Abstract
Free full text

The Staphylococcus aureus Global Regulator MgrA Modulates Clumping and Virulence by Controlling Surface Protein Expression
Abstract
Staphylococcus aureus is a human commensal and opportunistic pathogen that causes devastating infections in a wide range of locations within the body. One of the defining characteristics of S. aureus is its ability to form clumps in the presence of soluble fibrinogen, which likely has a protective benefit and facilitates adhesion to host tissue. We have previously shown that the ArlRS two-component regulatory system controls clumping, in part by repressing production of the large surface protein Ebh. In this work we show that ArlRS does not directly regulate Ebh, but instead ArlRS activates expression of the global regulator MgrA. Strains lacking mgrA fail to clump in the presence of fibrinogen, and clumping can be restored to an arlRS mutant by overexpressing either arlRS or mgrA, indicating that ArlRS and MgrA constitute a regulatory pathway. We used RNA-seq to show that MgrA represses ebh, as well as seven cell wall-associated proteins (SraP, Spa, FnbB, SasG, SasC, FmtB, and SdrD). EMSA analysis showed that MgrA directly represses expression of ebh and sraP. Clumping can be restored to an mgrA mutant by deleting the genes for Ebh, SraP and SasG, suggesting that increased expression of these proteins blocks clumping by steric hindrance. We show that mgrA mutants are less virulent in a rabbit model of endocarditis, and virulence can be partially restored by deleting the genes for the surface proteins ebh, sraP, and sasG. While mgrA mutants are unable to clump, they are known to have enhanced biofilm capacity. We demonstrate that this increase in biofilm formation is partially due to up-regulation of SasG, a surface protein known to promote intercellular interactions. These results confirm that ArlRS and MgrA constitute a regulatory cascade, and that they control expression of a number of genes important for virulence, including those for eight large surface proteins.
Author Summary
Staphylococcus causes a wide range of diseases, ranging from skin infections to deadly invasive condition like endocarditis, septicemia, osteomyelitis, and pneumonia. In this work we examine the ArlRS two-component regulatory system, which controls interactions with the host plasma protein fibrinogen. S. aureus normally forms large aggregates called clumps in the presence of fibrinogen, but the arlRS mutant is unable to clump. We demonstrate that ArlRS activates expression of the DNA-binding protein MgrA, and that mgrA is also required for clumping. Transcriptional analysis of an mgrA mutant shows that MgrA regulates expression of eight surface proteins. Expression of these surface proteins affects clumping, possibly by physically interfering with fibrinogen binding. Strains lacking mgrA are less virulent in an endocarditis model, and virulence can be partially restored by deleting genes for three of these surface proteins. An mgrA mutant is also known to have enhanced biofilm formation, and we show that this is partially due to increased production of one of these surface proteins. These results demonstrate that ArlRS and MgrA constitute a regulatory cascade in S. aureus that is crucial for pathogenesis and may be a good candidate to target for drug development.
Introduction
Staphylococcus aureus is a human commensal that asymptomatically colonizes the nares, throat, and skin of ~30% of the population [1,2]. It is also a pervasive opportunistic pathogen that is the most common infectious agent isolated from hospital inpatients in the US [3]. S. aureus causes a range of diseases, from skin and soft tissue infections to life-threatening conditions like pneumonia, osteomyelitis, sepsis and infective endocarditis. Antibiotic resistance has been increasing among S. aureus isolates in the past few decades [4], limiting the available treatment options. For example, invasive infections caused by methicillin-resistant S. aureus (MRSA) have mortality rates approaching 20% [5], highlighting the need for innovative therapies.
S. aureus strains encode a wide variety of virulence factors, including up to 24 different cell wall anchored proteins that are covalently attached to the peptidoglycan layer by the transpeptidase sortase [6]. A subset of these, termed Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs), are critical for attaching to components of the host extracellular matrix, such as fibrinogen, fibronectin, and collagen. Two of these MSCRAMMs, Clumping Factors A and B (ClfA and ClfB), facilitate S. aureus binding to fibrinogen and lead to agglutination or clumping of cells [7–10], and for simplicity, we will use the term clumping throughout. Fibrinogen is an abundant, soluble, 340 kDa elongated glycoprotein present in plasma that is processed by thrombin to form insoluble fibrin clots, a property that is crucial for blood clotting and platelet aggregation. S. aureus facilitates this conversion of fibrinogen to fibrin by secreting two coagulases that activate prothrombin, coagulase (Coa) and von Willibrand factor binding protein (vWbp). Notably ClfA can interact with both soluble fibrinogen and fibrin cables with similar affinities [11].
Clumping is thought to have a number of functions in the context of staphylococcal infections. Clumps are likely to be more resistant to clearance by the immune system, in part because they may be too large to be phagocytosed by neutrophils [12]. In addition, the fibrinogen coating on cells within a clump may impede antibody binding and complement deposition [13–15]. S. aureus can also form similar aggregates in the presence of synovial fluid that are dependent on ClfA/ClfB binding to fibrin [16], and these aggregates appear to be more resistant to antibiotic treatment, analogous to the recalcitrance to antibiotics seen with surface-attached biofilms [16]. Finally, it has been demonstrated that the cell density-dependent agr quorum sensing system, which controls expression of many virulence factors, is turned on in clumps, likely due to the increased local concentration of autoinducing peptide [17].
Clumping factors A and B are necessary for forming clumps and play an essential role in pathogenesis. Their genes are differentially regulated, with clfB being primarily expressed during exponential phase [18,19], and clfA expression increasing in later growth stages [20]. Strains lacking both clfA and clfB are unable to bind to fibrinogen, and because of this they fail to clump with fibrin and platelets in vitro [21]. Adhesion to platelets and fibrin is particularly important in the growth of vegetations on heart valves at the onset of infective endocarditis. These vegetations consist of bacteria, platelets, and fibrin, and, as expected, strains lacking clfA are less virulent in a rat model of endocarditis [22]. In addition, expressing clfA exogenously in normally nonpathogenic Lactococcus lactis significantly enhances its ability to generate heart valve vegetations [23]. clfA mutants also cause fewer septic arthritis symptoms [24] and are less lethal in bacteremia models [11,24–26]. In support of this, mice engineered to express a modified version of fibrinogen lacking the ClfA binding site are less susceptible to S. aureus bacteremia [27].
Until recently, clumping was assumed to be a passive property of S. aureus that was not subject to regulation. Yet other virulence factors of S. aureus are highly regulated, suggesting that clumping may also be modulated in response to environmental cues. S. aureus has 16 two-component systems (TCS) that respond to environmental signals and alter transcription accordingly [28], and these systems typically consist of a membrane-bound histidine kinase sensor protein and a DNA-binding response regulator. One of these TCSs is the agr quorum sensing system, which regulates a large number of virulence related secreted products, including cytolysins, proteases, phenol-soluble modulins, lipases, and superantigens [29]. Likewise, the SaeRS TCS regulates expression of a variety of virulence-related secreted proteins, such as the coagulases, hemolysins, and matrix binding proteins [30]. A third TCS, ArlRS, has been linked to virulence [31–33], but there is still relatively little known about which gene(s) it regulates and what signal activates the system. We have previously demonstrated that an arlRS mutant has a clumping defect and that it is attenuated in a rabbit model of endocarditis [33]. This failure to clump appears to be due, in part, to overproduction of the large surface protein Ebh, which may interfere with fibrinogen binding through steric hindrance [33]. Ebh, also known as the Giant Staphylococcal Surface Protein (GSSP), is an ~1.1 MDa protein of unknown function anchored at its C terminus in the cell membrane [34,35]. In this work we show that ArlR regulates Ebh production indirectly; we demonstrate that ArlR activates expression of the global regulator MgrA, which in turn represses ebh. We used RNA-seq to identify genes regulated by MgrA in USA300 strain LAC, and found eight genes for surface proteins that are repressed by MgrA, including ebh. We show that mgrA mutants are unable to clump, and that clumping can be restored by also deleting genes for the surface proteins Ebh and SraP in strain LAC. These results indicate that ArlRS and MgrA constitute a regulatory cascade that controls expression of a large number of genes, including those for Ebh and seven cell wall anchored proteins.
Results
MgrA is required for clumping
We have previously shown that the ArlRS TCS is required for S. aureus clumping, and that this is due in part to ArlRS suppressing production of the large surface protein Ebh [33]. However, we were unable to show direct binding of purified ArlR to the ebh promoter, despite several attempts with electrophoretic mobility shift assays (EMSAs), with unphosphorylated or phosphorylated ArlR. This led us to hypothesize that ArlRS modulates expression of ebh indirectly, presumably through the action of another regulator. In support of this idea, ArlRS was previously shown to regulate capsule expression indirectly by up-regulating expression of the global regulator MgrA [36]. MgrA modulates >10% of the S. aureus genome, and indeed one of largest differences in gene expression was the upregulation of ebh in an mgrA mutant [37].
S. aureus strains lacking mgrA show increased autolysis [38] and biofilm formation [39], but MgrA has not been linked to clumping. We constructed an mgrA deletion in the USA300 strain LAC. Compared to USA300 WT, the mgrA mutant had a pronounced clumping defect with both human plasma (Fig 1A) and purified human fibrinogen (Fig 1B). This defect could be partially complemented by expressing mgrA under the control of its own promoters at the phage 11 integration site on the chromosome (Fig 1A and 1B), or by expression from a plasmid (used in epistasis studies shown below). Scanning electron microscopy (SEM) images of the wild type and ΔmgrA mutant after incubation with fibrinogen show a distinct difference in cell packing that can be complemented (Fig 1C–1H). It is somewhat surprising that mgrA mutants have a clumping defect, given their increased capacity to form a biofilm [39,40].

Washed S. aureus cells were incubated statically with either human plasma (A) or fibrinogen (B) and clumping was monitored by measuring clearing of the solution over time, as described in the methods section. Strains tested were the wild type USA300 LAC (black), an isogenic mgrA deletion (red), and the mgrA mutant complemented with a copy of mgrA expressed from an exogenous site on the chromosome (blue). Values represent averages and standard deviations of three separate experiments. Scanning electron micrographs of the same strains after incubation with fibrinogen for 2 h are shown in panels C-H. The white boxes in the top panels show the region magnified in the lower panels. Magnification is 5,000X in panels C, E, and G, and 45,000X in panels D, F, and H.
ArlRS and MgrA constitute a regulatory pathway
The similar clumping defects of arlRS and mgrA mutants led us to propose a model in which the ArlRS TCS activates expression of MgrA (Fig 2). MgrA in turn represses ebh and possibly other clumping-related genes, allowing the wild type strain to interact with fibrinogen and clump. There is evidence that MgrA and ArlRS work together, although the exact mechanism is unclear. It was initially reported that MgrA controlled arlRS expression [38], but a later study used qRT-PCR to show that ArlRS regulated MgrA [36]. To assess whether ArlRS acts upstream of MgrA as depicted in our model (Fig 2), we tested if overexpressing mgrA could complement an arlRS mutant. Indeed, the expression of mgrA from a multicopy plasmid restored clumping to an arlRS mutant, but expressing arlRS in an mgrA mutant had no effect (Fig 3A). As an additional epistasis test, we looked at production of Ebh, which is elevated in arlRS and mgrA mutant strains. Ebh could be restored to wild type levels in an arlRS mutant by expressing either arlRS or mgrA (Fig 3B). Similar to the clumping results, expressing arlRS did not restore Ebh production to an mgrA mutant (Fig 3B). These results demonstrate that ArlRS and MgrA form a regulatory cascade in which MgrA acts downstream of ArlRS.
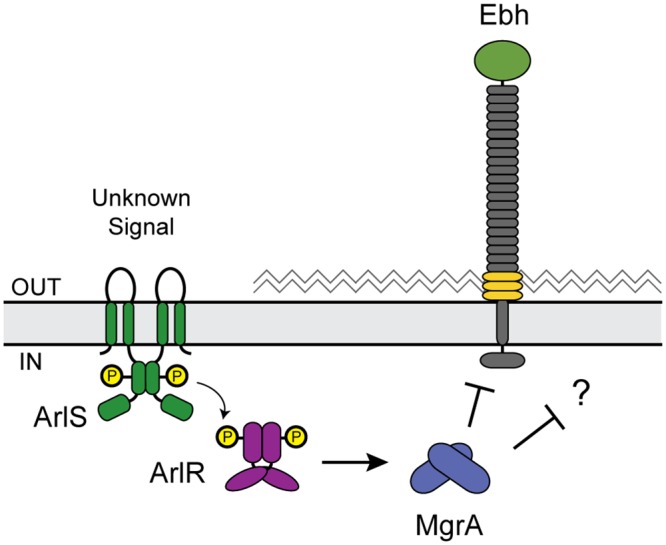
In response to an unknown signal ArlS either phosphorylates or dephosphorylates ArlR. When ArlR is phosphorylated it binds to the mgrA promoter, activating expression of mgrA. MgrA in turn represses ebh and other unknown targets. When either arlRS or mgrA is disrupted then ebh is de-repressed, and production of Ebh presumably blocks clumping.

LAC WT, arlRS, and mgrA mutants contained either the empty vector or plasmids for constitutive expression of arlRS or mgrA. (A) Clumping ability was measured after two hours of incubation with human plasma, and results from three separate experiments were averaged. (B) Dot blot showing Ebh protein levels in 2-fold dilutions of culture supernatants from overnight cultures. Strains were identical to those used in Fig 3A except that they lacked spa, to prevent non-specific antibody binding.
The ArlRS two-component system regulates MgrA
To confirm that ArlRS regulates MgrA, we used a combination of transcriptional reporters and Western blots. The mgrA gene has two promoters located 302 nucleotides (P2) and 124 nucleotides (P1) upstream of the start codon [38]. We amplified each of these promoters and fused them to GFP to generate transcriptional reporters (Fig 4C). Expression from mgrA P2 was entirely dependent on ArlRS, whereas expression from mgrA P1 was unchanged in the arlRS mutant compared with LAC (Fig 4A). In addition, under the conditions of this assay it appears that mgrA P2 may be ~10-fold stronger than mgrA P1. Detection of MgrA protein levels by Western blot confirmed that there was ~80–95% less MgrA produced in the arlRS mutant throughout the growth curve (Fig 4B and 4D). This sharp decrease in MgrA protein levels in the arlRS mutant likely explains why arlRS and mgrA mutants have similar phenotypes, despite the observation that ArlRS only regulates one of the mgrA promoters.
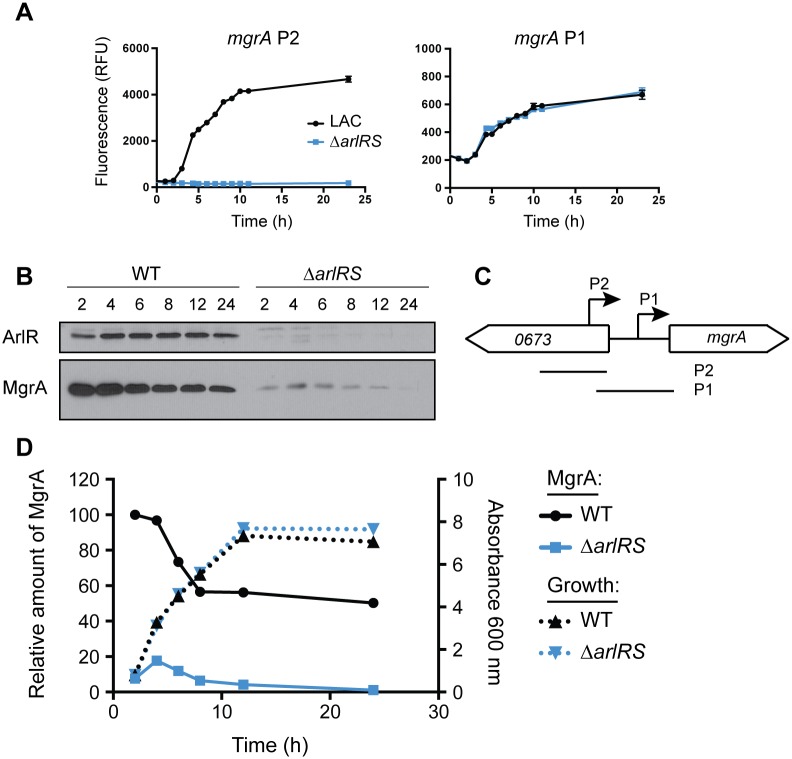
The mgrA gene has two promoters, depicted in (C). Lines labeled P2 and P1 show the fragments used to make transcriptional reporters. (A) Each mgrA promoter was independently cloned upstream of a promoterless copy of GFP in plasmid pCM11. Expression from the upstream promoter mgrA P2 (left) and downstream mgrA P1 (right) was assessed in the WT strain LAC (black) and the arlRS mutant (blue). (B) Western blot showing ArlR and MgrA protein levels at various time points (hours). (D) Quantification of MgrA protein levels in LAC (black, solid line) and the arlRS mutant (blue, solid line), and growth curves of the same strains (dotted lines). Quantification is representative of three separate experiments.
MgrA represses ebh
To understand how MgrA controls clumping, we used qRT-PCR to investigate if MgrA regulates expression of genes known to affect clumping and coagulation. Although several cell wall associated proteins in S. aureus have been reported to interact with fibrinogen [6], under the conditions of these in vitro experiments it appears that ClfA is the dominant adhesin required for clumping with both plasma and fibrinogen ([33], S1 Fig). Genes for the clumping factors clfA and clfB showed modest increases in expression in the mgrA mutant (Fig 5A), which would be expected to enhance rather than inhibit clumping. Notably, expression of ebh increased 28-fold in the mgrA mutant, whereas changes in all other genes tested were <2.5-fold. There was very little change in expression of the coagulase genes coa and vWbp, the plasminogen activator staphylokinase (sak), or in the sortase gene srtA. Thus, of the genes tested, it seems that MgrA is most likely to affect clumping through repression of ebh.
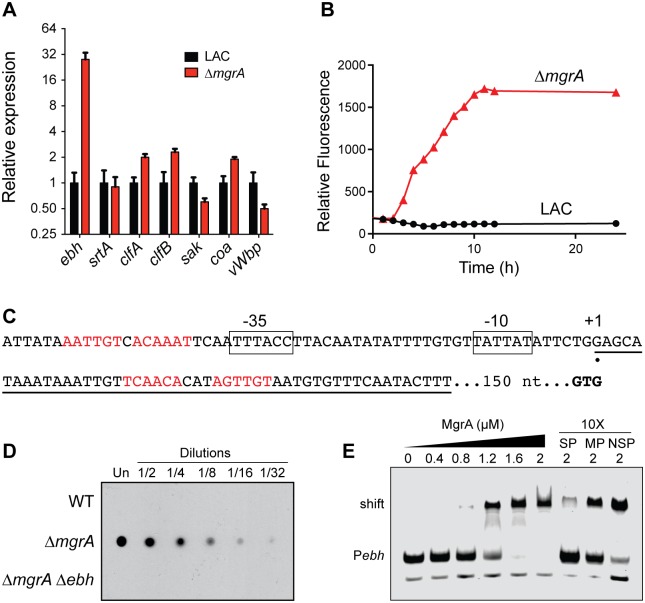
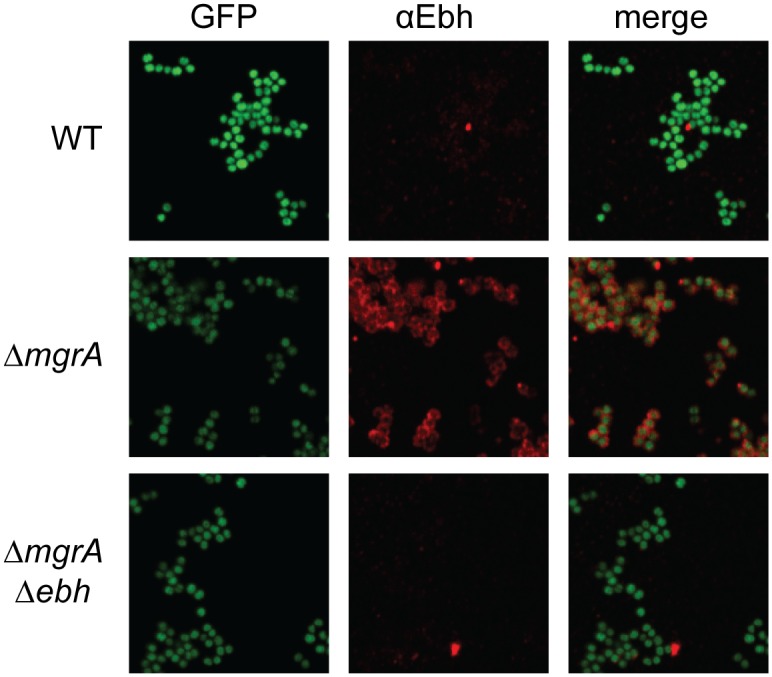
(A) Quantification of transcripts of genes related to clumping in LAC and the mgrA mutant using qRT-PCR. Values are averages and standard deviations of three biological replicates, normalized to expression in LAC for each gene. (B) Expression of ebh was measured in LAC and the mgrA mutant using a transcriptional fusion of the entire intergenic region upstream of ebh to GFP. (C) The putative ebh transcription start site, indicated by the black circle, was determined by 5’ RACE. Possible -10 and -35 promoter elements are boxed, and the ebh start codon (GTG) is shown in bold. Potential MgrA binding sites are shown in red text, and the sequence of the DNA probe used for EMSA experiments is underlined. (D) Ebh protein levels were measured by dot blot in 2-fold dilutions of supernatants from overnight cultures of the indicated strains. All strains also lacked the spa gene. (E) EMSA showing MgrA binding to the ebh promoter. Increasing concentrations of MgrA were incubated with an IRDye-labeled probe (Pebh, sequence underlined in C) before separation by PAGE. Unbound probe (Pebh) and MgrA-probe complex (shift) are indicated. The last three lanes show competition experiments, where the binding reaction included a 10-fold excess of unlabeled specific probe (SP), non-specific probe (NSP), or a version of the specific probe in which the proposed MgrA binding site was mutated (MP).
We confirmed that MgrA regulates ebh by measuring expression of an ebh transcriptional reporter in which the ebh promoter region was fused to GFP. As seen previously [33], expression of ebh was very low in the wild type strain when growing in rich media (Fig 5B). In contrast, expression of the Pebh-GFP fusion was much higher in the mgrA mutant, suggesting that MgrA represses ebh. Likewise, detection of Ebh protein levels by dot blot showed a substantial increase in the mgrA mutant (Fig 5D) that was specific for Ebh, as there was no signal in the mgrA ebh double mutant.
To investigate MgrA binding in greater detail, we mapped the ebh promoter using 5’ RACE (Fig 5C). We identified one putative transcription start site located 200 nucleotides upstream of the GTG start codon. Putative -10 and -35 promoter elements are shown in Fig 5C. MgrA was previously shown to bind to the six-nucleotide sequence (A/T)GTTGT [41]. As a member of the MarR/SlyA family of dimeric DNA binding proteins [42], MgrA likely binds to closely spaced inverted repeats of this hexameric sequence. There are at least two potential MgrA binding sites in the vicinity of the ebh promoter, shown in red in Fig 5C. One site, centered 24 nucleotides downstream of the putative transcription start site, matches the consensus sequence perfectly. A second potential MgrA binding site is centered 11 nucleotides upstream of the putative -35 element. We tested if MgrA is able to interact directly with the ebh promoter using a 50 bp DNA probe spanning the downstream potential MgrA binding site (Fig 5E). We found that MgrA was able to bind to the ebh promoter probe, and this binding could be outcompeted with a 10-fold excess of an identical unlabeled probe. A version of this competitor probe with a mutated potential MgrA binding site was poor competitor, however, and a 10-fold excess of a non-specific probe was not able to compete for MgrA binding. Taken together, these results indicate that MgrA represses ebh by directly interacting with the ebh promoter.
Lastly, we used immunofluorescence microscopy to visualize Ebh presence and localization in the wild type and mgrA mutant (Fig 6). All strains also lacked spa, the gene encoding Protein A, to avoid potential non-specific antibody binding. There was essentially no Ebh visible in the wild type strain or in the mgrA ebh double mutant. The mgrA mutant, however, showed abundant Ebh localized to the cell surface (Fig 6), consistent with our observations that ebh is up-regulated in an mgrA mutant.
Comparison of clumping in a variety of S. aureus strains
Many, but not all, strains of S. aureus encode a full-length copy of ebh. Truncated versions of ebh lack the trans-membrane domain, meaning the resulting protein should no longer be membrane anchored (Fig 7A). Because of this strain variability, we were interested in comparing behavior of mgrA and arlRS mutants across of range of strains. We hypothesized that truncations in Ebh might mask mgrA and arlRS mutant clumping phenotypes. Sequenced strains with full-length copies of ebh included 502a, the USA200 strain MRSA252, and the USA400 strain MW2. Strains with truncations in ebh included Newman, the USA100 strain N315, and the USA200 strain MN8. Most strains clumped with similar kinetics to LAC, although MRSA252 and N315 were noticeably slower, which could be due to a number of factors, potentially including decreased expression of clumping factors. In general, strains with full-length copies of ebh had a large clumping defect when either mgrA or arlRS was inactivated (Fig 7B). Newman had an intermediate phenotype, similar as previously noted [33], consistent with it expressing a version of Ebh that is nearly full-length but not membrane-anchored. Lastly, N315 and MN8, which encoded the shortest variants of ebh that we tested, had essentially no clumping defect when mgrA or arlRS were deleted (Fig 7B). These results suggest that there is a correlation between expression of an intact version of Ebh and inhibition of clumping. In light of this observation, we constructed an LAC strain lacking both mgrA and ebh. Surprisingly, however, deleting ebh did not restore clumping to an mgrA mutant, suggesting that additional factors were involved (Fig 7C).

(A) Schematic of predicted Ebh proteins from a collection of S. aureus strains, showing only the product of the first predicted ebh ORF for each strain. Key domains, as well as the secretion signal sequence and transmembrane (TM) domain are indicated. (B) Clumping of arlRS and mgrA mutants in each of the strains shown in (A) after 1 h of incubation with human plasma. (C) Clumping time course of LAC, mgrA mutant and mgrA ebh double mutants incubated with human plasma. All clumping results represent averages of three separate experiments.
Identification of genes regulated by MgrA
We hypothesized that the mgrA mutant was overproducing multiple surface proteins, including Ebh, that were interfering with clumping. Luong et al. [37] used microarray analysis to identify 355 genes that were regulated by MgrA in Newman. These included genes for the surface proteins FmtB and Protein A (Spa), in addition to Ebh. Initial tests with triple mutants lacking mgrA, ebh, and either spa or fmtB did not restore clumping (discussed in more detail below). Thus we decided to identify genes regulated by mgrA in LAC using RNA sequencing (RNA-seq), speculating that there may be variation between genes regulated by MgrA in Newman and LAC.
LAC and the isogeneic mgrA mutant were grown in rich medium to an optical density of 1.5, a time point at which MgrA is highly expressed (Fig 4D). We identified 104 genes whose expression was ≥4-fold different at 95% confidence, listed in S1 Table. There were 55 genes with increased expression in the mgrA mutant and 49 genes with decreased expression, including mgrA itself. In agreement with previous studies [37,38,43], expression of the capsule genes cap5A, cap5B, cap5C, and cap5E was modestly decreased in the mgrA mutant. In addition, ebh was upregulated ~45-fold in the mgrA mutant, consistent with our qRT-PCR measurements (Fig 5A). Whether MgrA directly or indirectly regulates expression of these genes remains to be determined. While the half-site consensus binding sequence for MgrA is thought to be (A/T)GTTGT [41], the preferred spacing between half-sites and the tolerance for variability remain unknown, making it difficult to distinguish MgrA binding sites without experimental validation. Some of the genes identified by RNA-seq are likely to be indirectly regulated by MgrA, though, because MgrA represses expression of at least two transcriptional regulators, SarV and AtlR (S1 Table).
The genes for eight known and putative surface proteins were significantly upregulated in the mgrA mutant, including ebh (Table 1). All except Ebh contain an LPXTG motif and are predicted to be cell wall anchored. Several are fairly well studied, including protein A (Spa), which binds IgG, and the fibronectin-binding protein FnbB. SdrD, a member of the serine-aspartate repeat family, and SasG are both reported to contribute to adherence to desquamated nasal epithelial cells [44,45]. There is limited information about FmtB and SasC, which both contain multiple repeats of the domain of unknown function DUF1542 (a domain also present in Ebh), although SasC has been shown to promote biofilm formation [46]. Finally, SraP is a member of the serine-rich repeat (SRR) glycoprotein family involved in adhesion to platelets, likely through binding to sialylated glycoproteins [47–49].
Table 1
| Gene | Fold Change in ΔmgrA vs LAC | Function |
|---|---|---|
| ebh | +45 | extracellular matrix binding protein homolog |
| fmtB | +41 | truncated FmtB protein |
| spa | +22 | immunoglobulin G binding protein A |
| sdrD | +20 | serine-aspartate repeat-containing protein D |
| sasG | +8.3 | truncated surface protein G |
| sasC | +7.7 | surface protein C |
| fnbB | +5.4 | fibronectin binding protein B |
| sraP | +5.0 | serine rich adhesin for platelets |
We mutated each of these genes individually in the ΔmgrA Δebh background using transposon insertions from the Nebraska Transposon Mutant Library (NTML) [50] and tested for restoration of clumping (Fig 8A). The mgrA ebh sraP triple mutant was able to clump almost as well as the LAC wild type strain, suggesting that SraP is involved in blocking clumping. Comparison of mgrA ebh and mgrA sraP double mutants showed that only the mgrA ebh sraP triple mutant was able to restore clumping (Fig 8B). This suggests that Ebh and SraP are redundant in this situation, and up-regulation of either SraP or Ebh is sufficient to block clumping in LAC. Therefore genes for both surface proteins must be disrupted to restore clumping to an mgrA mutant. However, in MW2 it was not sufficient to delete ebh and sraP to restore clumping in the mgrA mutant (Fig 8C). Unlike LAC, MW2 encodes a full-length copy of SasG, which is also predicted to be regulated by MgrA (Table 1). We hypothesized that up-regulation of SasG in the MW2 mgrA mutant may also be contributing to inhibition of clumping, and indeed an mgrA ebh sraP sasG quadruple mutant was able to clump as well as the wild type parent strain (Fig 8C). These results suggest that mgrA mutants are unable to clump due to up-regulation of surface proteins such as Ebh, SraP, and SasG that interfere with clumping.
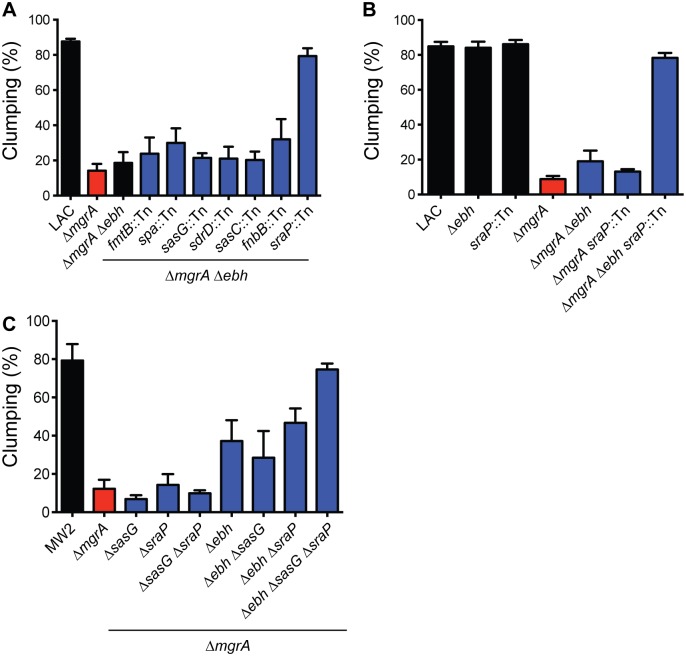
(A) Genes encoding surface proteins predicted to be regulated by MgrA were inactivated in an mgrA ebh double mutant background, and clumping was measured after 2 h of incubation with human plasma. (B) Comparison of clumping in LAC strains lacking combinations of mgrA, ebh, and sraP, measured after incubation with human plasma for two hours. (C) Clumping of MW2 strains lacking combinations of mgrA, ebh, sraP, and sasG. Graph shows clumping after 1 h of incubation with human plasma. All clumping results represent averages of three separate experiments.
sraP is regulated by MgrA
SraP is unusual in that it is glycosylated and exported by its own secretion system, consisting of the secretory proteins SecY2 and SecA2, three accessory secretory proteins (Asp1-3), and two putative glycosyltransferases, GtfA and GtfB [47,51]. All eight genes are co-localized on the chromosome (Fig 9B), but little is known about their expression. Our RNA-seq data indicated that the first five genes in the cluster (sraP, secY2, asp1, asp2, and asp3) were all up-regulated between 4.7 and 5.5-fold in the mgrA mutant (S1 Table). qRT-PCR analysis confirmed that expression of these five genes, as well as secA2, was increased ~4-fold in the mgrA mutant, whereas there was essentially no change in expression of the last two genes, gtfA, and gtfB (Fig 9E). Thus, MgrA appears to regulate expression of sraP and its secretion apparatus, but not the two downstream glycosyltransferases that are believed to modify SraP.

(A) The sraP promoter region, showing the two putative transcription start sites (marked with black circles) determined using 5’ RACE. Potential -10 and -35 promoter elements are boxed, and possible MgrA binding sites are shown in red text. The sraP ATG start codon is indicated in bold font. The promoter probe used for EMSA experiments is underlined. (B) Schematic of the sraP gene cluster, including its dedicated secretory system (secY2, asp1-3, and secA2) as well as the putative glycosyltransferases gtfA and gtfB. Inset below shows the two promoters mapped in (A) and the regions used to make transcriptional reporter plasmids (labeled rep 1 and rep 3). (C) Expression of a GFP transcriptional fusion containing both P1 and P2 (labeled rep 1 in panel B) in LAC and the isogenic mgrA mutant. (D) Expression of transcriptional reporter 3, containing just sraP P2 driving production of GFP. (E) Expression of each gene in the sraP gene cluster in LAC and the mgrA mutant, measured by qRT-PCR. Values are averages and standard deviations of three biological replicates and are normalized to expression in LAC for each gene. (F) EMSA assessing MgrA binding to the sraP P2 promoter. Increasing concentrations of purified MgrA were incubated with the probe underlined in panel A. Unbound probe and shifted probe are indicated on the left. In the last three lanes competition with a 10-fold excess of unlabeled probes was assessed. SP, specific probe identical to the labeled probe; MP, specific probe in which the putative MgrA binding site has been mutated; NSP, non-specific probe.
We used 5’ RACE to map the sraP transcription start site(s) and identified two putative promoters (Fig 9A and 9B). The first promoter, P1, initiates transcription 343 nucleotides upstream of the sraP start codon, and the second promoter, P2, is located 33 nucleotides upstream of the start codon. Interestingly, there are potential MgrA binding sites overlapping the putative -35 sequence of P2 and centered 35 bp downstream of the P1 transcription start site (shown in red in Fig 9A). To test if MgrA regulates both sraP promoters, we constructed two transcriptional fusions to GFP. The first fusion included both P1 and P2 (Fig 9C), whereas the second fusion contained only P2 (Fig 9D). Both fusions showed increased expression in the mgrA mutant, although overall expression from the fusion containing P1 and P2 was ~10-fold higher. These results suggest that P1 is the major promoter under these conditions, and that both P1 and P2 are controlled by MgrA.
We performed EMSAs with purified MgrA to see if it could bind to the putative MgrA binding site overlapping P2 (Fig 9F). MgrA was able to bind in a dose-dependent manner, producing a distinct shifted band. This binding could be outcompeted by adding a 10-fold excess of unlabeled probe, but not by adding the same unlabeled probe in which the putative MgrA binding site had been mutated (Fig 9F). Likewise, addition of a 10-fold excess of a non-specific probe from within the sraP coding sequence did not affect MgrA binding.
MgrA is required for endocarditis in MW2 and 502a
We have previously shown that an arlRS mutant is less virulent in a rabbit model of endocarditis [33]. Compared to the wild type strain MW2, the arlRS mutant was less lethal, formed smaller heart valve vegetations, and had lower bacterial burdens within the vegetations [33]. We hypothesized that this was due in part to the arlRS mutant’s defects in clumping and fibrinogen binding, and indeed, virulence could be partially restored by also deleting ebh. Since MgrA is also required for clumping, we predicted that an mgrA mutant would be less able to cause endocarditis. S. aureus strain Newman mgrA mutants have been shown to be less virulent in mouse sepsis, septic arthritis, and abscess models [42,52,53]. However, the requirement for MgrA to cause endocarditis is not clear, as a rat endocarditis model of a Newman mgrA mutant had lower bacterial burdens within vegetations [54], but in a rabbit model COL and MW2 mgrA mutants behaved like the wild type strains [55].
To revisit if MgrA is required for endocarditis, we constructed mgrA deletions in MW2 and the recently sequenced strain 502a [56]. 502a was used in the 1960s to deliberately colonize newborns, which provided some protection from other virulent S. aureus strains, but was halted after a child died of pneumonia caused by 502a [57]. These strains were chosen because mgrA mutants in both backgrounds had clumping defects similar to that seen with LAC (Fig 7B). Unlike LAC and other USA300 isolates, both MW2 and 502a cause endocarditis in this model without immediately killing the rabbits.
In studies of sepsis and infective endocarditis, we damage the aortic valves for 2 hours, then remove the catheters, followed by intravenous administration of microbes by the marginal ear vein. This procedure allows us to study infection development more resembling human disease with the absence of biofilm formation on catheters, in contrast to models that leave the catheters in place [54,55]. In our model, the wild type 502a was significantly more lethal than the mgrA knockout, with all 4 rabbits receiving 502a succumbing by day 2 post infection (Fig 10A). In contrast, all 5 animals receiving the same dose of 502a mgrA mutant survived the entire 4 day test period (Fig 10A). Total vegetation weights in the wild type-treated animals was greater than in animals given the mgrA mutant (Fig 10B); however the differences were not statistically significant, likely because vegetation sizes were being compared between animals that succumbed by day 2 compared to not dying. Total vegetation CFUs were significantly higher in 502a-treated animals (Fig 10C) than in rabbits given the mgrA mutant.
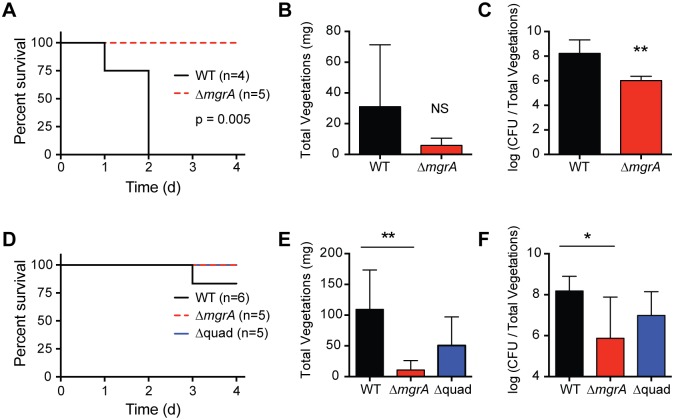
Prior to infection, aortic valve damage was induced by temporary placement of plastic catheters in the carotid artery. Catheters were removed and rabbits were infected intravenously with either strain 502a (A-C), or strain MW2 (D-F). WT strains were compared to the mgrA single mutant, and for MW2 the mgrA ebh sraP sasG quadruple mutant (Δquad) was also tested. Rabbits were monitored for up to four days, and survival (A, D), total heart valve vegetation weights (B, E), and CFUs per total vegetations (C, F) were monitored. Statistical significance was determined using a log-rank (Mantel-Cox) test (survival), or two-tailed t test (vegetation weights and CFU counts). ** p<0.005, * p<0.05, NS not significant.
When rabbits were infected with strain MW2, one of the animals treated with wild type succumbed over the 4 day period, whereas none of the animals infected with the mgrA mutant died (Fig 10D). Both total vegetation weights (Fig 10E) and total CFUs (Fig 10F) were significantly higher in animals treated with MW2 than in animals treated with the MW2 mgrA mutant. We also tested if virulence was restored in the MW2 mgrA ebh sraP sasG quadruple mutant (Δquad). All rabbits infected with the quadruple mutant survived the four day time course, similar to the wild type and mgrA single mutant (Fig 10D). Total vegetation weights were higher than in the animals treated with the mgrA mutant (Fig 10E), although this difference was not statistically significant. Likewise, the total CFUs were higher in rabbits infected with the quadruple mutant compared to treatment with the mgrA mutant (Fig 10F). These results suggest that overexpression of surface proteins contributes to the lack of fitness of the mgrA mutant in vivo, although other factors likely contribute to its decreased virulence.
MgrA controls biofilm formation through regulation of surface proteins
Differential expression of surface proteins is also likely to affect biofilm formation by S. aureus. In contrast to our observations that mgrA mutants have a clumping defect, strains lacking mgrA show increased biofilm formation [39,40,58]. This increase in biofilm formation appears to be particularly pronounced in strains that produce SasG, such as MW2 and SH1000 [39]. SasG is known to promote biofilm formation by facilitating intercellular adhesion [59,60], and we predicted that increased production of SasG in mgrA mutants could explain the observed enhancement in biofilm formation. In agreement with previous results, we observed increased biofilm formation in the MW2 mgrA mutant (Fig 11). To test if this enhanced biofilm formation was due to increased production of extracellular polysaccharide PIA (polysaccharide intracellular adhesin), we constructed a double mutant lacking both mgrA and the icaADBC locus responsible for production of PIA. The mgrA ica mutant produced as much biofilm as the mgrA single mutant, indicating that the increase in biofilm formation is PIA-independent. However, the mgrA sasG mutant formed significantly less biofilm than the mgrA single mutant. Deletion of ebh or sraP had no effect, suggesting that the enhanced biofilm formation in the MW2 mgrA mutant is primarily due to increased expression of SasG.
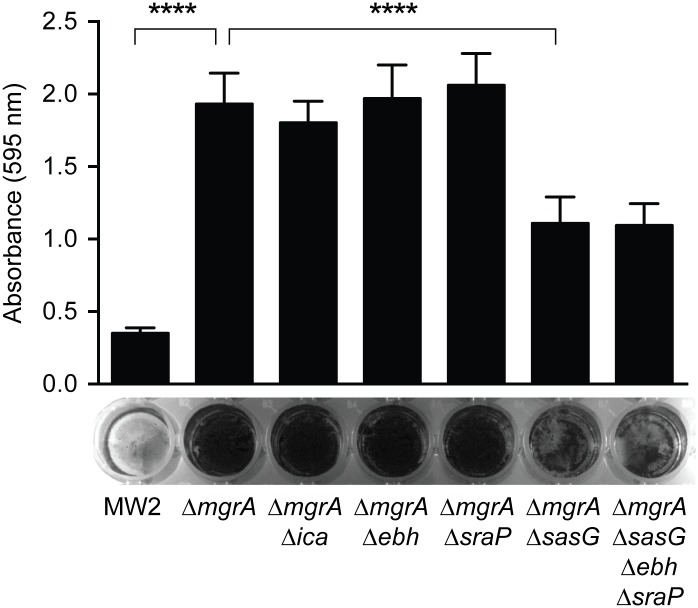
Cultures were incubated statically for 16 h in BHI + 0.25% glucose, and biofilm biomass was assessed by staining with crystal violet. Solubilized crystal violet was quantified by measuring absorbance at 595 nm. Values represent averages and standard deviations of two separate experiments with six replicate wells each. Statistical significance was determined using a two-tailed t test; **** p<0.0001.
Discussion
The ArlRS TCS has been linked to virulence phenotypes multiple times [31–33], but we are only beginning to understand how this regulatory system functions. We demonstrate here that ArlR activates expression of the global regulator MgrA, and that mgrA mutants, like arlRS mutants, show a pronounced defect in clumping in the presence of fibrinogen. Numerous studies, including this one, have shown that MgrA is important for virulence [42,52–54,58], although the reason for this has been largely unclear. Our RNA-seq findings indicate MgrA affects the expression of >100 genes, including eight surface proteins that are likely to be important for adhesion and immune evasion within the host. These results support the idea that ArlRS and MgrA constitute a regulatory cascade that, in response to an unknown signal, profoundly changes expression of cell wall associated proteins, perhaps allowing S. aureus to adapt to a new niche within the host or progress to a different disease stage. A proposed model for how the ArlRS-MgrA cascade affects clumping and biofilm formation is shown in Fig 12.
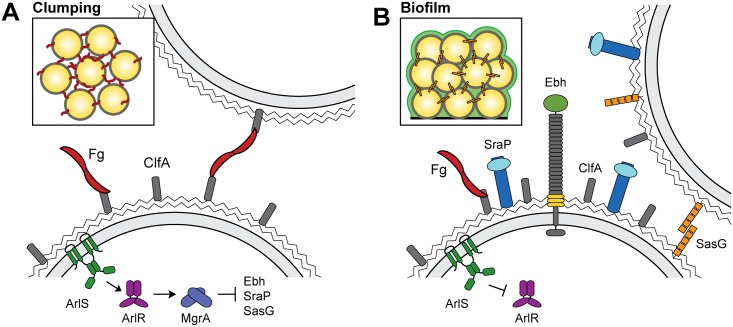
In response to an unknown signal, the ArlRS two component system activates expression of MgrA, which in turn represses expression of the large surface proteins Ebh, SraP, and SasG. ClfA interacts with fibrinogen, allowing clumping to occur (inset). When ArlRS is inhibited or inactivated, MgrA production is diminished and expression of Ebh, SraP, and SasG is de-repressed (B). These surface proteins interfere with clumping, while up-regulation of SasG can also promote biofilm formation (inset). Fg, fibrinogen.
Both arlRS and mgrA mutants in a variety of strain backgrounds fail to clump in the presence of fibrinogen or plasma. We show that this is due to increased expression of surface proteins such as Ebh, SraP, and SasG, which may interfere with clumping by steric hindrance. The ability of S. aureus to interact with fibrinogen and form clumps appears to be important for establishing infections such as septicemia and infective endocarditis [22,23,25,26]. Using a rabbit model of infective endocarditis, we show that mgrA mutants form smaller vegetations, and that the bacterial burden within these vegetations is lower. This virulence defect can be partially restored by deleting the genes for Ebh, SraP, and SasG, suggesting that up-regulation of surface proteins contributes to the virulence defect of the mgrA mutant. We predict that clumping may also be important for abscess formation. There is growing evidence that staphylococcal abscesses consist of a dense core of bacterial cells, called the staphylococcal abscess community, surrounded by a fibrin “pseudocapsule” [61–63]. Formation of this fibrin layer is dependent on the action of the staphylococcal coagulases and likely protects the bacteria from clearance by the immune system [61,63]. These abscess communities are likely to be held together by fibrin/fibrinogen, similar to the dense clumps generated in vitro with purified fibrinogen. Indeed, mgrA mutants are attenuated in a kidney and liver abscess model [42].
Among the genes repressed by MgrA are those encoding eight surface proteins: Ebh, FmtB, Spa (protein A), SdrD, SasG, SasC, FnbB, and SraP. By size, Ebh and SraP are two of the largest extracellular proteins made by S. aureus, at 1.1 MDa for Ebh, and 228 kDa for SraP before glycosylation. We propose that increased production of Ebh and SraP at the cell surface physically interferes with the fibrinogen cross-linking between cells that is required for clumping. Unlike LAC, strain MW2 encodes a full-length copy of SasG that also appears to contribute to blocking clumping. A similar phenomenon was observed previously when SasG was overproduced from a plasmid [45,59]. SasG is normally undetectable under in vitro growth conditions, but when it was artificially produced at high levels it was able to interfere with adhesion of cells to fibrinogen and fibronectin, without affecting expression of other surface proteins like ClfA, ClfB, and the fibronectin binding proteins. The mechanism of physically interfering with binding to matrix proteins has also been seen with Pls, a ~230 kDa cell wall associated protein related to SasG that is encoded within the type I SCCmec element of some MRSA strains. Pls producing strains have defects in fibronectin binding and cellular invasion that can be mitigated by deleting the pls gene [64,65]. This inhibition of fibronectin binding has been attributed to either steric hindrance or competition between Pls and the fibronectin binding proteins for anchoring sites on the cell wall [65]. It has also been observed that under some conditions expression of capsular polysaccharide can interfere with ClfA-mediated binding to fibrinogen, again likely through steric hindrance [66].
Altering expression of large surface proteins also has implications for biofilm formation. Increased biofilm production has been observed previously for both arlRS [67,68] and mgrA [39,40,58] mutants, and it appears to be polysaccharide independent [39,67]. In agreement with these results, we observed increased biofilm production in the MW2 mgrA mutant, which was unaffected by deletion of the ica locus encoding the machinery for polysaccharide synthesis (Fig 11). The increased biofilm formation in the mgrA mutant appears to be largely due to up-regulation of SasG, a protein known to promote biofilm accumulation [60]. SraP is also reported to contribute to biofilm formation [69], although we did not see a decrease in biofilm biomass when sraP was deleted. Deletion of sasG in an mgrA mutant only partially reduces biofilm production, suggesting that other proteins, such as SasC [46], may be involved. Alternatively, mutation of mgrA may promote release of extracellular DNA through increased rates of autolysis, which could contribute to the biofilm matrix. The fact that SasG-producing strains such as the MW2 mgrA mutant show both increased biofilm formation and decreased clumping demonstrates that these are different modes of intercellular interaction that are governed by distinct mechanisms.
We show here that regulation of surface proteins like Ebh, SraP, and SasG is likely more important than previously realized, and that production of these proteins can dramatically impact interactions with matrix proteins like fibrinogen. MgrA also regulates various other genes thought to be involved in virulence. For example, it has been suggested that mgrA mutants are more susceptible to phagocytosis because they produce less capsular polysaccharide [52,54]. Yet some particularly virulent strains of S. aureus, including the USA300 lineage, do not produce capsule [70,71]. Alternatively, there are multiple reports that MgrA regulates production of α-toxin, although whether MgrA represses or activates hla expression is debated [43,54,72]. In agreement with Gupta et al. [54] we did not observe significant changes in hla expression in the LAC mgrA mutant by RNA-seq. However, we did see an ~20-fold decrease in expression of Panton-Valentine leukocidin (PVL) in the mgrA mutant. Several other immune evasion proteins were also down-regulated in the mgrA mutant, including the LukAB leukocidin, chemotaxis inhibiting protein CHIP, and staphylococcal complement inhibitor SCIN. Whether these factors contribute to the virulence phenotypes seen with mgrA mutants remains to be investigated.
In summary, we have shown that the ArlRS-MgrA regulatory cascade controls expression of a variety of genes, including those for up to eight surface proteins, depending on the strain. Apart from protein A and the fibronectin binding protein FnbB, relatively little is known about these surface proteins, likely because expression of some of them is low in wild type strains in vitro. The signal for the ArlRS TCS is still unknown, making it difficult to predict when this system is active. We did identify the first promoter activated by ArlR, the mgrA P2 promoter (Fig 4A), which could aid future studies on unraveling the ArlRS regulatory mechanism. Since the ArlRS-MgrA cascade alters the expression of many surface proteins, we would expect to see large changes in behaviors such as clumping, biofilm formation, and adhesion to host tissues. It is possible that expression of these surface proteins is associated with moving into a new environment within the host, or a means to disseminate from a vegetation or abscess community in the later stages of disease. Although there is still much left to learn about the ArlRS-MgrA cascade, it is clear that it is a major switch controlling virulence determinants in S. aureus, and will likely be a good target for novel therapeutics.
Materials and Methods
Ethics statement
The animal studies were reviewed and protocol approved by the University of Iowa Institutional Animal Care and Use Committee. The approved protocol was assigned number 4071100. Animals were anesthetized with the combination of ketamine (10 mg/kg subcutaneously) and xylazine (10 mg/kg subcutaneously). Animals were administered pain relieving medications (buprenorphine; 0.05 mg/kg twice daily subcutaneously) throughout experimentation. Additionally, animals that could not simultaneously maintain upright positions and exhibit normal escape behavior were prematurely euthanized; these criteria are 100% predictive of death in the model used. The University of Iowa is AAALAC accredited, and the centralized facilities meet and adhere to the standards in the “Guide and Care of Laboratory Animals.”
Reagents and growth conditions
S. aureus strains and plasmids used in this work are listed in Table 2. For most experiments S. aureus was cultured in tryptic soy broth (TSB) at 37°C with shaking; for assessment of clumping, strains were grown in brain heart infusion broth (BHI). E. coli was cultured in lysogeny broth (LB). Antibiotics were added to the media at the following concentrations: chloramphenicol (Cam), 10 μg/mL; erythromycin (Erm), 5 μg/mL; and tetracycline (Tet), 0.625 μg/mL. E. coli strains with plasmids were maintained on media supplemented with ampicillin at 100 μg/mL; kanamycin, 50 μg/mL; or spectinomycin at 50 μg/mL.
Table 2
| Strain/plasmid | Genotype/properties | Reference |
|---|---|---|
| E. coli | ||
 DH5α DH5α | Cloning strain | Protein Express |
 ER2566 ER2566 | Overexpression strain | NEB |
| S. aureus | ||
 RN4220 RN4220 | Restriction deficient cloning host | [73] |
 AH1263 AH1263 | USA300 CA-MRSA ErmS (LAC*) | [74] |
 AH3150 AH3150 | LAC* Δebh | This work |
 AH3455 AH3455 | LAC* ΔmgrA::tetM | This work |
 AH3481 AH3481 | LAC* Δebh ΔmgrA::tetM | This work |
 AH3485 AH3485 | LAC* ΔmgrA::tetM Φ11::LL29erm mgrA | This work |
 AH3007 AH3007 | LAC* spa::ΦNΣ | [33] |
 AH3488 AH3488 | LAC* spa::ΦNΣ ΔmgrA::tetM | This work |
 AH3487 AH3487 | LAC* spa::ΦNΣ ΔmgrA::tetM Δebh | This work |
 AH3052 AH3052 | LAC* Δspa | [75] |
 AH3056 AH3056 | LAC* Δspa ΔarlRS | This work |
 AH3458 AH3458 | LAC* Δspa ΔmgrA::tetM | This work |
 AH1975 AH1975 | LAC* ΔarlRS | [33] |
 AH3520 AH3520 | LAC* ΔarlRS::tetM | This work |
 AH3577 AH3577 | LAC* Δebh ΔmgrA::tetM fmtB::ΦNΣ | This work |
 AH3738 AH3738 | LAC* Δebh ΔmgrA::tetM sdrD::ΦNΣ | This work |
 AH3739 AH3739 | LAC* Δebh ΔmgrA::tetM sasG::ΦNΣ | This work |
 AH3792 AH3792 | LAC* Δebh ΔmgrA::tetM sasC::ΦNΣ | This work |
 AH3797 AH3797 | LAC* Δebh ΔmgrA::tetM fnbB::ΦNΣ | This work |
 AH3798 AH3798 | LAC* Δebh ΔmgrA::tetM sraP::ΦNΣ | This work |
 AH3808 AH3808 | LAC* sraP::ΦNΣ | This work |
 AH3811 AH3811 | LAC* ΔmgrA::tetM sraP::ΦNΣ | This work |
 MW2 MW2 | USA400 CA-MRSA | [76] |
 AH3060 AH3060 | MW2 ΔarlRS::tetM | This work |
 AH3456 AH3456 | MW2 ΔmgrA::tetM | This work |
 AH3422 AH3422 | MW2 ΔmgrA | This work |
 AH3494 AH3494 | MW2 ΔmgrA Δebh::tetM | This work |
 AH3934 AH3934 | MW2 ΔmgrA ΔsraP | This work |
 AH3945 AH3945 | MW2 ΔmgrA ΔsraP Δebh::tetM | This work |
 AH3976 AH3976 | MW2 ΔmgrA ΔsraP ΔsasG | This work |
 AH3977 AH3977 | MW2 ΔmgrA ΔsraP ΔsasG Δebh::tetM | This work |
 AH3989 AH3989 | MW2 ΔmgrA ΔsasG | This work |
 AH4032 AH4032 | MW2 ΔmgrA ΔsasG Δebh::tetM | This work |
 AH4473 AH4473 | MW2 ΔmgrA Δica::tet | This work |
 502a 502a | [56] | |
 AH3624 AH3624 | 502a ΔarlRS::tetM | This work |
 AH3625 AH3625 | 502a ΔmgrA::tetM | This work |
 Newman Newman | MSSA | [77] |
 AH3062 AH3062 | Newman ΔarlRS::tetM | This work |
 AH3472 AH3472 | Newman ΔmgrA::tetM | This work |
 MRSA252 MRSA252 | USA200 HA-MRSA | [78] |
 AH3608 AH3608 | MRSA252 ΔarlRS::tetM | This work |
 AH3483 AH3483 | MRSA252 ΔmgrA::tetM | This work |
 N315 N315 | USA100 | [79] |
 AH3082 AH3082 | N315 ΔarlRS::tetM | This work |
 AH3473 AH3473 | N315 ΔmgrA::tetM | This work |
 MN8 MN8 | USA200 toxic shock isolate | [80] |
 AH3063 AH3063 | MN8 ΔarlRS::tetM | This work |
 AH3480 AH3480 | MN8 ΔmgrA::tetM | This work |
| Plasmids | ||
 pJB38 pJB38 | Mutation generation vector, CamR | [81] |
 pJMB202 pJMB202 | arlRS deletion vector, CamR | [33] |
 pHC02 pHC02 | arlRS::tetM deletion vector, CamR | This work |
 pHC12 pHC12 | ebh deletion vector, CamR | This work |
 pHC34 pHC34 | mgrA deletion vector, CamR | This work |
 pHC75 pHC75 | mgrA::tetM deletion vector,CamR | This work |
 pHC76 pHC76 | sraP deletion vector, CamR | This work |
 pHC77 pHC77 | sasG deletion vector, CamR | This work |
 pCM28 pCM28 | S. aureus–E. coli shuttle vector, CamR | [74] |
 pHC66 pHC66 | mgrA complementing vector, CamR | This work |
 pArl pArl | arlRS complementing vector, CamR | [33] |
 pCM29 pCM29 | sGFP expression vector, CamR | [82] |
 pLL29 pLL29 | S. aureus integration vector, TetR | [83] |
 pLL29-erm pLL29-erm | S. aureus integration vector, ErmR | This work |
 pHC67 pHC67 | mgrA chromosomal complementing vector, ErmR | This work |
 pCM11 pCM11 | S. aureus sGFP expression plasmid, ErmR | [84] |
 pCM11-ebh pCM11-ebh | Pebh-sGFP fusion, ErmR | [33] |
 pHC68 pHC68 | mgrA P2-sGFP fusion, ErmR | This work |
 pHC70 pHC70 | mgrA P1-sGFP fusion, ErmR | This work |
 pHC71 pHC71 | sraP (P1 and P2)-sGFP fusion, ErmR | This work |
 pHC73 pHC73 | sraP P2-sGFP fusion, ErmR | This work |
 pHC07 pHC07 | pET28a-arlR, overexpression, KanR | This work |
 pHC74 pHC74 | pKLD66-mgrA, overexpression, AmpR | This work |
Human plasma (HP) was obtained from donors at the University of Iowa Inflammation Program with all necessary approvals. HP was diluted 1:1 with heparin/dextran sulfate to prevent clotting, and for the purposes of this study, this level of HP was considered a final concentration of 100%. Purified human fibrinogen was purchased from Sigma-Aldrich.
Recombinant DNA and genetic techniques
E. coli DH5α was used as a cloning host for plasmid constructions. Restriction enzymes, DNA ligase, and Phusion DNA polymerase were purchased from New England Biolabs. The plasmid mini-prep and gel extraction kits were purchased from Invitrogen. Lysostaphin, used for S. aureus DNA extractions, was purchased from AMBI Products LLC. Plasmids were electroporated into S. aureus RN4220 as described previously [85]. Bacteriophage transductions between S. aureus strains were performed with phage 80α or 11 as described previously [86]. All oligonucleotides were ordered from IDT (Coralville, IA) and are listed in S2 Table. DNA sequencing was performed at the University of Iowa DNA Core Facility.
Clumping assay
Measurements of S. aureus clumping in the presence of fibrinogen or plasma were performed essentially as described by Walker et al. [33]. Briefly, cultures were grown in BHI to an OD600 of 1.5, harvested by centrifugation, and resuspended in the same volume of phosphate buffered saline (PBS). Human plasma was added to a final concentration of 2.5% (vol/vol), and clearing of the suspension was measured over two hours by periodically removing small aliquots from the top of the tube and measuring the OD600. Alternatively, clumping was initiated by adding purified fibrinogen to a final concentration of 18.5 μg/mL. Relative clumping values were calculated using the equation % clumping = ((ODtime0-ODtimeT)/ODtime0)x100.
Preparation of clumps for SEM
The LAC wild type strain, mgrA::tet mutant, and chromosomally complemented strain were allowed to clump for 2 hr with fibrinogen as described above. Slides were then prepared for SEM and imaged as described previously [33].
Immunofluorescence microscopy
Immunofluorescence microscopy was used to visualize Ebh on individual cells. All strains lacked the gene for protein A (spa), and contained plasmid pCM29, which constitutively expresses sGFP [82]. Cultures were grown overnight in TSB, washed three times with PBS, and adhered to poly-L-lysine coated glass chamber slides (Nunc). Cells were fixed for 20 min with 4% paraformaldehyde and then washed three times with PBS. Slides were blocked with Superblock (Pierce) containing 5% bovine serum albumin for 30 min and washed three times with PBS. Rabbit anti-Ebh serum [33] was diluted 1:100 in superblock plus 1% BSA and allowed to incubate on slides overnight at 4°C. The slides were then washed five times with Superblock and incubated with a 1:500 dilution of Alexa 568-conjugated goat anti-rabbit antibody (Molecular Probes) for 1 hr at room temperature. Slides were washed an additional five times with PBS, chamber slide wells were removed, and cover slips were mounted with Prolong Diamond Antifade mountant (Molecular Probes). Images were obtained with a Leica DM5500 Q confocal microscope.
Generation of gene deletions
To construct the mgrA deletion plasmid, ~700 bp regions flanking the gene were amplified with primer pairs HC116/HC117 and HC118/119. The products were column purified and fused in a second PCR using primers HC116 and HC119. This fusion product was gel purified, digested with SacI and SalI, and ligated into pJB38 [81] to generate pHC34. This plasmid was electroporated in RN4220, selecting on TSA plates containing Cam at 30°C. The plasmid was then transduced into S. aureus strains LAC and MW2. Individual colonies were streaked on TSA Cam plates incubated at 42°C to select for integration into the chromosome. Single colonies were grown in TSB at 30°C and diluted 1:500 in fresh media for four successive days before diluting to 10−6 and plating on TSA containing 0.2 μg/mL (LAC) or 0.6 μg/mL (MW2) anhydrotetracycline to select for loss of the plasmid. Colonies were screened for resistance to Cam, and CamS colonies were screened by PCR for deletion of mgrA.
The ebh, sraP, and sasG gene deletions were constructed in a similar manner. Sequences flanking ebh were amplified with primers HC28/HC53 and HC54/HC55, fused, digested with EcoRI and SalI, and ligated into pJB38 to generate pHC12. For the sraP deletion construct, flanking regions were amplified from S. aureus MW2 genomic DNA using primer pairs HC338/HC339 and HC340/HC341. The fusion product was digested with SacI and SalI and ligated into pJB38 to generate pHC76. The plasmid for deleting sasG in MW2 was generated similarly, using primer pairs HC246/HC247 and HC248/HC249. The fusion product was digested with EcoRI and SalI and ligated into pJB38 to generate pHC77.
To generate the arlRS::tetM deletion plasmid pHC02, the tetracycline resistance cassette was amplified from pTET [87] using primers HC3 and HC4. The resulting product was digested with NheI and ligated into the NheI site located between the arlRS flanking segments in pJMB202. Likewise, the mgrA::tetM deletion vector pHC75 was constructed by ligating the same tetracycline resistance cassette into the NheI site of pHC34.
Construction of the mgrA chromosomal complement
The S. aureus chromosomal integration vector pLL29 [83], which confers resistance to tetracycline, was modified to generate pLL29erm, an erythromycin resistant variant. The ermC gene was amplified from pCM11 [84] using primers HC156 and HC157, and the resulting product was digested with BsrGI and NheI. pLL29 was digested with the same enzymes to remove the tetK gene, and the ermC cassette was ligated in its place. The ligation reaction was transformed into E. coli DH5α, selecting for spectinomycin resistant colonies. To generate pHC67 (pLL29erm mgrA), a 910-bp fragment containing the mgrA gene and both of its promoters was amplified using primers HC148 and HC169. The product was digested with BamHI and HindIII and ligated into the same sites in pLL29erm. This plasmid was electroporated into RN4220 containing the helper plasmid pLL2787 [83], and integration into the chromosome was confirmed by PCR using primer sets HC172/scv10 and scv8/scv9. The integrated construct was then transduced into the LAC ΔmgrA::tet strain, selecting for Erm resistance.
Construction of the mgrA complementing vector
The same 910-bp fragment described above, containing the mgrA gene and its native promoters, was amplified from LAC chromosomal DNA using primers HC169 and HC187. The product was digested with BamHI and SalI and ligated into the same sites in pCM28 [74] to generate pHC66. This plasmid was electroporated into RN4220 and subsequently transduced into LAC ΔmgrA::tet and LAC ΔarlRS::tet.
mgrA and sraP promoter fusions
All promoter-sGFP transcriptional reporters were generated in the shuttle vector pCM11 [84]. We generated transcriptional reporters for each of the mgrA promoters separately, based on the mgrA promoter mapping reported by Ingavale et al. [38]. A fragment containing the upstream promoter, P2, was amplified from LAC genomic DNA using primers HC184 and HC191. Likewise, a fragment containing only the downstream promoter, P1, was amplified using primers HC185 and HC194. The PCR products were digested with HindIII and KpnI, and subsequently ligated upstream of the sGFP gene in pCM11, to generate plasmids HC68 and HC70, respectively.
The sraP-sGFP transcriptional fusions were generated by amplifying fragments of increasing length from the region upstream of the sraP gene and cloning them into pCM11. To construct pHC73, a 143-bp fragment upstream of the sraP start codon was amplified with primers HC290 and HC291, digested with HindIII and KpnI, and ligated into pCM11. pHC71 was constructed in a similar fashion, except that primers HC288 and HC291 were used to amplify a 518-bp fragment that was then cloned into pCM11. All transcriptional fusion plasmids were electroporated into RN4220 and subsequently transduced into the LAC strains of interest.
To assess expression, overnight cultures were diluted 1:100 in TSB in a black 96-well plate, and plates were incubated at 37°C with shaking in a humidified microtiter plate shaker (Stuart). A Tecan Infinite M200 plate reader was used to periodically measure OD600 and fluorescence intensity with excitation at 495 nm and emission at 515 nm. Values from quadruplicate wells were averaged, and the experiment was repeated at least once.
RNA purification
Cultures were grown in TSB to an OD600 of 1.5, at which point cells were pelleted and washed briefly with RNAprotect Bacterial Reagent (Qiagen). Cells were lysed with lysostaphin for 1 h at room temperature, and RNA was purified using the RNeasy Mini Kit (Qiagen). Following purification, genomic DNA was removed using the Turbo DNA free kit (Ambion).
Quantitative polymerase chain reaction
DNase-treated RNA was used as a template to generate cDNA with the High-Capacity Reverse Transcription Kit (Applied Biosystems). Primers for coa, sak, vWbp, srtA, clfA and ebh have been described previously [33]. Primers specific for the sraP gene cluster were designed using the PrimerQuest tool on the IDT website (see S2 Table for primer sequences). qPCR was performed by amplifying 10 ng of cDNA with Power SYBR Green Master Mix (Applied Biosystems) under the following conditions: 10 minutes at 95°C, 40 cycles of 15 seconds at 95°C and 1 minute at either 53°C (coa, sak, vWbp, srtA, clfA, and ebh primers) or 57°C (sraP gene cluster), followed by a dissociation curve. Expression was normalized to that of DNA gyrase (gyrB), and values represent averages of three biological replicates.
Promoter mapping using 5’RACE
Mapping of the ebh and sraP promoters using rapid amplification of 5’ cDNA ends (5’ RACE) was performed as described [88]. Template RNA was obtained from an LAC arlRS mutant to identify the ebh promoter, and from an mgrA mutant for the sraP promoters. Gene specific primers are listed in S2 Table, and reactions were performed using SuperScript III reverse transcriptase (Invitrogen Life Technologies) and terminal transferase (New England Biolabs).
RNA-seq
RNA was prepared from triplicate cultures of LAC and LAC ΔmgrA::tet, treated with DNase, and assessed for quality using a Bioanalyzer (Agilent). rRNA was depleted using the Ribo-Zero rRNA Removal Kit for Gram-positive bacteria (Illumina). cDNA libraries were generated at the University of Iowa Genomics Division using the TruSeq Stranded mRNA Library Prep Kit (Illumina). Samples were barcoded, pooled, and sequenced in 100x100 paired end reads using a HiSeq 2000 sequencer (Illumina). The resulting sequences were aligned to the USA300_FPR3757 genome sequence using SeqMan NGen (DNASTAR) and the alignment data were analyzed using ArrayStar (DNASTAR). Genes were considered differentially expressed if they showed a ≥4-fold change in expression with 95% confidence as evaluated using the student’s t-test with a false discovery rate (FDR) correction applied for multiple t-tests.
Cloning and purification of ArlR and MgrA
The arlR gene was amplified from LAC genomic DNA using primers HC41 and HC42. The PCR product was digested with NdeI and Xho, and ligated into pET28a to generate pHC07, which expresses arlR with an N-terminal His6 tag. pHC07 was transformed into the E. coli overexpression strain ER2566. Cells were grown in LB supplemented with kanamycin at 37°C to an OD600 of 0.6, at which point expression was induced by adding 0.1 mM IPTG and shifting to 30°C overnight. Cells were harvested by centrifugation and stored at -20°C. To purify His6-ArlR, cells were resuspended in bind buffer (50 mM Na phosphate, 300 mM NaCl, pH 8) and lysed by two passages through a Microfluidics LV1. Cell debris was removed by centrifugation and the soluble fraction was passed over a HIS-Select nickel affinity (Sigma-Aldrich) column. The column was washed with bind buffer supplemented with 10 mM imidazole, and purified ArlR was eluted with elute buffer (50 mM Na phosphate, 300 mM NaCl, 250 mM imidazole, pH 8). Fractions containing ArlR were pooled and dialyzed twice against storage buffer (50 mM Na phosphate, 150 mM NaCl, pH 8), with 0.5 mM EDTA added to the first dialysis buffer. The dialyzed protein was concentrated and glycerol was added to 20% (vol/vol) before freezing in an ethanol/dry ice bath and storage at -80°C.
The mgrA coding sequence was amplified from LAC genomic DNA using primers HC148 and HC190. The product was digested with KpnI and HindIII and ligated into the overexpression vector pKLD66 [89] to generate pHC74. This plasmid expresses mgrA with sequential His6 and maltose binding protein tags at the N-terminus, both of which can be removed by cleavage with Tev protease. pHC74 was transformed into E. coli expression strain ER2566. For overexpression of mgrA, cells were grown with shaking at 37°C in LB supplemented with ampicillin. When the OD600 reached ~0.5, expression was induced with 0.5 mM IPTG and the culture was shifted to 30°C overnight. Cells were harvested by centrifugation and the pellet was stored at -80°C. To purify tagged MgrA, the cell pellet was resuspended in bind buffer and cells were lysed by adding lysozyme and sonicating in four ~1 min pulses at 50% duty. Tagged MgrA was purified as described above for ArlR. Fractions containing MgrA were pooled and incubated with a 1:20 molar ratio of His6-Tev protease [90] to MgrA at room temperature for 3 h, while dialyzing against bind buffer containing 0.5 mM EDTA. The cleaved protein was dialyzed two additional times against bind buffer at 4°C. To purify MgrA away from the His6-MBP tag and His6-Tev protease, the cleaved protein was passed over the nickel affinity column again, where MgrA eluted in the flow-through and wash steps. Purified, untagged MgrA was dialyzed against storage buffer and frozen as described for ArlR.
Western and dot blots
All Western and dot blots were performed using strains lacking protein A (spa). Ebh production was assessed by dot blot using antibodies raised against the H2 peptide within the G/A repeat region of Ebh as previously described [33]. Antibodies specific for ArlR and MgrA were generated in rabbits using the purified proteins described above.
To monitor MgrA and ArlR protein production, cultures were diluted 1:100 in TSB and aliquots were removed at each time point. The OD600 was measured, and 1 OD600 unit of cells was centrifuged, washed once with tris-buffered saline (TBS), and frozen at -20°C. Cell pellets were resuspended in 200 μL TBS and incubated for 1 h at 37°C with 5 μg lysostaphin and 1 U DNase (New England Biolabs). Cell debris was removed by centrifugation and the soluble fraction was heated to 95°C before loading on a 15% SDS-PAGE gel. Proteins were separated by electrophoresis and transferred to a nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with TBS containing 0.05% Tween 20 (TBST) and 5% milk, and then incubated for 1 hr with ArlR or MgrA antiserum (diluted 1:1000 in TBST + 5% milk). The membrane was washed three times with TBST and incubated with HRP-conjugated goat anti-rabbit antibodies (diluted 1:20,000 in TBST + 5% milk). The membrane was washed three times in TBST before incubation with SuperSignal West Pico chemiluminescent substrate for 5 min and exposure to X-ray film. Band intensities were quantified using Image Studio Lite (LI-COR). Results are representative of two (ArlR) or three (MgrA) separate experiments.
Electrophoretic mobility shift assays
MgrA binding to the ebh promoter was observed using a probe labeled at one end with IRDye 700 (LI-COR), synthesized by Integrated DNA Technologies. 50-mer oligos HC487 and HC489 were combined in PBS + 1 mM EDTA and annealed by heating to 95°C for 5 min and then gradually cooling by 1°C per minute to 25°C. Three additional unlabeled probes were prepared in a similar fashion: a specific competitor with the same sequence (primers HC488/HC489), a competitor in which the putative MgrA binding site had been mutated (primers HC492/HC493), and a non-specific competitor from within the sraP coding sequence (primers HC349/HC350). For sraP P2 EMSAs an IRDye 700 labeled probe was generated with 50-mer oligos HC336 and HC346. An unlabeled specific competitor probe with the same sequence was generated with oligos HC336 and HC337, and a competitor in which the putative MgrA binding site had been mutated was made with oligos HC347 and HC348. The nonspecific competitor probe was the same as for the ebh EMSA described above. Binding reactions contained 50 nM labeled probe, 500 nM competitor probe, 25 mM HEPES pH 7.4, 50 mM KCl, 1 mM DTT, and MgrA (0–2 μM). Reactions were incubated for 20 min at room temperature, and then separated on a pre-run 5% TBE-acrylamide gel at 100 V for 1 h in the dark at 4°C. Images were obtained using an Odyssey CLx imaging system (LI-COR).
Biofilm assay
S. aureus overnight cultures were diluted 1:40 in BHI supplemented with 0.25% glucose in a 48-well polystyrene microtiter plate. The plate was incubated statically at 37°C in a humidified plate incubator (Stuart) for 16 h. At this point the media was removed and remaining biomass was stained with 0.1% crystal violet. The wells were then washed with sterile distilled water and the plate was photographed with a GelDoc XR+ (Biorad). For quantification, the crystal violet stain was resuspended with isopropanol and the absorbance was measured at 595 nm.
Infective endocarditis and sepsis
New Zealand white rabbits (approximately 2–3 kg), either sex, were purchased from Bakkom Rabbitry, Red Wing, MN and used according to University of Iowa IACUC approved protocol 4071100. Rabbits were anesthetized with ketamine (25 mg/kg) and xylazine (25 mg/kg) (Phoenix Pharmaceuticals, Burlingame, CA). Their necks were shaved, and 5 cm incisions were made to expose the left carotid arteries. Hard plastic catheters were inserted into the carotid arteries until the catheters just abutted against the aortic valves. The catheters were then tied in place and allowed to cause damage to the aortic valves for 2 h. Subsequently, the catheters were removed and carotid arteries tied off, and the animals were closed. Animals were injected intravenously through the marginal ear veins with S. aureus strains in 1 ml PBS (approximately 2.5 x 107 CFU/ml for MW2; 1.3 x 108 CFU/ml for 502a). The rabbits were monitored for health status for up to 4 days; during this time, animals that simultaneously failed to exhibit escape behavior and failed to be able to right themselves, 100% predictive of lethal infection, were prematurely euthanized with 1 ml/kg of Beuthanasia D (Shering-Plough, Westlake, TX). After 4 days (or at the time of premature euthanasia), the animals were euthanized, hearts removed, and vegetation formation determined. Vegetations, cauliflower-like clumps of bacteria and host cells, were removed, weighed, and homogenized for CFU determination. Statistical differences in vegetation weights and CFUs were determined by Student’s t test analysis of normally-distributed, non-paired data. Differences in animal survival rates were determined by log-rank (Mantel-Cox) test.
Supporting Information
S1 Fig
Clumping of LAC clfA clfB fnbAB mutant strains.Clumping of LAC clfA clfB fnbAB mutants in the presence of human plasma (A) or purified human fibrinogen (B). Values represent averages and standard deviations of three separate experiments.
(PDF)
Acknowledgments
We thank S. Nakada, L. Ackermann and J. Kavanaugh for technical assistance. We thank J. Bair and E. Snir at the University of Iowa Genomics Division of the Institute of Human Genetics for the RNA-seq library preparation and sequencing. We thank T. Moninger of the University of Iowa Central Microscopy Research Facility, a core resource supported by the Vice President for Research & Economic Development, the Holden Comprehensive Cancer Center and the Carver College of Medicine. We thank W. Nauseef at the University of Iowa Inflammation Program for providing human plasma.
Funding Statement
HAC was supported by NIH T32 AI007511 and an American Heart Association postdoctoral fellowship; ARH was supported by project 3 of NIH grant AI083211; PMS was supported by a University of Iowa start-up grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Articles from PLOS Pathogens are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.ppat.1005604
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plospathogens/article/file?id=10.1371/journal.ppat.1005604&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.ppat.1005604
Article citations
Staphylococcus aureus adapts to exploit collagen-derived proline during chronic infection.
Nat Microbiol, 9(10):2506-2521, 12 Aug 2024
Cited by: 1 article | PMID: 39134708 | PMCID: PMC11445067
Response of Staphylococcus aureus physiology and Agr quorum sensing to low-shear modeled microgravity.
J Bacteriol, 206(9):e0027224, 09 Aug 2024
Cited by: 0 articles | PMID: 39120147
Elucidating the potential of isorhapontigenin in targeting the MgrA regulatory network: a paradigm shift for attenuating MRSA virulence.
Antimicrob Agents Chemother, 68(9):e0061124, 24 Jul 2024
Cited by: 0 articles | PMID: 39046236
Exploring the potential of isorhapontigenin: attenuating Staphylococcus aureus virulence through MgrA-mediated regulation.
mSphere, 9(6):e0031724, 05 Jun 2024
Cited by: 0 articles | PMID: 38837389 | PMCID: PMC11332347
MarR family proteins sense sulfane sulfur in bacteria.
mLife, 3(2):231-239, 15 May 2024
Cited by: 0 articles | PMID: 38948149
Review
Go to all (89) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Staphylococcus aureus adhesion in endovascular infections is controlled by the ArlRS-MgrA signaling cascade.
PLoS Pathog, 15(5):e1007800, 22 May 2019
Cited by: 33 articles | PMID: 31116795 | PMCID: PMC6548404
The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA.
Mol Microbiol, 113(1):103-122, 06 Nov 2019
Cited by: 52 articles | PMID: 31618469 | PMCID: PMC7175635
Host-derived protease promotes aggregation of Staphylococcus aureus by cleaving the surface protein SasG.
mBio, 15(4):e0348323, 21 Mar 2024
Cited by: 0 articles | PMID: 38511930 | PMCID: PMC11005337
Regulation of Staphylococcus aureus Virulence.
Microbiol Spectr, 7(2), 01 Apr 2019
Cited by: 237 articles | PMID: 30953424 | PMCID: PMC6452892
Review Free full text in Europe PMC
Funding
Funders who supported this work.
American Heart Association (1)
Grant ID: 15POST25720016
NIAID NIH HHS (2)
Grant ID: P01 AI083211
Grant ID: T32 AI007511
NIDDK NIH HHS (1)
Grant ID: P30 DK054759
National Institute of Allergy and Infectious Diseases (2)
Grant ID: AI083211
Grant ID: T32 AI007511