Abstract
Unlabelled
Studies of the cancer genome have demonstrated that tumors are composed of multiple subclones with varied genetic and phenotypic properties. However, little is known about how metastases arise and evolve from these subclones. To understand the cellular dynamics that drive metastasis, we used multicolor lineage-tracing technology in an autochthonous mouse model of pancreatic cancer. Here, we report that precursor lesions exhibit significant clonal heterogeneity but that this diversity decreases during premalignant progression. Furthermore, we present evidence that a significant fraction of metastases are polyclonally seeded by distinct tumor subclones. Finally, we show that clonality during metastatic growth-leading to either monoclonal or polyclonal expansion-differs based on the site of metastatic invasion. These results provide an unprecedented window into the cellular dynamics of tumor evolution and suggest that heterotypic interactions between tumor subpopulations contribute to metastatic progression in native tumors.Significance
Studies of tumor heterogeneity indicate that distinct tumor subclones interact during cancer progression. Here, we demonstrate by lineage tracing that metastases often involve seeding by more than one clone and that subsequent cellular outgrowth depends on the metastatic site. These findings provide insight into clonal diversity and evolution in metastatic disease.Free full text

Pancreatic Cancer Metastases Harbor Evidence of Polyclonality
Abstract
Studies of the cancer genome have demonstrated that tumors are comprised of multiple sub-clones with varied genetic and phenotypic properties. However, little is known about how metastases arise and evolve from these sub-clones. To understand the cellular dynamics that drive metastasis, we used multi-color lineage tracing technology in an autochthonous mouse model of pancreatic cancer. Here, we report that precursor lesions exhibit significant clonal heterogeneity but that this diversity decreases during pre-malignant progression. Furthermore, we present evidence that a significant fraction of metastases are polyclonally seeded by distinct tumor sub-clones. Finally, we show that clonality during metastatic growth – leading to either monoclonal or polyclonal expansion – differs based on the site of metastatic invasion. These results provide an unprecedented window into the cellular dynamics of tumor evolution and suggest that heterotypic interactions between tumor subpopulations contribute to metastatic progression in native tumors.
Introduction
Metastasis is thought to occur as a linear sequence of events involving cellular invasion, bloodstream entry, seeding, and colonization of host organs (1). Classically, this process has been conceptualized as the end result of genetic and epigenetic events involving a single cell, leading to clonally-derived lesions at distant sites (1–4). Alternatively, it has been proposed that interactions between distinct tumor sub-clones may promote tumor dissemination (5–8), and recent data from implantable breast cancer models support the view that metastases can arise from collective migration and colonization of tumor cells (6, 9). Importantly, these observations have been based primarily on aggregate sequencing data and cell transplantation assays, and thus the extent to which they reflect the behavior of cancer cells in their native environment (and in solid tumors other than breast cancer) remains unclear.
Pancreatic ductal adenocarcinoma (PDAC) is predicted to soon become the second leading cause of cancer-related death in the US, with an overall five-year survival rate of 5.8% (10). Many factors influence this poor outcome, but the presence of metastatic disease at the time of diagnosis is a pivotal contributor. Genetic sequencing studies of human PDAC have demonstrated the presence of extensive clonal heterogeneity in primary tumors (11, 12). However, the manner in which these sub-clones contribute to metastases is unclear, as sequencing-based approaches are limited in their ability to assess cellular dynamics.
Lineage labeling has proven to be a key tool in addressing questions of cell fate during tumor progression. We previously demonstrated the utility of lineage tracing to understand the role of early cellular dissemination in an autochthonous model of metastatic pancreatic cancer (13). With the advent of multiplexed labeling, it is now possible to track the contributions of multiple distinct cellular populations, including cancer stem cells, to tumor growth (14, 15). We reasoned that these methods could be exploited to study the clonality of metastases in vivo in the native tumor environment. Accordingly, we used a multi-color reporter system to investigate clonal evolution during metastasis in a model driven by stochastic events.
Results
Modeling tumor heterogeneity
To track tumor subpopulations during malignant progression, we employed the “Confetti” lineage-labeling system (14, 15), in which Cre mediated recombination leads to stochastic expression of one of four fluorescent proteins in any given cell. A series of crosses were performed to generate “KPCX” mice (Fig. 1A) in which a tamoxifen (TAM)-inducible Cre recombinase (Pdx1-CreERTM; “C”) (16) simultaneously activates an oncogenic KrasG12D allele (“K”) (17), deletes a single p53 allele (“P”) (18), and generates a color-producing recombination event within the RosaConfetti locus (“X”). The efficiency of recombination in Pdx1-CreERTM; RosaConfetti (“CX”) mice following TAM administration was found to be ~65% (Supplementary Fig. S1A). All four colors were represented in pancreatic parenchymal cells (Fig. 1B), although GFP cells were significantly under-represented compared to CFP, RFP, and YFP cells, as has been previously reported (15). Importantly, cell marking was stable, as clonally-isolated cells maintained in culture for weeks to months exhibited no evidence of color-switching (Supplementary Fig. S1B).
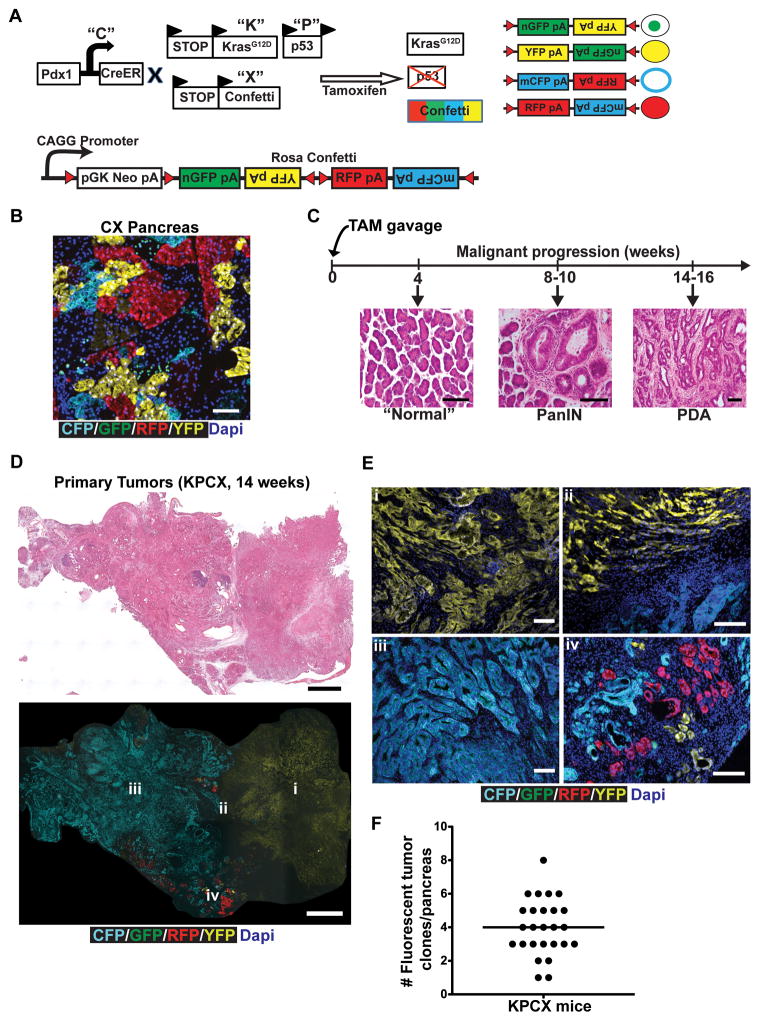
A, Schematic of the KPCX mouse model of pancreatic cancer used in this study, which employs the KrasG12D (“K”), p53fl/+ (“P”), Pdx1-CreER (“C”), and RosaConfetti (“X”) alleles. Tamoxifen inducible expression of the pancreas-specific Cre leads to expression of activated mutant KrasG12D, deletion of one allele of the p53 tumor suppressor, and recombination of the multi-color RosaConfetti Cre-reporter. Recombination of loxP sites within the Confetti locus results in labeling of pancreatic cells with one of 4 possible colors: nuclear GFP (green), cytoplasmic YFP (yellow), membrane CFP (cyan), and cytoplasmic RFP (red).
B, Representative confocal fluorescent image of a section from a 10-week-old CX mouse pancreas depicting expression of the different RosaConfetti fluorescent labels. Labeling is principally seen in acinar cells, with negligible labeling of duct cells.
C, H&E images of malignant progression in KPCX mice following tamoxifen (TAM) administration at birth. The pancreata of KPCX mice are initially normal but develop ADMs, PanINs, and PDAC with a reproducible time-course.
D, Representative tile scan images of a cross-section through a KPCX pancreas. H&E staining (top panel) and fluorescence imaging (bottom panel) demonstrate the presence of two anatomically distinct monochromatic primary tumors within an apparent single pancreatic mass.
E, Magnified fluorescent images from the tumor center (i, iii), periphery (iv) and border between adjacent clones (ii) of the pancreatic tumors depicted in D.
F, A mean of 4 distinct monochromatic tumor lesions (as depicted in D, E) were found in each KPCX mouse (●). Data pooled from 26 tumor-bearing KPCX mice.
Scale bars 100 μm for B, C, and E and 300μm for D.
Two other models of PDAC – “KPC” and “KPCY” (which carries a YFP allele instead of the Confetti allele employed here) – recapitulate most salient aspects of the human disease (13, 17). Because oncogenic mutations in the KPCX model occur postnatally, we compared features of tumor formation in KPCX mice to these other, more well-established, PDAC models. Following TAM administration to pups, KPCX mice developed PanIN lesions within 8–10 weeks and invasive PDAC within 14–16 weeks, a time-course consistent with that described for KPC and KPCY mice (Fig. 1C and Supplementary Fig. S1C). Moreover, KPCX tumors were indistinguishable from KPC and KPCY tumors at a histological level (Supplementary Fig. S1D) and exhibited a similar pattern of metastatic spread (predominantly liver, lung, parietal peritoneum, and diaphragm). Thus, KPCX mice behaved similarly to KPC and KPCY mice, even though Kras and p53 mutations were generated after birth.
Despite having what appeared at a gross level to be a solitary dominant tumor mass (Fig. 1D, top panel), examination of color distributions revealed that most pancreatic lesions were actually a conglomerate of multiple tumors arising from independent initiating events (Fig. 1D, bottom panel and Fig. 1E). Individual tumor foci contained a single-colored (“monochromatic”) population of cells, which shared borders with adjacent tumor, PanIN, or normal tissue (Fig. 1E). Quantification following serial sectioning revealed that each pancreatic mass contained, on average, 4 of these monochromatic lesions (Fig. 1F). Retrospectively, it was sometimes possible to recognize these multifocal tumors by virtue of regions with distinctive histology, a phenomenon also observed with KPC and KPCY mice (data not shown).
PanIN lesions are associated with a clonal bottleneck
Multiple lines of evidence suggest that PDACs arise from PanINs, which themselves arise from acinar-to-ductal metaplasia (ADM) (13, 19). We thus sought to understand how and when clonality changes during pre-malignant stages of tumor progression. To this end, we examined serial pancreatic sections from 8–10 week-old KPCX mice, a time-point at which only PanINs and ADMs were present. We found that roughly a fourth (24%) of all ADMs were polychromatic, indicating that they arose from multiple distinct acinar cells (Fig. 2A and C). By contrast, almost all PanIN lesions (97%) were found to be monochromatic (Fig. 2B and C). In the context of an ADM → PanIN → PDA model, therefore, these results suggest that premalignant progression is associated with a loss of clonal diversity during the transition from ADM to PanIN.
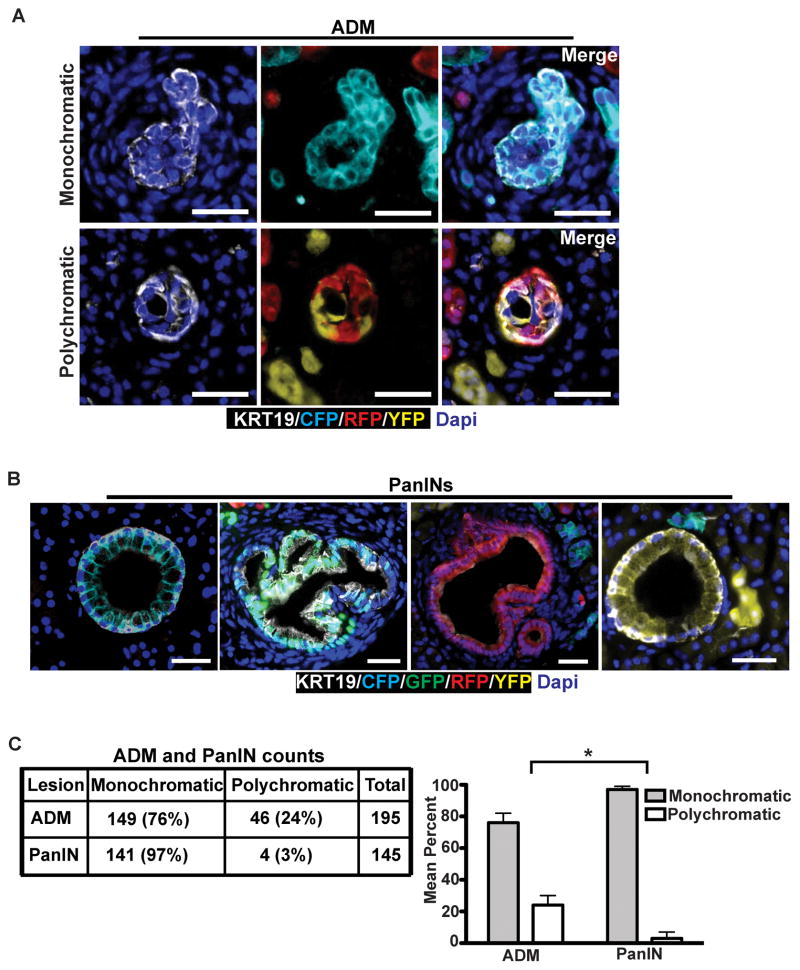
A and B, Fluorescent images of acinar-to-ductal metaplasia (ADM; A) and pancreatic intraepithelial neoplasia (PanIN; B) of 10 week-old KPCX mice. In A, ADMs are positive for the ductal maker Krt19 (white). Representative examples of monochromatic (top) or polychromatic (bottom) ADMs are shown. In B, representative images of monochromatic PanINs are shown.
C, Quantification of monochromatic and polychromatic ADMs and PanINs in 10 week-old KPCX mice. Data were pooled from n=3 mice, and the total number of lesions counted is shown. The bar graph on the right shows the mean percentage of ADMs and PanINs in each group. Error bars represent 95% confidence intervals. * p<0.001 by Fisher’s exact test between lesion type in ADMs and PanINs.
Scale bars 25 μm for A and 50μm for B.
Polyclonality in peritoneal and diaphragmatic metastases
We next sought to understand how clonality evolves during metastasis. To this end, we used the multifocal nature of KPCX tumors to model sub-clonal heterogeneity. In human cancers, sub-clones are defined genetically as distinct tumor populations that share a set of identical founder mutations (2, 11, 20). In this regard, each fluorescent KPCX clone shares an identical KRAS and p53 mutation but otherwise evolves independently and can thus be used to represent a distinct sub-clonal population. Using the fluorescent marker in each lesion, we then tracked their contribution to metastasis formation.
We initially examined metastases to the peritoneal wall and diaphragm. Each KPCX animal had, on average, 2–4 distinct large metastatic foci at each of these sites (Supplementary Fig. S2A, S2B, and S2C). Importantly, these metastases were spatially well-separated from the pancreas and from each other, eliminating the possibility of direct local invasion as a route of spread. Surprisingly, nearly 80% of these large lesions were polychromatic (Fig. 3A, B, and C), indicating that they originated from more than one source in the primary tumor. Likewise, examination of peritoneal and diaphragmatic micro-metastases also revealed the presence of polychromatic lesions (Fig. 3D, Supplementary Fig. S2A), demonstrating that polyclonality is present in small as well as large metastatic lesions. Interestingly, polychromatic metastases were comprised of at most two distinct populations (i.e. tri-colored metastases were not observed even if the pancreas harbored three distinctly-colored tumors). Thus, pancreatic cancer metastases to the peritoneum and diaphragm are frequently polyclonal.
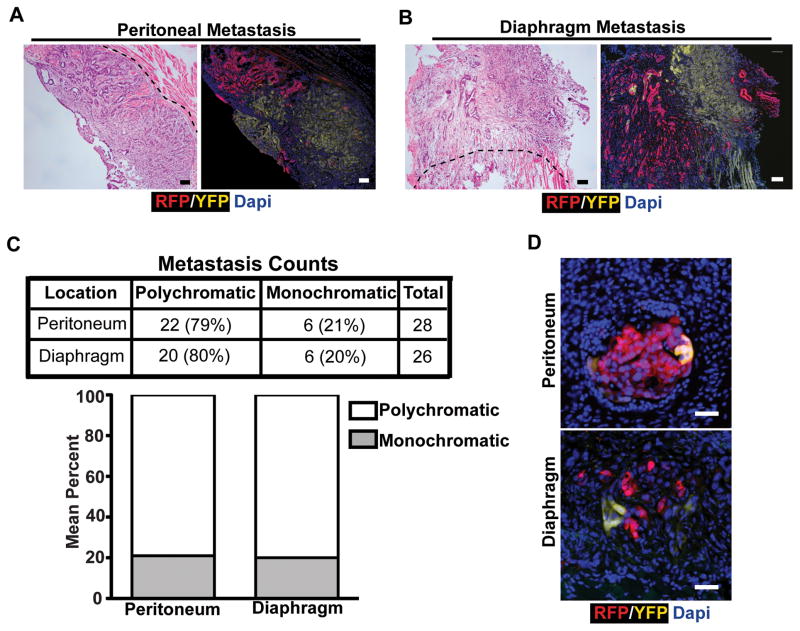
A, Representative fluorescent images and H&E stains of adjacent sections depicting polychromatic metastasis in the peritoneum.
B, Representative fluorescent images and H&E stains of adjacent sections depicting polychromatic metastasis in the diaphragm.
C, Total counts and percentages of metastases in each site. Data pooled from n= 7 (peritoneum) and n=9 (diaphragm) 14–16 week old tumor-bearing KPCX mice.
D, Fluorescent images of microscopic metastases from the peritoneal lining (top panel) and diaphragm (bottom panel).
Scale bars 100 μm for A and B, and 25μm for D.
Evidence that polyclonal diaphragmatic metastases come from polyclonal clusters
Polyclonal metastases could arise from the outgrowth of lesions that were polyclonal at the time of seeding or through a two-step mechanism involving seeding by one clone and subsequent recruitment (re-seeding) by another (21). Examination of ascites fluid from tumor-bearing KPCX mice revealed the presence of bi-chromatic cellular aggregates ranging in size from 2 to 50 cells in 5 out of 5 KPCX mice (Fig. 4A), suggesting that polyclonality might arise through the former mechanism.
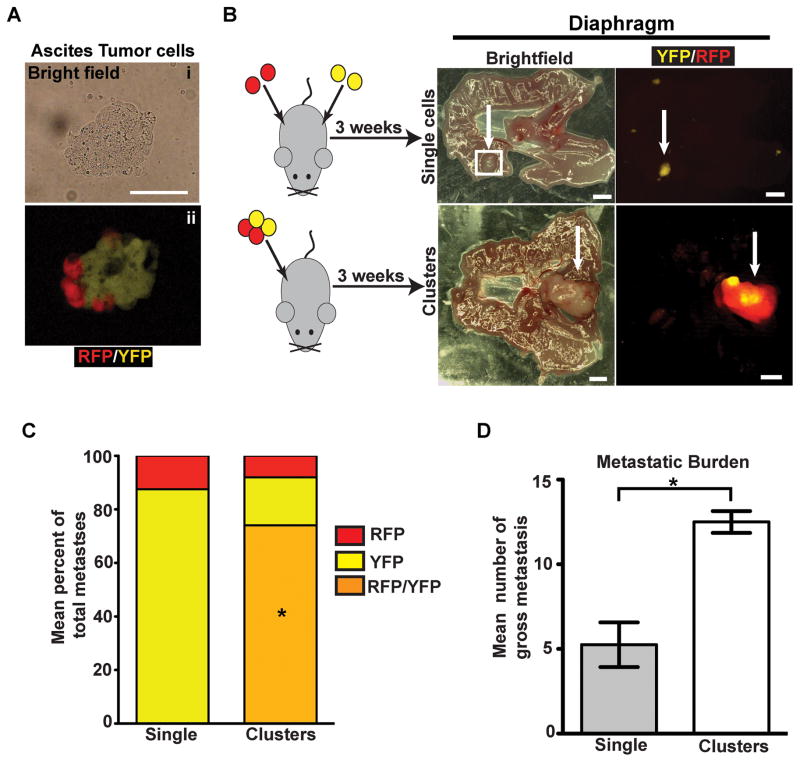
A, Bright field (i) and fluorescent images (ii) of a multi-colored cluster of disseminated tumor cells in the ascites.
B, Intraperitoneal injection of 458d_R and 458d_Y cells (30,000) either as a suspension mixture of single cells (top panel) or multi-color clusters (bottom panel) into NOD.SCID mice. Images are paired brightfield (left panel) and fluorescent (right panel) stereomicroscope images from mice 3 weeks following injection with n=4 mice for each group. Monochromatic lesions are either YFP or RFP positive and polychromatic lesions are both YFP and RFP positive.
C, Bar graph depicting mean percentage of total gross monochromatic (RFP or YFP only) or polychromatic (positive for both YFP and RFP) metastases between single cell and cluster injection groups. Data pooled from n=4 mice for each group (a total of 24 lesions were counted in the single cell group and 50 lesions were counted in the cluster group). No lesions from the single cell injection group were polychromatic. *p<0.001 by Fisher’s exact test comparing multi-color metastases between single cell and cluster injections.
D, Bar graph depicting the mean number of gross metastases in the single cell and cluster injection groups (n=4 mice in each group). Error bars represent SEM. *p=0.0026 by Student’s t-test.
Scale bars 25 μm for A and 1mm for B.
To further understand the dynamics of polyclonal metastasis formation, we performed a series of in vivo cell mixing experiments in which low passage cell lines were derived from a multi-color (RFP/YFP) diaphragmatic metastasis and then FACS sorted into their RFP (458d_R) and YFP (458d_Y) components (Supplementary Fig. S3A and S3B). We then injected 30,000 cells intraperitoneally into 6–8 week-old NOD.SCID mice, either as a 1:1 suspension of single 458d-R and 458d-Y cells or after the cells were allowed to form multi-colored clusters (Fig 4B and Supplementary Fig. S3B). After 3 weeks, diaphragmatic tissue was harvested and the number of gross monochromatic and polychromatic lesions was examined using fluorescent stereomicroscopy. Significantly, although polychromatic lesions constituted the majority (70%) of metastases following injection of cell clusters, only monochromatic lesions were seen following injection of the single cell suspension (Fig. 4B and C). Furthermore, cell clusters were more efficient than single cells at forming metastases (Fig. 4D). Taken together, these data suggest that polyclonal peritoneal metastases develop from multi-clonal aggregates shed from the primary tumor.
Polyclonality in liver and lung metastases
We then turned our attention to liver and lung, the most common sites of metastasis in human PDAC (12), which are thought to arise through hematogenous dissemination. To characterize the size distribution of metastatic foci, we counted individual lesions and binned them according to size: single cell, nano- (2–10 cells), micro- (11–100 cells), and macro-metastases (>100 cells) (Supplementary Fig. S4A and S4B). This analysis revealed that the majority of liver and lung metastases in KPCX mice were single cells or nano-metastases, with only 10–26% having more than 10 cells (Supplementary Fig. S4C). We then analyzed metastatic deposits in each size category for number and color composition. This revealed that 11–14% of nano- and micro-metastases contained two clones (Fig. 5A, B, and C); tri-colored metastases were never observed. Surprisingly, and in contrast to our findings in peritoneum and diaphragm, large polyclonal metastases were also not observed (Fig. 5B and C).
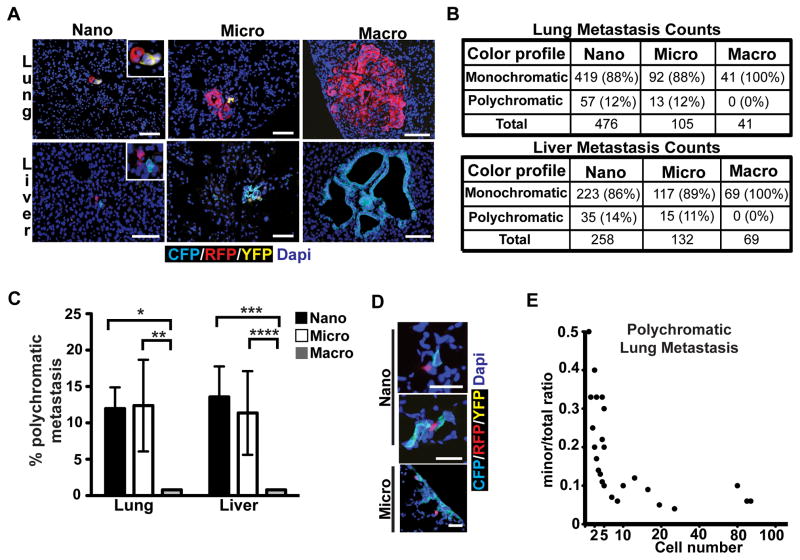
A, Representative fluorescent images of metastases in lung (top panels) and liver (bottom panels).
B, Quantification of monochromatic and polychromatic lesions in lung (top) and liver (bottom) binned according to lesion size (nano: 2–10 cells; micro: 11–100 cells; and macro: >100 cells). Percentages are relative to the total number of metastases counted for each size category. Data are pooled from n=6 (lung) and n=5 (liver) tumor-bearing animals.
C, Bar graph depicting the data presented in B. Error bars represent 95% confidence intervals. *p=0.016, **p=0.02, ***p=0.0003, and ****p=0.0016 by Fisher’s exact test.
D, Fluorescent images of bi-chromatic lung metastases depicting an increase in the ratio of the major component (CFP) relative to the minor component (RFP) as a function of lesion size.
E, Curve showing the trend depicted in (C). The ratio of the number of cells in the minor fraction relative to the total number of fluorescent cells in a metastatic lesion (y-axis) is plotted as a function of the total fluorescent cell number in the lesion (x-axis). Each (●) represents a single lung metastasis. Data were taken from 63 individual lung metastases pooled from n=6 mice (lesions with identical ratios and size are represented as a single data point). p<0.0001 by Wald Chi-square test.
Scale bars 50 μm for A and D
This result raised the possibility that the development of large metastases in the lung and liver might involve preferential outgrowth of one clone over another. To test this possibility, we quantified changes in the color composition of polychromatic liver and lung metastases as a function of cell number. Consistent with the notion of selective outgrowth, we found that lesions became increasingly dominated by a single clonal population as they increased in size (Fig. 5D and Supplementary Fig. S5A). This process was evident early, with ~90% of each polychromatic lesion dominated by one clone by the 10-cell stage (Fig. 5E and Supplementary Fig. S5B). These results indicate that while small metastatic lesions in the lung and liver may be polyclonal, there is an early drift towards monoclonality during metastatic growth in these tissues.
Evidence that polyclonal clusters seed polyclonal lung metastases
As with diaphragmatic and liver metastases, polyclonal liver and lung metastases could arise via seeding of polyclonal lesions present in the circulation or by seeding of one clone followed by subsequent seeding by a second clone. Examination of blood smears from tumor bearing KPCX mice revealed that clusters of fluorescent tumor cells were abundantly present in the bloodstream, where they comprised approximately 7–20% of all circulating tumor cells (Fig. 6A). Furthermore, in 2 out of 6 mice multi-color clusters were detected in the circulation (Fig. 6B).
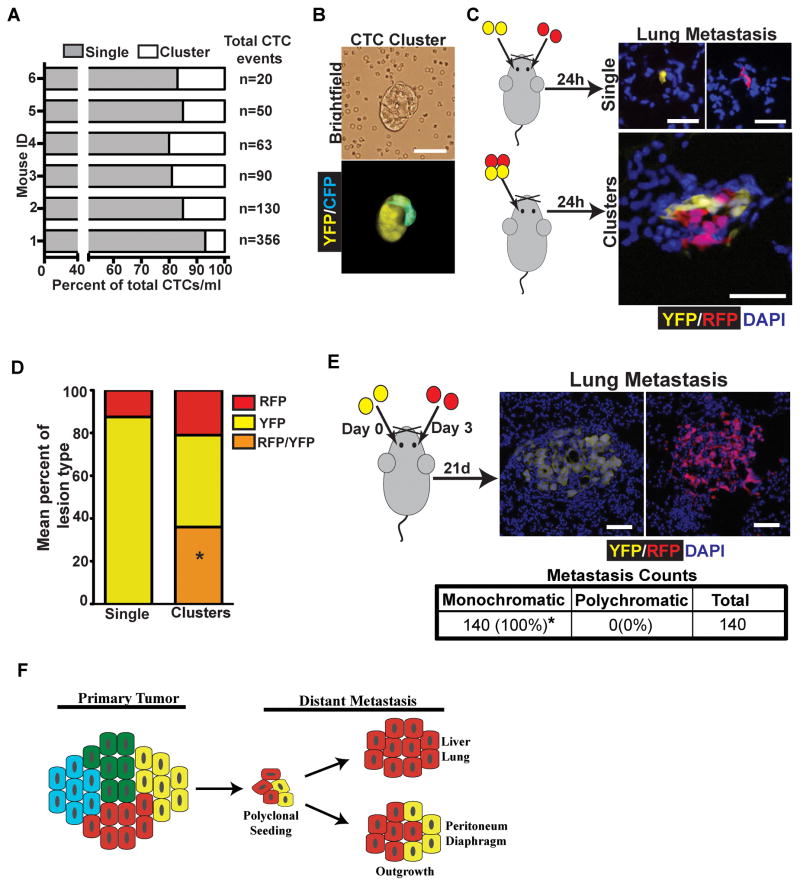
A, Bar graph depicting the mean percentage of single cell CTC and CTC cluster events per ml of blood in 6 individual KPCXY tumor-bearing mice. Total CTC events/ml are shown for each mouse are listed to the right of the graph. Percentage of CTC clusters ranged from 7–20%.
B, Brightfield (top panel) and fluorescent (bottom panel) image of a multi-color CTC cluster isolated from the blood of a KPCXY mouse.
C, Retro-orbital injection of 458d_R and 458d_Y cells (20,000) either as mixture of single cells (top panel) or multi-color clusters (bottom panel) into NOD.SCID mice. Right panels are representative fluorescent images of resulting metastatic lung lesions in the two injection groups.
D, Quantification of the data in C. The mean percentage of monochromatic (RFP or YFP only) or polychromatic (positive for both RFP and YFP) metastases are indicated in stacked graph format for each injection group (single cell or cluster). Data are pooled from n=4 mice for each group (a total of 208 lesions were counted for the single cell group and 607 lesions were counted for the cluster group). *p<0.001 by Fisher’s exact test comparing the frequency of polychromatic metastases between single cell and cluster injections.
E, Retro-orbital injection of 458d_R and 458d_Y cells as sequential injections of single cells into NOD.SCID mice separated by 3 days. Right panels are representative fluorescent images of lung metastatic lesions detectable 21 days later. Table shows total metastatic counts and the percentage of monochromatic and polychromatic lesions. Data pooled from n=4 mice. *p<0.001 by Fisher’s exact test.
F, Model for the development of polyclonal metastases. Polyclonal cell clusters derived from the primary tumor give rise to polyclonal seeding events, followed by either monoclonal or multi-clonal outgrowth depending on tissue site.
Scale bars 25 μm for B, C, and E.
To further resolve the cellular mechanisms by which polyclonal metastases form, we again performed a series of cell-mixing experiments with the 458d_R and 458d_Y cells in a retro-orbital injection model of lung metastasis (22). First, we injected 458d_R and 458d_Y cells simultaneously (either as a single cell suspension or as clusters) and then examined the lungs 24 hours post-injection (Fig 6C). Similar to our results with peritoneal metastasis, metastatic burden was markedly higher following injection of tumor cell clusters compared to single cells when the same number of cells was introduced (Supplementary Fig. S6A and S6B).
We then examined the color makeup of the metastatic lesions following injection of single cells or clusters (Fig. 6C). While roughly 36% of metastatic lesions following the injection of clusters were polychromatic, all lesions seen following injection of single cells were monochromatic (Fig. 6D). This result suggests that cellular aggregates capable of giving rise to polychromatic metastases do not readily form in the bloodstream. Finally, to assess whether multi-colored lung metastases could arise via recruitment, we injected 20,000 458d_R or 458d_Y cells retro-orbitally on day 0, followed by injection of 20,000 cells of the other population on day 3, and then assessed the color makeup of the resulting metastases (Fig. 6E). Consistent with our previous results, all resulting metastases were monochromatic (Fig. 6E), suggesting that multi-colored metastases in this model are likely to come from seeding by polyclonal clusters of tumor cells rather than sequential rounds of seeding.
Discussion
Efforts to understand the clonal origins of metastases often invoke cell autonomous factors as primary determinants of spread (1). Using an unbiased lineage-labeling approach, we have observed that metastases can have polyclonal origins and that further outgrowth can differ based on the location of the lesion. Importantly, such evidence comes from an autochthonous model, making it likely that this phenomenon reflects cellular behavior in the context of naturally-evolving tumors. Taken together, our results are consistent with a model in which heterotypic interactions between tumor sub-clones, in conjunction with site-dependent selective pressures, influence metastatic initiation and progression (Fig. 6F).
Genomic sequencing of human primary pancreatic tumors and their associated metastases has failed to identify a consistent pro-metastatic gene signature or “metastatic driver” mutations (11, 12). These findings suggest a role for non-genetic mechanisms during metastasis formation, and recent data from implantable tumor models suggest that cell-cell interactions may facilitate metastasis development by enhancing collective invasion, altering the tumor microenvironment, and/or cooperatively enhancing the fitness of different tumor subsets (6, 7, 9, 23). Our data are consistent with this notion of cooperativity, as the high frequency of polyclonal metastasis in the KPCX model (11–14% in lung and liver and 80% in peritoneum and diaphragm) and the enhanced metastatic phenotype associated with disseminated cell clusters observed by us and others (9) suggests that clonal interactions may provide a metastatic advantage. While the molecular nature and functional consequences of such putative interactions remain to be determined, it is enticing to imagine that clonal collaboration improves the efficiency of metastatic spread by enhancing cellular invasion, facilitating cell survival in the bloodstream, and/or improving seeding/colonization at distant sites.
One can envision several mechanisms by which polyclonal metastases could arise, including (i) seeding by polyclonal clusters shed from the primary tumor, (ii) seeding by polyclonal clusters that form within ascites fluid or the circulation, or (iii) independent seeding by distinct clones at the same location, either simultaneously or sequentially. Using low passage cell lines derived from a polyclonal YFP/RFP metastasis, we found that injection of polychromatic clusters, but not single cells, resulted in polychromatic metastases. Because either of the latter mechanisms should have resulted in polychromatic lesions following the injection of single cells, the failure to find such lesions supports the first possibility (i.e. polyclonal lesions arise from pre-existing clusters). Consistent with this idea, we did not observe tri-color metastases in the KPCX model, even in animals that harbored three or more differently colored pancreatic tumors, a result that argues against re-seeding or latent (post-shedding) aggregation.
Metastatic progression is influenced by a combination of tumor cell-intrinsic properties and environmental factors that are unique to each metastatic site (4). This interplay is reflected in our system as evidenced by the differences in clonal outgrowth between organ sites, whereby peritoneal and diaphragmatic metastases remain polyclonal and lung and liver lesions drift towards monoclonality. While this observation may in part reflect differences in the behavior of lesions derived from early versus late dissemination, it also suggests that the peritoneum and diaphragm provides a more permissive state for multi-clonal expansion. Conversely, factors intrinsic to lung and liver may exert significant selective pressures during metastatic growth.
One advantage of lineage-tracing is that it permits the recognition of selective “bottlenecks” at discrete stages during the life of a stochastically-evolving tumor. Specifically, we observed that nearly a quarter of ADMs are polyclonal while almost all PanINs are monoclonal. As ADMs are widely viewed as the precursors to PanINs (19), this observation suggests that PanIN formation is associated with clonal expansion. For example, such bottlenecks could reflect the outgrowth of a Kras/p53-mutant cell from a field of wild-type cells that have undergone ADM. Alternatively, this reduction in clonal diversity might reflect the outgrowth of a cell with additional genetic and/or epigenetic changes from a field of cells that are already mutant for Kras and p53. Likewise, the transition to frank carcinoma is associated with a further decrease in clonal diversity, as only a few dominant lesions emerge from the large number of PanIN precursors present in each KPCX pancreas. These results indicate that selective pressures acting throughout tumorigenesis can influence the clonal progression through each of these bottlenecks. In the future, it should be possible to use this system to delineate the mechanistic underpinnings of these hurdles to progression in native tumor environments, providing new therapeutic avenues.
Methods
Mice
To perform lineage labeling, a series of backcrosses were performed to introduce the RosaConfetti (“X”) reporter allele into mutant strains bearing Pdx1CreER (“C”), KrasG12D (“K”), and p53fl/+ (“P”) alleles to obtain Pdx1CreER; KrasG12D; p53fl/+; RosaConfetti (“KPCX”) and Pdx1CreER; RosaConfetti (“CX”) mice. For some experiments, animals were homozygous for the reporter allele or contained the RosaYFP (“KPCXY”) in lieu of the second confetti allele. To induce recombination, a suspension of TAM (MP Biomedicals) in corn oil (Sigma Aldrich) was administered to pups via lactation following oral gavage of the mother with 6 mg of the drug on post-natal day 0, 1, 2, and 4. All experiments involving control CX mice were performed at 8–10 weeks of age following TAM administration as described above. For studies involving PanIN lesions, KPCX mice were sacrificed at 10 weeks of age, a time point at which there was no evidence of carcinoma based on histological evaluation. PDAC-bearing KPCX mice were examined three times per week for evidence of morbidity and euthanized accordingly. On average, tumor bearing KPCX mice were 14–16 weeks of age at time of sacrifice (as shown in Supplementary Fig. S1C). Pdx1Cre; KrasG12D;p53fl/+; RosaYFP (“KPCY”) mice used in this study were generated as previously described (13). All experiments were performed in accordance with the National Institutes of Health policies on the use of laboratory animals and approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Cell Sorting and Culture
Pancreatic tumors were dissociated into single-cell suspensions through mechanical separation and enzymatic digestion as described (13). FACS sorting was performed using the FACSAriaII (BD biosciences) sorter at the Penn PathBioResource Flow Cytometry Core. Excitations of confetti colors were performed using the 488 nm argon laser for YFP, 405 nm violet laser for CFP, and the 532 nm red laser for RFP. Detection was performed using bandpass filters at 530/40 nm for YFP, 450/50 nm for CFP, and 575/25 nm for RFP. Cells were collected into a 2% BSA/PBS solution and cultured in pancreatic ductal cell media (13). For monitoring of fluorescent cell marker stability, >5000 cells were counted from passage 1 and passage 5 cultures following a 3–4 week interval. Total numbers of each fluorescent cell type was counted and averaged from 5 replicates.
For in vivo cell mixing experiments, cells were isolated as described above form a polychromatic diaphragmatic metastasis (mouse 458d) containing RFP and YFP populations. Cells were FACS sorted and cultured as described. No cell line authentication was performed, as all cell lines used in this paper were obtained from primary cultures of tumors derived from KPCX mice.
Image acquisition and quantification
Immunofluorescent (IF) images were acquired using both confocal and inverted fluorescent microscopes. The Zeiss LSM 710 confocal microscope employed the argon laser 488 nm line to excite nuclearGFP (nGFP), 514 nm line for YFP, 561 nm red diode laser for RFP, laserline at 458 nm for membraneCFP (mCFP), and 405 nm laser line for Dapi. Fluorescence was collected between 495–514, airy 0.41 for nGFP; 524–563, airy 0.41 for YFP; 585–622, airy 0.41 for RFP; 462–498, airy 0.41 for mCFP; and 417–494, airy 0.41 for Dapi. Images were taken with Zen 2011 software and spectral imaging coupled with image analysis using linear unmixing was performed. Imaging was performed with the Olympus IX71 inverted multicolor fluorescent microscope equipped with the following dry objective lenses: 4x (UPlanFLN NA0.13), 10x (UPlanFLN NA0.30), 20x (UPlanFLN NA0.50), 40x (UPlanFLN NA0.75). A mercury short arc lamp source (Osram) and the following excitation\emission filter sets (Chroma) were used to detect fluorescence: 535/500 nm for YFP/nGFP, 480/436 nm for CFP, 645/560 nm for RFP, 406/350 nm for Dapi, and 690/640 nm for Far Red. Images were captured with the DP71 camera (Olympus). Cytoplasmic YFP was distinguished from nuclear GFP based on cellular localization; GFP recombination events were rare compared to YFP. Histological images were captured using the Olympus BX1 light microscope attached to the DP25 camera (Olympus). Montage H&E images were taken with the EVOS FL auto cell imaging system (Thermo Fischer) with a 10x objective. Stereomicroscope images were obtained using the Lecia M216FA fluorescent microscope. Assembly and analysis of all CMYK images were performed using ImageJ 1.47v software.
Multicolor immunofluorescence and histological analysis
Tumors and tissue samples were fixed in 4% paraformaldehyde at room temperature for 30–60 minutes followed by an overnight incubation in 30% weight/volume sucrose solution. Samples were then embedded in O.C.T. (Tissue-Tek) and frozen on dry ice. Once solid, 10 μm sections were cut with a Microm HM550 cryostat (Thermo Scientific). Serial sectioning was performed on all tissue samples with 70–100 μm between sections. All sections were laid out in a similar orientation on each slide so that any given lesion could be tracked over multiple levels based on anatomical tissue landmarks and XY coordinates on the slide. Sections were stained with the nuclear marker Dapi (Life Technologies, 1:1000) and the ductal marker cytokeratin-19(24) (KRT-19, rabbit, 1:1000). For immunostaining, frozen tissues were blocked in StartingBlock (Thermo) with 2% donkey serum and 0.1% Tween-20 for 1 h, and incubated for 1h in primary antibody diluted in blocking buffer in a humidified chamber. Sections were washed three times in PBS containing 0.1% Tween-20 and incubated with DAPI and Alexa flour 633-conjugated antibodies at a 1:250 dilution for 1 h at room temperature followed by an additional 3 washes, and mounted on slides with Aqua polymount mounting reagent (Polysciences). Histological analyses were performed on adjacent frozen sections with heamtoxylin and eosin staining using standard protocols. Recombination efficiency of the RosaConfetti allele was examined in CX mice by counting the fluorescent cells of each color and dividing by the total Dapi positive cells. Data was gathered from 5 random pancreas fields in 5 adjacent levels. Dapi positive cells were counted using ImageJ 1.47v software resulting in >16,000 Dapi positive cells counted per pancreas. ADMs and PanINs were identified in pancreatic tissue based on typical histologic features (13) and positive staining for KRT-19.
Quantification was performed by counting the total number of ADMs and PanINs in one section from each level throughout the entirety of the pancreas. Lesions were categorized as monochromatic if >95% of cells in the lesion were of a single color and polychromatic if they contained cells of more than one color. To ensure accuracy, all PanINs that were identified as monochromatic were tracked in adjacent levels in their entirety. Analysis of metastases was performed by counting all distinct fluorescent lesions in one section from each level throughout the organ. To avoid counting larger lesions more than once, all lesions >50 cells in size were tracked in all adjacent levels in their entirety. Lung and liver metastases were binned based on the number of Dapi positive fluorescent cells in each lesion. Groupings included single (1 cell), nano (<10 cells), micro (11–100 cells), and macro (>100 cells). Lesions were scored as monochromatic if >95% of the cells in a lesion were the same color, otherwise they were considered polychromatic. Counting of lung metastases from in vivo mixing assays was performed on multiple sections taken from each organ. Diaphragmatic lesions were assessed based stereomicroscopic images. To quantitate the number of distinct lesions within each primary tumor mass, we defined a “tumor clone” as an anatomically contiguous region of monochromatic cells that shared distinct histological and IF boarders with adjacent clones as examined in multiple levels throughout the pancreatic mass.
In vivo cell mixing experimental metastasis assay
For intraperitoneal injections, equal number of cells (30,000) were injected either as a mixture of single cells or as clusters of 458d_R and 458d_Y cells into the peritoneal cavity of 6–8 week old NOD.SCID mice using an 27 gauge needle. For lung metastasis equal number of cells (20,000) were injected retro-orbitally, as previously described (22), either as a mixture of single cells or as clusters of 458d_R and 458d_Y cells into 6–8 week old NOD.SCID mice. Clusters were generated by mixing equal number of RFP and YFP cells in a low-attachment petri dish (Corning) and placed on a rocker for 8–12hrs in a 37° incubator.
Analysis of cell clusters from ascites and blood
Ascitic fluid was isolated from the abdominal cavity of euthanized tumor bearing mice with a 3 cc insulin syringe containing 100 μl of 1mg/ml heparin sulfate (Sigma Aldrich, H3149) to prevent coagulation. Fluid was immediately placed on a 100cm plate (BD Falcon) containing PBS with minimal manipulation. Monochromatic and polychromatic disseminated tumor cell clusters were identified by direct visualization on a fluorescent microscope and imaged. Similarly, blood was obtained via cardiac puncture and immediately placed on a 100 cm plate (BD falcon) containing PBS with minimal manipulation and then visually examined for cell clusters.
Acknowledgments
We thank E. Dekleva, E. Mirek, A. Sahmoud, and C. Yang for technical assistance and N. Aiello, N. Bhagwat, D. Balli, and A. Penzo for helpful discussions. We are grateful to K. Polyak, A. Rustgi, and R. Vonderheide for comments on the manuscript, to A. Ewald for sharing unpublished data and K. Forde for assistance with statistical analysis.
Grant Support: This work was supported in part by National Institutes of Health grant CA169123 (B.Z.S.) and CA076931 (R.M.), the Penn Center for Molecular Studies in Digestive and Liver Diseases (NIH P30-DK050306), and the Abramson Family Cancer Research Institute.
Abbreviations
| PDAC | pancreatic ductal adenocarcinoma |
| PanIN | pancreatic intraepithelial neoplasia |
| ADM | acinar-to-ductal metaplasia |
Footnotes
Conflict of Interest: The authors have no conflicts to disclose.
Author Contributions: R.M. and B.Z.S designed and interpreted the experiments and wrote the manuscript. R.M. performed the experiments.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/2159-8290.cd-15-0120
Read article for free, from open access legal sources, via Unpaywall:
https://cancerdiscovery.aacrjournals.org/content/candisc/5/10/1086.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Claudin 7 suppresses invasion and metastasis through repression of a smooth muscle actin program.
J Cell Biol, 223(12):e202311002, 25 Sep 2024
Cited by: 0 articles | PMID: 39320351
Circulating tumor cells: from new biological insights to clinical practice.
Signal Transduct Target Ther, 9(1):226, 02 Sep 2024
Cited by: 0 articles | PMID: 39218931 | PMCID: PMC11366768
Review Free full text in Europe PMC
IDH1 Inhibition Potentiates Chemotherapy Efficacy in Pancreatic Cancer.
Cancer Res, 84(18):3072-3085, 01 Sep 2024
Cited by: 1 article | PMID: 38843355 | PMCID: PMC11403292
Cancer tissue of origin constrains the growth and metabolism of metastases.
Nat Metab, 6(9):1668-1681, 19 Aug 2024
Cited by: 1 article | PMID: 39160333
Mapping the transcriptional evolution of human metastatic breast cancer.
J Clin Invest, 134(17):e183971, 03 Sep 2024
Cited by: 0 articles | PMID: 39225091 | PMCID: PMC11364401
Go to all (171) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics.
Mol Cell Proteomics, 13(11):2803-2811, 03 Jun 2014
Cited by: 39 articles | PMID: 24895378 | PMCID: PMC4223473
Distant metastasis occurs late during the genetic evolution of pancreatic cancer.
Nature, 467(7319):1114-1117, 01 Oct 2010
Cited by: 1562 articles | PMID: 20981102 | PMCID: PMC3148940
Cancer-associated fibroblast heterogeneity is associated with organ-specific metastasis in pancreatic ductal adenocarcinoma.
J Hematol Oncol, 14(1):184, 02 Nov 2021
Cited by: 21 articles | PMID: 34727952 | PMCID: PMC8561960
Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing.
Clin Cancer Res, 18(16):4257-4265, 01 Aug 2012
Cited by: 83 articles | PMID: 22896692 | PMCID: PMC3422771
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: CA076931
Grant ID: K12 CA076931
Grant ID: CA169123
Grant ID: R01 CA169123
NIDDK NIH HHS (3)
Grant ID: P30 DK050306
Grant ID: P30-DK050306
Grant ID: T32 DK007066





