size distributions and the Smoluchowski approximation for determining zeta potential.
Protein assays
Whole cells, OMVs and subcellular fractions were assayed for Bla, LacZ and OPH activity using nitrocefin (Sigma), ONPG (Sigma) and paraoxon (Sigma), respectively, according to standard spectrophotometric assays.55; 56; 57 Cells expressing anti-digoxin scFv were labeled with Dig-BODIPY and analyzed by flow cytometry as described.58 Total protein concentration was assayed using the BCA Protein Assay kit (Pierce). Protease accessibility assays were performed as described but with Proteinase K.33 Briefly, vesicles were treated at 37°C for 30 min in 20 mM Tris HCl (pH 8.0) with PK (0.1 mg/ml) in either the absence or presence of 1% SDS. In parallel control experiments, ClyA-GFP and GFP-ClyA that had been purified by IMAC were similarly treated with PK. Following the incubation, all samples were placed on ice and 1 mM PMSF was added to quench all proteolysis, and the samples were analysed by SDS-PAGE. Western blotting was performed as described by Chen et al.48 using the following primary antibodies: anti-ClyA (kindly provided by Sun Nyunt Wai, Umeå University, Sweden), anti-GFP (Sigma), anti-GroEL (Sigma), anti-polyhistidine (Sigma), anti-OmpA and anti-DsbA (kindly provided by Jon Beckwith, Harvard Medical School). Membranes were developed on film using Immuno-Star™ HRP Substrate Kit (Bio-Rad).
Surface plasmon resonance (SPR)
The SPR configuration consisted of a sensor chip, an optical measuring unit, a flow cell and a syringe pump and was similar to that developed previously.59; 60 The SPR chip was SF10 glass with a thin layer (50 nm) of gold that was attached to a prism made of SF10 using index matching oil. Two microfluidic channels (reference and test) made of polydimethylsiloxane (PDMS) were placed on the SPR sensor chip and screw-clamped to seal the channels. Single-wavelength light was obtained by passing white light from a Xe-lamp (Oriel) through a monochromater (Oriel). Light with a bandwidth less than 1 nm passed through the polarizer, where only p-polarized light was transmitted. The reflected light intensity (RI) was measured with a CCD camera (Sony) that showed high sensitivity around 600 nm. When the incident angle of the beam was fixed to 60 degrees, treatment of the sensor chip with PBS resulted in an SPR wavelength of about 600 nm. The SPR wavelength was obtained at each pixel by fitting the RI versus wavelength data to a second-order polynomial equation and the resulting SPR wavelengths, covering a pre-defined region of interest, were averaged. Before each measurement, the RI of s-polarized light was recorded at each pixel for reference. The reliable detection limit of the SPR sensor was measured to be less than 0.2 nm.
Vesicle and vesicle antigen detection using SPR
An SPR chip for detection of vesicles and vesicle-associated antigens was performed as follows. First, an alkanethiol monolayer was self-assembled on the 50-nm gold layer of the sensor chip surface with a mixed solution (1:2 molar ratio) of 10 mM 11-mercaptoundecanoic acid (11-MUA) and 6-mercapto-1-hexanol (6-MCH) as described previously.61; 62 The hydroxyl-terminated self-assembled monolayer (SAM), which cannot be activated by N-hydroxysuccinimide (NHS) and N-ethyl-N’-(3-diethylaminopropyl) carbodiimide (EDC), was used as a spacer to construct the sensor surface. Second, following activation of the terminal carboxylic groups of the mixed SAM with a 1:1 mixture of 0.1 M NHS and 0.4 M EDC for 10 min, streptavidin (SA; 200 µg/ml; MP Biomedicals) in 10 mM sodium acetate buffer (pH 5.5) was injected and allowed to covalently couple for 15–20 min followed by PBS rinsing and blocking with 1.0 M ethanolamine hydrochloride (pH 8.5) for 10 min. This resulted in a large increase in the SPR wavelength (data not shown). Because of the close relationship that exists between electrostatic binding and covalent binding of SA with carboxylic terminated SAMs, this increase in SPR signal is a decent estimate of the extent of covalent binding of SA to the SAMs.61 Third, following a thorough PBS wash, biotinylated rabbit anti-E. coli antibody (140 µg/ml in PBS; Cortex Biochem) was injected over the SA surface for 20 min and unbound biotinylated anti-E. coli antibody was removed by washing with PBS for 10 min. This resulted in an exponential increase in the SPR wavelength that was characteristic of electrostatic binding between SA and biotin-conjugated proteins (data not shown). As control, bovine serum albumin (BSA; 140 µg/ml in sodium acetate buffer) was added in place of anti-E. coli antibody to the SA-coated reference channel of the SPR sensor chip and, as expected, no detectable SPR wavelength shift was observed following introduction of BSA or, subsequently, anti-E. coli antibody (data not shown).
Fluorescence microscopy
For immunofluorescence studies, E. coli cells that had been induced to express GFP or ClyA-GFP were washed three times in PBS, incubated at 4°C overnight with mouse anti-GFP (or anti-polyhistidine) diluted 1:500, pelleted, washed three times with PBS, incubated for 1 h with rhodamine-labeled goat anti-mouse IgG (Molecular Probes) diluted 1:100, pelleted and washed three more times with PBS. Finally, cells were examined by a Zeiss Axioskop 40 fluorescent microscope with Spotflex color digital camera and filter sets for GFP (485 nm for excitation and 505 nm for emission) and Rhodamine (540 nm for excitation and 600 nm for emission). For fluorescent studies of OMV interactions with eukaryotic cells, HeLa cells grown on glass coverslips were washed in OptiMEM (Life Technologies) without serum and then treated as described under Results. Following treatment, cells were fixed with 3.7% formalin in PBS, washed three times in PBS, permeabilized in PBS/0.1% Triton X-100, stained with 0.5 mg/mL ethidium bromide in PBS, and finally washed three times in PBS. Coverslips were mounted onto glass slides using Vectashield Hardset mounting medium (Vector Laboratories) prior to wide-field epifluorescence analysis. For WGA studies, non-permeabilized cells were incubated with 1 µg/ml Texas red WGA (Molecular Probes) for 1 h at 4°C. For GM1 experiments, ~150 µg of vesicles were preincubated with 10 µg GM1 (Sigma) for 30 min at 25°C.
Electron microscopy
Ultrastructural analysis of vesicles was performed by negative staining technique as described previously.63 For immunogold labeling, a 10-µL suspension of induced E. coli cells was collected, washed and applied to 400-mesh Formvar- and carbon-coated copper grids (Electron Microscopy Sciences) and incubated for 1 h with anti-GFP diluted 1:500. Cells were washed with PBS, incubated for 1 h with 25 nm colloidal gold-conjugated goat anti-mouse IgG (Electron Microscopy Sciences) diluted 1:100, washed again, negatively stained with 0.25% phosphotungstic acid (PTA, Electron Microscopy Sciences) with 0.01% BSA in water and viewed using a FEI/Philips Morgagni transmission electron microscope.
Cytotoxicity assay
Vesicles were prepared in PBS and total protein in vesicle fractions was quantified by the Coomassie Plus Assay (Pierce) using BSA protein standards. HeLa cells were grown in clear, flat-bottom tissue culture polystyrene 96-well plates (Costar) at an initial density of 5,000 cells per well in 200 µL of growth medium. After 24 h, the growth medium was removed and replaced with 110 µL of Opti-MEM I® (Invitrogen) serum-free medium and 40 µL of undiluted (1×; ~90–150 µg/ml total protein) or 1:1 diluted (0.5×; ~40–60 µg/ml total protein) OMV samples in PBS. Cells were incubated in the presence of vesicle samples for 4 h; afterwards the OMV-containing medium was removed and replaced with 175 µL of phenol red-free growth medium. Following an additional 48 h of incubation, 35 µL of CellTiter 96® Aqueous One Solution Cell Proliferation Assay reagent (Promega) was added to the wells. This assay uses the tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H–tetrazolium, inner salt; MTS] and the electron coupling reagent phenazine methosulfate. MTS is chemically reduced by cells into formazan whose concentration and optical absorbance at 490 nm provide a measure of metabolically active live cells. Samples were incubated for 1 h and the absorbance was read in a microplate spectrophotometer at 490 nm. Cell viability is reported relative to PBS controls.
Acknowledgements
We thank Kyung Hun Yoonand Michael Shuler (Cornell University) and Sang Hun Lee, Tai Hyun Park and Sung June Kim (Seoul National University) for their assistance with the SPR studies. We also thank Harris Bernstein, Wilfred Chen, George Georgiou, Koreaki Ito, Roland Lloubes, Virginia Miller, Sun Nyunt Wai and Joel Weiner for their generous gifts of strains, plasmids and antibodies. This material is based upon work supported by the Ladies Auxiliary to the Veterans of Foreign Wars Cancer Research Fellowship (to A.M.D.), a New York State Health Research Science Board (HRSB) Breast Cancer Research Fellowship (to A.M.D.), the National Institutes of Health under Grant R21 NIBIB EB005669 (to D.A.P. and M.P.D), and a NYSTAR James D. Watson Young Investigator Award (to M.P.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.jmb.2008.03.076
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4617544?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.jmb.2008.03.076
Article citations
The two coin sides of bacterial extracellular membrane nanovesicles: atherosclerosis trigger or remedy.
Discov Nano, 19(1):179, 12 Nov 2024
Cited by: 0 articles | PMID: 39532781 | PMCID: PMC11557815
Review Free full text in Europe PMC
Genetically surface-modified Escherichia coli outer membrane vesicles targeting MUC1 antigen in cancer cells.
Biotechnol Rep (Amst), 44:e00854, 03 Sep 2024
Cited by: 0 articles | PMID: 39290790 | PMCID: PMC11406022
In Silico Design of CT26 Polytope and its Surface Display by ClearColi™-Derived Outer Membrane Vesicles as a Cancer Vaccine Candidate Against Colon Carcinoma.
Appl Biochem Biotechnol, 03 Jul 2024
Cited by: 1 article | PMID: 38958886
Outer membrane vesicles as a platform for the discovery of antibodies to bacterial pathogens.
Appl Microbiol Biotechnol, 108(1):232, 24 Feb 2024
Cited by: 0 articles | PMID: 38396192 | PMCID: PMC10891261
Review Free full text in Europe PMC
The proteome of bacterial membrane vesicles in Escherichia coli-a time course comparison study in two different media.
Front Microbiol, 15:1361270, 06 Mar 2024
Cited by: 1 article | PMID: 38510998 | PMCID: PMC10954253
Go to all (92) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Delivery of foreign antigens by engineered outer membrane vesicle vaccines.
Proc Natl Acad Sci U S A, 107(7):3099-3104, 27 Jan 2010
Cited by: 159 articles | PMID: 20133740 | PMCID: PMC2840271
Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin.
Cell, 115(1):25-35, 01 Oct 2003
Cited by: 308 articles | PMID: 14532000
Display of Recombinant Proteins on Bacterial Outer Membrane Vesicles by Using Protein Ligation.
Appl Environ Microbiol, 84(8):e02567-17, 02 Apr 2018
Cited by: 40 articles | PMID: 29439988 | PMCID: PMC5881043
Cytolysin A (ClyA): A Bacterial Virulence Factor with Potential Applications in Nanopore Technology, Vaccine Development, and Tumor Therapy.
Toxins (Basel), 14(2):78, 21 Jan 2022
Cited by: 9 articles | PMID: 35202106 | PMCID: PMC8880466
Review Free full text in Europe PMC





