Abstract
Free full text

Chromosomal context dependence of a eukaryotic recombinational hot spot.
Abstract
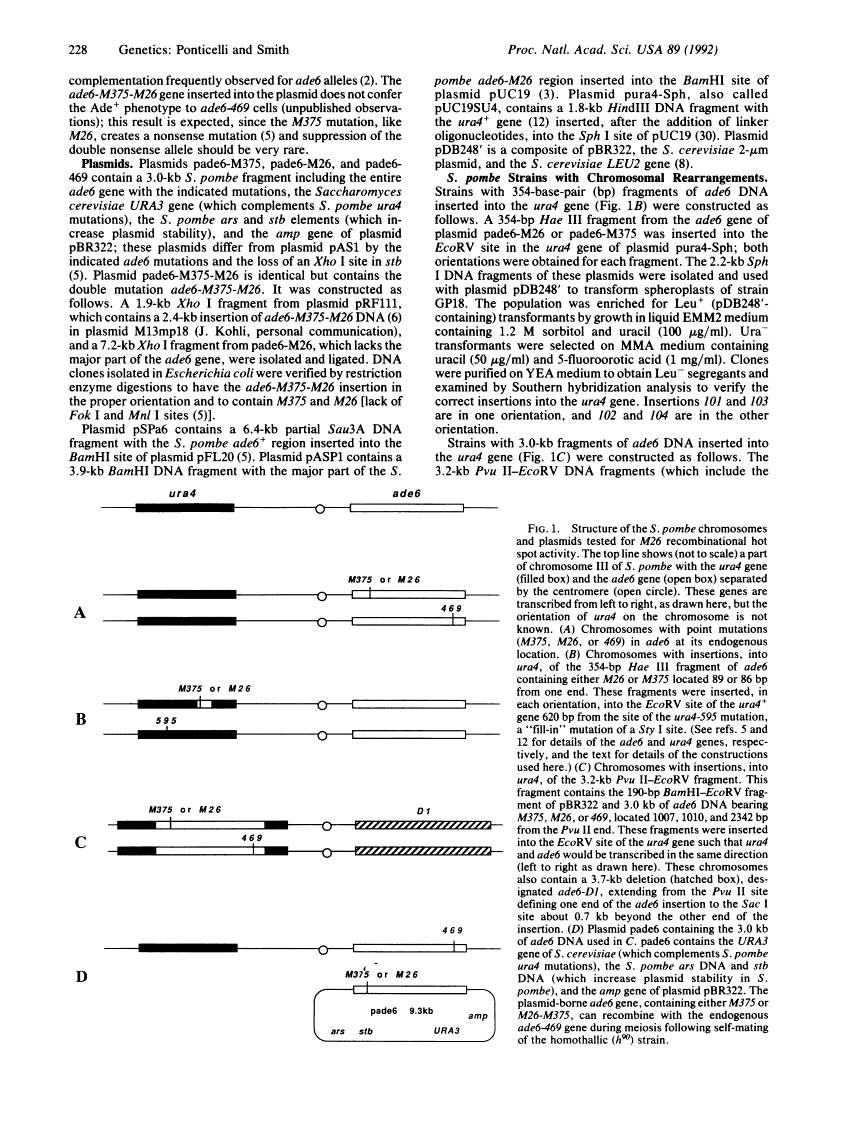
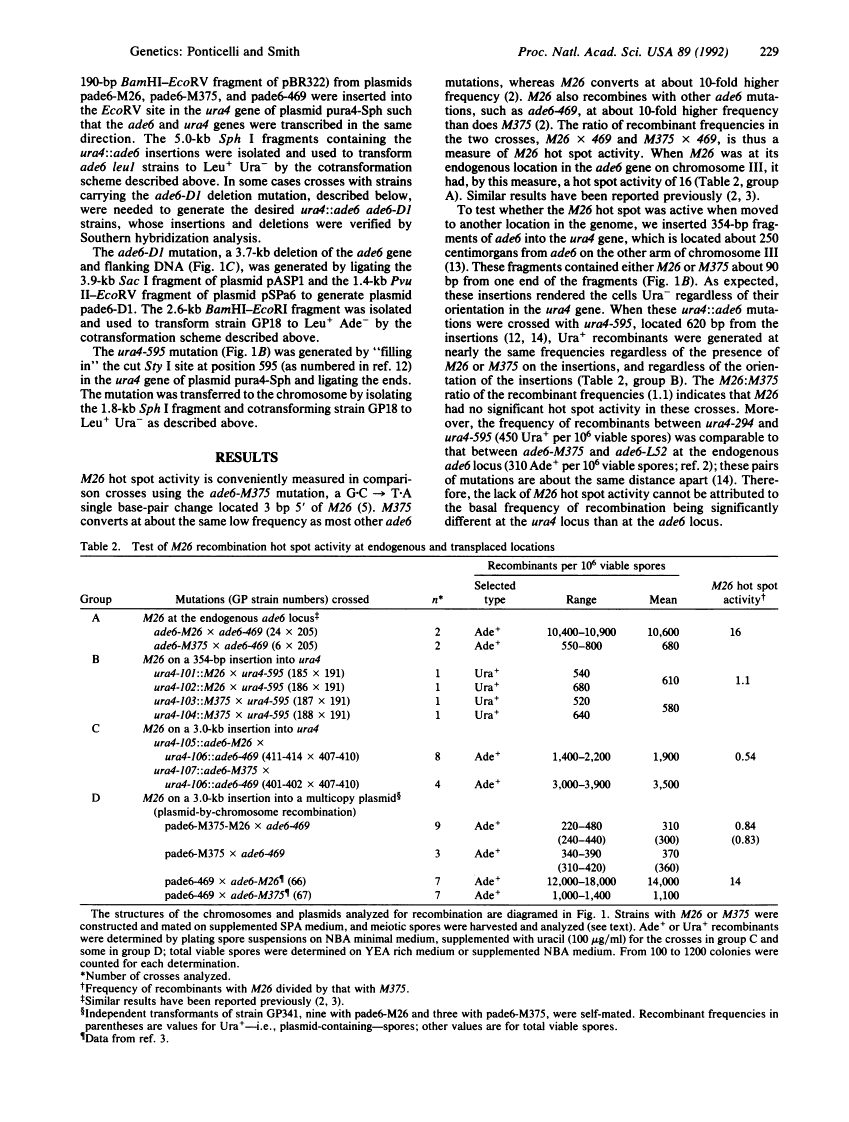
The single base-pair mutation M26 in the ade6 gene of the fission yeast Schizosaccharomyces pombe creates a hot spot for meiotic homologous recombination. When DNA fragments containing M26 and up to 3.0 kilobases of surrounding DNA were moved to the ura4 gene or to a multicopy plasmid, M26 had no detectable hot spot activity. Our results indicate that nucleotide sequences at least 1 kilobase away from M26 are required for M26 hot spot activity and suggest that, as for transcriptional promoters, a second site or proper chromatin structure is required for activation of this eukaryotic recombinational hot spot. We discuss the implications of these results for studies of other meiotic recombinational hot spots and for gene targeting.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gutz H. Site Specific Induction of Gene Conversion in SCHIZOSACCHAROMYCES POMBE. Genetics. 1971 Nov;69(3):317–337. [Europe PMC free article] [Abstract] [Google Scholar]
- Ponticelli AS, Sena EP, Smith GR. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics. 1988 Jul;119(3):491–497. [Europe PMC free article] [Abstract] [Google Scholar]
- Schuchert P, Kohli J. The Ade6-M26 Mutation of Schizosaccharomyces Pombe Increases the Frequency of Crossing over. Genetics. 1988 Jul;119(3):507–515. [Europe PMC free article] [Abstract] [Google Scholar]
- Szankasi P, Heyer WD, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6-M26. J Mol Biol. 1988 Dec 20;204(4):917–925. [Abstract] [Google Scholar]
- Schuchert P, Langsford M, Käslin E, Kohli J. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 1991 Aug;10(8):2157–2163. [Europe PMC free article] [Abstract] [Google Scholar]
- Beach D, Piper M, Nurse P. Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol Gen Genet. 1982;187(2):326–329. [Abstract] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. [Europe PMC free article] [Abstract] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975 Aug 14;256(5518):547–551. [Abstract] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989 Sep;123(1):45–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988 Dec;215(1):81–86. [Abstract] [Google Scholar]
- Kohli J. Genetic nomenclature and gene list of the fission yeast Schizosaccharomyces pombe. Curr Genet. 1987;11(8):575–589. [Abstract] [Google Scholar]
- Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991 Jan;11(1):289–298. [Europe PMC free article] [Abstract] [Google Scholar]
- Losson R, Lacroute F. Plasmids carrying the yeast OMP decarboxylase structural and regulatory genes: transcription regulation in a foreign environment. Cell. 1983 Feb;32(2):371–377. [Abstract] [Google Scholar]
- Heyer WD, Sipiczki M, Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986 Jan;6(1):80–89. [Europe PMC free article] [Abstract] [Google Scholar]
- Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985 May;41(1):145–151. [Abstract] [Google Scholar]
- Taylor AF, Schultz DW, Ponticelli AS, Smith GR. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell. 1985 May;41(1):153–163. [Abstract] [Google Scholar]
- Faulds D, Dower N, Stahl MM, Stahl FW. Orientation-dependent recombination hotspot activity in bacteriophage lambda. J Mol Biol. 1979 Jul 15;131(4):681–695. [Abstract] [Google Scholar]
- Kobayashi I, Murialdo H, Crasemann JM, Stahl MM, Stahl FW. Orientation of cohesive end site cos determines the active orientation of chi sequence in stimulating recA . recBC-mediated recombination in phage lambda lytic infections. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5981–5985. [Europe PMC free article] [Abstract] [Google Scholar]
- Kobayashi I, Stahl MM, Stahl FW. The mechanism of the chi-cos interaction in RecA-RecBC-mediated recombination in phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:497–506. [Abstract] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. [Abstract] [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989 Mar 2;338(6210):35–39. [Abstract] [Google Scholar]
- Schultes NP, Szostak JW. A poly(dA.dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):322–328. [Europe PMC free article] [Abstract] [Google Scholar]
- Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987 May 22;49(4):559–567. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.89.1.227
Read article for free, from open access legal sources, via Unpaywall:
http://www.pnas.org/content/89/1/227.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.89.1.227
Article citations
Regulation Mechanisms of Meiotic Recombination Revealed from the Analysis of a Fission Yeast Recombination Hotspot ade6-M26.
Biomolecules, 12(12):1761, 26 Nov 2022
Cited by: 1 article | PMID: 36551189 | PMCID: PMC9775316
Review Free full text in Europe PMC
Rec8 Cohesin-mediated Axis-loop chromatin architecture is required for meiotic recombination.
Nucleic Acids Res, 50(7):3799-3816, 01 Apr 2022
Cited by: 5 articles | PMID: 35333350 | PMCID: PMC9023276
Unique properties of multiple tandem copies of the M26 recombination hotspot in mitosis and meiosis in Schizosaccharomyces pombe.
Gene, 593(1):185-192, 14 Aug 2016
Cited by: 0 articles | PMID: 27535724
Unexpected DNA context-dependence identifies a new determinant of Chi recombination hotspots.
Nucleic Acids Res, 44(17):8216-8228, 21 Jun 2016
Cited by: 9 articles | PMID: 27330137 | PMCID: PMC5041463
High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae.
Genetics, 201(2):525-542, 05 Aug 2015
Cited by: 31 articles | PMID: 26245832 | PMCID: PMC4596667
Go to all (43) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim.
Genetics, 140(2):469-478, 01 Jun 1995
Cited by: 34 articles | PMID: 7498729 | PMCID: PMC1206627
Active and inactive transplacement of the M26 recombination hotspot in Schizosaccharomyces pombe.
Genetics, 141(1):33-48, 01 Sep 1995
Cited by: 24 articles | PMID: 8536980 | PMCID: PMC1206730
The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast.
Genes Dev, 11(7):876-886, 01 Apr 1997
Cited by: 69 articles | PMID: 9106659
Control of meiotic recombination in Schizosaccharomyces pombe.
Prog Nucleic Acid Res Mol Biol, 61:345-378, 01 Jan 1998
Cited by: 25 articles | PMID: 9752725
Review